Abstract
Objective: To monitor the bacterial load in newborns with proven infections on the day of admission, 48 h and 7 days after treatment.
Methods: Real-time PCR (qPCR) targeting the 16S rDNA.
Results: The study recruited 17 newborns and the bacterial load was in general low (<50 CFU/mL). In three of four deaths, the bacterial load values increased, and in 11 of the 13 survivors the values decreased until the third evaluation.
Conclusion: Considering the extreme sensitivity and high negative predictive value of qPCR, this test could help to monitor the treatment of neonatal sepsis and to assist in medical decision to discontinue antibiotics.
Introduction
Neonatal sepsis still accounts for 40% of all deaths in this period of life [Citation1]. Blood cultures are the gold standard for the diagnosis of sepsis, however, quantitative real-time amplification systems (qPCR) based on the bacterial 16S rDNA [Citation2,Citation3] have been suggested not as a replacement, but as a complementary diagnostic tool to blood cultures because they are faster and more sensitive than conventional amplifications, and when compared to blood cultures are particularly useful to rule out infection due to a high negative predictive value, and also because both, positive and negative results can be released within 24 h [Citation4]. Nevertheless, the qPCR has the disadvantage of not being able to identify the bacterium species, unlike blood cultures [Citation2,Citation3], is more prone to environmental contamination, and cannot discriminate between viable and non-viable bacteria [Citation5,Citation6].
In all studies with newborns and qPCR [Citation2–4], the amplification results were used only qualitatively (positive or negative) and were compared with the blood culture results. The aim of this study was to monitor the bacterial load levels of newborns with proven infections during treatment.
Methods
This research was approved by the Research Ethics Committee of the participating institutions (CAPPesq process number 0889/09 and 587/CEP-HUJM/08) and was carried out in a neonatal intensive care unit of out born patients, located in a tertiary hospital, from January to July 2012. To be recruited, newborns had to present with a positive blood culture at the time of admission to the neonatal intensive care unit, aside from one sign or symptom suggestive of sepsis [Citation7], and one altered laboratory parameter in the ancillary exams performed to investigate the potential infection. The reference values were: leukocytosis (>20 000 mm−3), leukopenia (<5000 mm−3), neutrophilia (>13 000 mm−3), neutropenia (<1000 mm−3), neutrophil index > 0.2 (immature neutrophil count/total), thrombocytopenia (<100 000 mm−3). Serum C-reactive protein (CRP) concentrations were determined by turbidimetry (CRP ultrasensitive kit – Turbitest AA, Wiener, Argentina) with reference values recommended by the manufacturer ≤5 mg/L [Citation8]. In the present study, only CRP values >10 mg/L were considered high.
The exclusion criteria were surgical procedures in the newborn; the presence of severe perinatal asphyxia defined by an Apgar score of less than 4 at 5 min of life [Citation9]; concurrent metabolic abnormalities such as hypoglycemia and hypocalcemia; congenital malformations and congenital infections.
One milliliter of blood was obtained from a peripheral venepuncture at the time of suspected neonatal infection and used for the culture of aerobic and anaerobic organisms (BACT ALERT 3D, BioMerieux®). The identification of microbial susceptibility was determined using the Vitek 2, BioMerieux® (BioMerieux Inc., Durham, NC). In addition, 1 mL of blood was obtained at each of the three time points to perform the qPCR. DNA extraction (QIAamp DNA mini kit, QIAGEN Inc., Hilden, Germany) was performed with the addition of a specific step for the extraction of bacterial DNA [Citation3].
PCR amplifications were only performed after blood cultures turned positive. Prior to amplification, the solutions were autoclaved, and the reagents were filtered through 0.22-μm filters (Merck-Millipore, Billerica, MA). Amplifications were performed by an experienced biologist under sterile conditions in an ABI StepOne Real Time PCR System (Applied Biosystems®, Foster City, CA). The mixture of reagents reached a final volume of 25 µL containing QuantiFast SybrGreen PCR Master Mix (Hot Start Taq Plus DNA, QuantiFast SybrGreen PCR Buffer, dNTP mix, SybrGreen) (QIAGEN Inc.), 0.1 μM of the forward and reverse primers (forward 5′-CAGCTCGT GTCGTGAGATGT-3′ and reverse 5′-CGTAAGGGCCATGATGAC T-3′), 100 ng of DNA extracted from the infected newborns, 100 ng of DNA from a known non-infected newborn, and sterile water instead of genomic DNA (non-template sample). To determine the detection range of the qPCR, a 10-fold dilution series ranging from 107 to 1 CFU/mL of blood was prepared. All samples were tested in duplicate. The primers generated a 150-bp fragment in positive samples.
The qPCR was performed in 40 cycles of amplification: an initial denaturation step at 95 °C for 5 min followed by 10 cycles at 95 °C for 30 s and 66 °C for 30 s, 10 cycles at 95 °C for 30 s and 64 °C for 30 s, and 20 cycles at 95 °C for 30 s and 62 °C for 30 s, ending with the melting curve. The threshold cycle of each duplicate sample could not show differences in more than one cycle. The limit of detection in all experiments was 1–10 CFU/mL, regardless of the type of bacteria (Gram-positive or Gram-negative).
Results
This is a pilot study, so that the sample size was limited by the period in which the collection and blood cultures could be performed in this particular neonatal intensive care unit. The study recruited 17 patients, 7 full-term and 10 preterm newborns (). In the group of seven full-term infants the median of gestational age was 38 weeks (min. 37 – max. 40), while among the 10 preterm infants the median value was 32.5 weeks (min. 26 – max. 35). Gestational ages were estimated at birth by using the ultrasound performed at 21–28 weeks, the ultrasound at 29 + weeks, the last menstrual period, and the Capurro method [Citation10]. With respect to the birth weight, in the group of seven full-term infants the median value was 2560 g (min. 1940 – max. 3760), while in the preterm group the median value was 1889 g (min. 1050 – max. 2350).
Table 1. Characterization of the 17 cases of neonatal sepsis included in the study, the bacterial load assessed by qPCR in the three time points (expressed in CFU/mL), the bacterium identified by culture, the antibiotics used for treatment and the clinical outcome.
On the day of admission (day 0), the median age of the newborns was 9 days (min. 2 – max. 15), the most frequent clinical signs/symptoms were respiratory distress and mild to moderate perinatal asphyxia, while the most frequent laboratory abnormalities were CRP >10 mg/L, neutrophilia, leukopenia and thrombocytopenia.
The 17 blood cultures showed a predominance of Gram-positive bacteria: 12 coagulase-negative Staphylococcus, two Staphylococcus aureus, one Streptococcus agalactiae, one Lysteria monocytogenes; the only Gram-negative isolated bacterium was a Klebsiella pneumoniae ().
All infants received at least 2 weeks of antibiotics. Of the 17 cases, 13 fully recovered and four died (two full-term and two preterm newborns). summarizes the data from the 17 newborns, with the bacterial load levels obtained in the three time points.
The bacterial load levels in all the time points were in general low (<50 CFU/mL). On day 0, there were two newborns with higher bacterial loads (2T – 120.9 CFU/mL, and 3T – 93.4 CFU/mL), S. aureus and S. agalactiae were isolated and both presented full recovery. In all the four infants who died, two full-term and two preterm (4T, 7T, 9PT, 13PT) coagulase negative Staphylococci were isolated. The median of the bacterial load levels in the three time points of the 17 newborns, or comparing the 13 survivors with the four deaths did not show statistical differences, probably due to the restricted number of cases. Regarding the variation of the bacterial loads, in three of the four deaths the values increased until the seventh day, except in the case 13PT that showed an undetectable bacterial load on the seventh day. In this newborn, the cause of death was multiple organ failure. In the group of 13 infants who evolved favorably, in 11 cases the bacterial load decreased from day zero until the third time point; in one case the bacterial load remained at the same level, and in one case there was an increment (though these two cases presented full recovery).
Discussion
Considering the high mortality rates of neonatal sepsis, the identification of a biomarker with high sensitivity, specificity, and also high negative predictive values, i.e. capable of ruling out the presence of infection would be ideal. In this study, our aim was not to apply qPCR to diagnosis but to show that this technique based on the bacterial 16S rDNA could be useful and cost-effective to monitor the treatment of neonatal sepsis. It is noteworthy that in three infants of the 13 who progressed favorably, the bacterial load was undetectable from the second time point, and in six cases, the bacterial load was undetectable on the seventh day after the introduction of antibiotics ().
Due to the impossibility of guaranteeing benefits to the participants, the Research Ethics Committee did not authorize us to compose a control group of infants without infection. However, in all experiments, two negative controls were added, one represented by the sample containing sterile water instead of DNA (non-template sample), and the other negative control represented by a DNA sample obtained in the delivery room from placentas (cord blood) of full-term newborns, whose mothers received prenatal care and did not present any evidence of infection. These newborns had negative blood cultures collected in the delivery room. Nevertheless, it is noteworthy that blood samples taken from placentas did not use the same disinfection protocol when compared to blood samples collected from newborns with suspected infection.
Because antibiotics are administered for at least two weeks during severe neonatal infections or for prolonged periods in sepsis, qPCR could be useful to reduce hospital costs and the length of hospital stay. Brozanski et al. [Citation11] previously showed that approximately eight antibiotic doses and 85 neonatal intensive care unit hours could be saved per infant based on negative PCR results. Therefore, because of the high negative predictive value of qPCR, negative results could rule out the presence of infection, directly influencing clinical decisions and practices, resulting in fewer antibiotic doses per patient and shorter hospital stays. shows a case in which the qPCR could help an early discontinuation of the treatment.
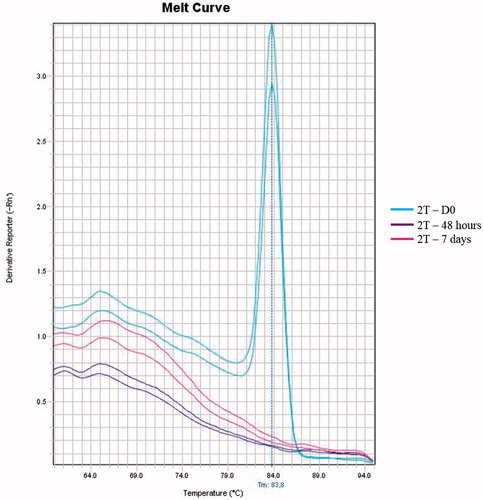
Figure 1. Melting curve of the qPCR corresponding to the three time points. The full-term newborn (2T) with confirmed sepsis caused by S. agalactiae presented a bacterial load of 120.9 CFU/mL in the first sample evaluated (day 0 – blue lines). The bacterial loads at 48 h (purple lines) and on the seventh day (pink lines) were undetected.
Although qPCR is not able to determine the bacterium species and to distinguish between viable and nonviable bacteria, some studies [Citation5,Citation6] have shown that this bias constitutes a problem only in samples of untreated patients, with no significant differences for the evaluation of patients receiving treatment. In the present study, our purpose was to highlight the role of a negative qPCR result in the context of neonatal bacterial infections under treatment precisely because the technique is extremely sensitive and susceptible to carry-over, so that a negative result should be valued. In addition, other authors claimed that most of the DNA from dead cells is actually lost during the DNA extraction procedure, probably together with cell debris [Citation12]. Recently, the DNA-intercalating agent propidium monoazide (PMA) which is able to promote cell membranes disruption was used in conjunction with qPCR to selectively detect live cells of pathogenic microorganisms [Citation13]. In future experiments, we plan to perform PMA-qPCR to reduce significantly the number of non-viable bacteria, bearing in mind that PMA may decrease qPCR signals [Citation14].
Taking into account the extreme sensitivity and the high negative predictive value of the qPCR technique, this test could help to monitor the treatment of neonatal sepsis and to assist in medical decision to discontinue antibiotics in infants with favorable clinical outcome and a negative qPCR result, shortening both, the antibiotic treatment and the hospital stay.
Acknowledgements
The authors are grateful to the mothers who kindly consented to the blood collection from their infants. The authors are also grateful to Paulo Roberto Bezerra de Melo, Associate Professor of the Department of Pediatrics, School of Medicine, Federal University of Mato Grosso, for his assistance. We would like to thank the nurses of the neonatal intense care unit of Hospital Santa Helena, Cuiabá, and the medical students Natalia de Carvalho Castro and Livia de Sousa Lima Pulcherio for their assistance during data collection.
Declaration of interest
The authors report no declarations of interest. This work was supported by FAPEMAT (Mato Grosso State Research Foundation, Brazil), process Universal 009/2011, grant number 752380/2011, and AUX-PE-DINTER/NF grant number 2535/2009 from CAPES – Ministry of Education, Brazil.
References
- Liu L, Johnson HL, Cousens S, et al. Global, regional, and national causes of child mortality: an updated systematic analysis for 2010 with time trends since 2000. Lancet 2012;379:2151–61
- Pammi M, Flores A, Leeflang M, Versalovic J. Molecular assays in the diagnosis of neonatal sepsis: a systematic review and meta-analysis. Pediatrics 2011;128:e973–85
- Ohlin A, Bäckman A, Ewald U, et al. Diagnosis of neonatal sepsis by broad-range 16S real-time polymerase chain reaction. Neonatology 2012;101:241–6
- Gosiewski T, Flis A, Sroka A, et al. Comparison of nested, multiplex, qPCR; FISH; SeptiFast and blood culture methods in detection and identification of bacteria and fungi in blood of patients with sepsis. BMC Microbiol 2014;14:313
- Rogers GB, Stressmann FA, Koller G, et al. Assessing the diagnostic importance of nonviable bacterial cells in respiratory infections. Diagn Microbiol Infect Dis 2008;62:133–41
- Jiang LJ, Wu WJ, Wu H, et al. Rapid detection and monitoring therapeutic efficacy of Mycobacterium tuberculosis complex using a novel real-time assay. J Microbiol Biotechnol 2012;22:1301–6
- Goldstein B, Giroir B, Randolph A. International pediatric sepsis consensus conference: definitions for sepsis and organ dysfunction in pediatrics. Pediatr Crit Care Med 2005;6:2–8
- Manroe BL, Weinberg AG, Rosenfeld CR, Browne R. The neonatal blood count in health and disease. J Pediatr 1979;95:89–98
- Leuthner SR, Das UG. Low Apgar scores and the definition of birth asphyxia. Pediatr Clin North Am 2004;51:737–45
- Pereira AP, Dias MA, Bastos MH, et al. Determining gestational age for public health care users in Brazil: comparison of methods and algorithm creation. BMC Res Notes 2013;6:60
- Brozanski BS, Jones, JG, Krohn MJ, et al. Use of polymerase chain reaction as a diagnostic tool for neonatal sepsis can result in a decrease in use of antibiotics and total neonatal intensive care unit length of stay. J Perinatol 2006;26:688–92
- Nocker A, Camper AK. Selective removal of DNA from dead cells of mixed bacterial communities by use of ethidium monoazide. Appl Environ Microbiol 2006;72:1997–2004
- Vendrame M, Iacumin L, Manzano M, Comi G. Use of propidium monoazide for the enumeration of viable Oenococcus oeni in must and wine by quantitative PCR. Food Microbiol 2013;35:49–57
- Barbau-Piednoir E, Mahillon J, Pillyser J, et al. Evaluation of viability-qPCR detection system on viable and dead Salmonella serovar enteritidis. J Microbiol Methods 2014;103:131–7
