Abstract
The human ejaculate contains subpopulations of sperm with distinct properties. Human X- and Y-bearing sperm were separated with fluorescence activated cell sorting. To avoid the use of UV light the quantitative DNA dyes DRAQ5® and Dyecycle™ Vybrant® Violet were used. Sorting efficiency was similar for both dyes, but lower than what is usually obtained with the classical method involving Hoechst 33342 and UV light (60-70% enrichment, versus 80-90%). A total of 2,739 spermatozoa were evaluated, from seven distinct samples using fluorescence in situ hybridization (FISH) chromosomal probes. No differences were found in sorted and unsorted populations in terms of chromosome positioning, and numeric chromosomal anomalies were not more evident following cell sorting. Furthermore in both sorted and unsorted populations the sex chromosomes were clearly located in the anterior portion of the sperm head, while a control autosome (chromosome 18) showed no such tendency, confirming previous findings. These results suggest that other quantitative DNA dyes may be used for sex chromosome-based human sperm sorting, but with lower efficiency than the standard UV-Hoechst based assay.
Introduction
The human ejaculate contains subpopulations of sperm with distinct properties, and functional abilities [Sousa et al. 2011]. Two obvious subpopulations are those defined by the sex chromosomes, theoretically dividing an ejaculate in half, with 50% of Y- and 50% of X- bearing cells. These subpopulations may have distinct biochemical properties, besides a clear role in embryonic sex determination. Indeed, many different methodologies have been described to separate X and Y-bearing sperm for study, animal production, or clinical purposes, such as avoiding the transmission of X-linked diseases [Cran and Johnson 1996; Garner 2006; Johnson et al. 1993; Sills et al. 1998]. However, the most effective protocol to efficiently separate these subpopulations is using quantitative DNA dyes and flow cytometry coupled to cell sorting [Cran and Johnson 1996; Garner 2006; 2009; Seidel 2012]. This methodology relies on the different size/DNA content of the sex chromosomes (and thus on distinct DNA content in X- and Y-bearing spermatozoa), and its efficiency in different mammalian species therefore depends on the relative size difference of the sex chromosomes [Cran and Johnson 1996; Garner 2006; Sills et al. 1998]. This difference is quite modest in humans (about 3%), leading to controversial results, but also to a few reported successes, which may have ethical implications in terms of possible sex-selection [Fugger et al. 1998; Sills et al. 1998; Vidal et al. 1998]. Despite its successful use in many species, a frequent point of contention is the common use of the UV-excited DNA dye Hoechst 33342 for sperm separation based on DNA content [Garner 2009].
Interestingly it has been well established that chromosome positioning in mammalian sperm in general, and of sex chromosomes in particular, is non-random. Although the nucleus of the male gamete is less studied and understood than nuclei from interphase somatic cells, it is believed that chromosome positions may have a physiological significance in terms of early transcriptional activity in the embryo, and possible consequences for male infertility or in the use of assisted reproduction technologies [Ioannou and Griffin 2011; Zalensky and Zalenskaya 2007]. Chromosome positions in the mammalian sperm head may vary in different ways, both in terms of where the chromosome tends to be predominantly located (in the anterior, equatorial, or posterior regions of the head, more centrally, or more peripherally) or where certain chromosome structures (centromeres, telomeres) tend to reside [Finch et al. 2008; Foster et al. 2005; Haaf and Ward 1995; Hazzouri et al. 2000; Luetjens et al. 1999; Manvelyan et al. 2008; Millan et al. 2012; Mudrak et al. 2005; Olszewska et al. 2008; Tilgen et al. 2001; Zalenskaya and Zalensky 2004].
Specific attention has been paid to the position of the sex chromosomes in the human sperm head (specifically the X chromosome), with several early studies using multiple samples pointing to its preferential location on the anterior portion of the sperm head [Hazzouri et al. 2000; Luetjens et al. 1999].
In this work we separated human sperm cells by flow cytometry and cell sorting using DNA content to study specific populations enriched in Y- and X- bearing chromosomes. To circumvent the use of UV light, we have used the quantitative DNA dyes DRAQ5® and Dyecycle™ Vybrant® Violet, which are excited at visible (i.e., higher) wavelengths [Mari et al. 2010; Zhao et al. 2009].
Results and Discussion
We attempted to find alternatives to the classically used Hoechst 33342 and UV light for sex chromosome-based sperm sorting. DRAQ5® and Dyecycle™ Vybrant® Violet yielded statistically identical results (data not shown). No clear separate peaks in terms of DNA content were ever visible, and thus we sorted two populations on the edges of the main peak (). This resulted in a modest but significant 60-70% enrichment of a specific sex chromosome-bearing sperm, rather than the 50% random distribution found in non-sorted cells (p < 0.05; ). However this is well below what is usually reported using the Hoechst-UV based approach, suggesting that this method remains the standard for this purpose, with consistent efficiency in the 80-90% range [Garner 2006; 2009; Seidel 2012]. Thus the dyes tested here may not be particularly useful for practical purposes. Furthermore, these results precluded more detailed functional and biochemical analysis of X- and Y- bearing sperm subpopulations, as enrichment was clearly not sufficient to ensure reliable results. It should be noted that human sperm are more difficult to separate than sperm from other species due to the small difference in DNA content, and this may hamper the search for alternative methodologies [Garner 2006; Johnson et al. 1993].
Figure 1. Separating X and Y- bearing human sperm. When separating human sperm according to DNA content using Dyecycle™ Vybrant® Violet a strongly labeled sperm population was visible (A, elipse), but when samples were scanned according to fluorescence intensity only one peak was visible, showing no distinct subpopulations clearly definable by DNA content (B). If the extremes of this peak were gated (defined in B) sperm subpopulations could be obtained enriched in either Y (green spots) or X (red spots) bearing chromosomes, depending on whether the ascending (lower DNA content; C) or descending (higher DNA content; D) arm of the peak was selected, respectively. Panels C and D are representative FISH images, with DAPI (blue) used as a nuclear counterstain. Results with DRAQ5®were essentially identical (data not shown).
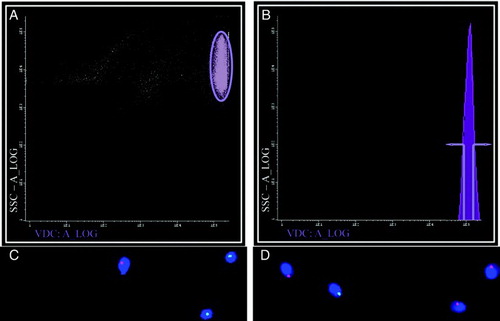
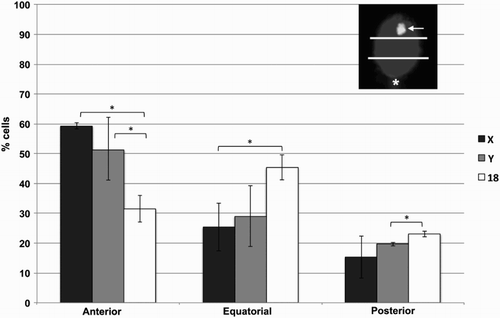
Given the nature of the assay, however, we were able to perform fluorescence in situ hybridization (FISH) analysis on the sorted subpopulations. No statistically significant differences in terms of chromosome position were observed when sorted samples were compared with unsorted samples using probes for the X and Y chromosomes, suggesting that sex chromosome position does not influence DNA content-based sorting (data not shown). Also no obvious numerical chromosomal anomalies were more prevalent in sorted subpopulations (data not shown). However a clear anterior distribution of both sex chromosomes was evident, while chromosome 18 showed no such tendency (). This was especially clear for the X chromosome, and is in accordance with previous data for this chromosome [Hazzouri et al. 2000; Luetjens et al. 1999]. This has been confirmed more recently for the Y chromosome, albeit in this case anterior positioning occurs to a lesser degree [Millan et al. 2012]. Interestingly, the position of X and Y chromosomes relative to the sperm head and to other chromosomes do not always coincide, suggesting distinct behaviors of sex chromosomes. This is perhaps related to their distinct transcriptional activities in the early embryo and in somatic interphase nuclei [Millan et al. 2012; Zalenskaya and Zalensky 2004]. This specific positioning may be relevant when using intracytoplasmic sperm injection (ICSI) given that in this case sperm head decondensation may be delayed in the anterior portion, due to the persistence in this region within the remnants of sperm structures (the acrosome, the perinuclear theca) that are normally discarded during fertilization [Hewitson et al. 1999; Ramalho-Santos et al. 2000].
Figure 2. Chromosome positions in sorted human sperm. Human sperm were sorted according to DNA content and analyzed by fluorescence in situ hybridization (FISH) using probes for chromosomes X, Y, and 18. For localization purposes, and as shown in an example in the top right corner, the sperm head was divided into three equal quadrants (anterior, equatorial, posterior), with the tail insertion (asterisk) used as a landmark for the posterior side, and the localization of the FISH spot (arrow) classified accordingly. A total of 2,739 spermatozoa were evaluated, from seven distinct samples, and percentages calculated for each sample. Error bars represent standard deviation. *statistically significant differences (p < 0.05)
Materials and Methods
Samples
Patients undergoing routine semen analysis or fertility treatment were recruited from the Fertility Clinic (University Hospitals of Coimbra, Portugal). All patients signed informed consent forms, and all human material was used in accordance with the appropriate ethical and Internal Review Board (IRB) guidelines provided by the University Hospitals of Coimbra. Fresh semen samples were obtained by masturbation after 3 to 5 d of sexual abstinence and semen analysis was carried out in conformity to the World Health Organization Guidelines [WHO 2010]. Semen samples were prepared by density gradient centrifugation as described elsewhere [Amaral et al. 2007; Sousa et al. 2011]. All samples used were obtained from healthy individuals and were normozoospermic for concentration (i.e., ≥ 15 million sperm/mL), motility (i.e., ≥ 40% motile sperm), and morphology (i.e., ≥ 4% normal forms), and had no leukocytes (or any other round cells) after semen preparation or other obvious indications of health problems that might affect sperm quality.
DNA dyes and cell sorting
Live sperm suspensions (20 million of sperm/mL) were incubated with 10 µM of DRAQ5® (BioStatus Limited, UK) or 9 µM of Dyecycle™ Vybrant® Violet (Molecular Probes, Eugene, OR, USA), for 5 min at room temperature and 30 min at 37°C, respectively. The sorting process was carried out with a BD FACSAria™ cell-sorting system (BD FACSAria III; BD Biosciences, NJ, USA) at 635 nm (DRAQ5® ) and 407 nm (Dyecycle™ Vybrant® Violet ) wavelength with the following settings: laser power – 13 mW, nozzle – 70 µm, and sort setup – medium. After separation, the sperm cell suspensions were dropped onto clean glass slides, air dried, and kept at −20°C until used.
Fluorescence in situ hybridization (FISH)
FISH analysis was carried out with modifications from previous protocols [Almeida Santos et al. 2002; Ramalho-Santos et al. 2004]. Probes used were CEP X red and CEP Y green (Vysis, Abbott Laboratories, IL, USA). ILM or CEP Y green and CEP 18 red (Vysis). Slides were counterstained with 10 µL DAPI in antifade solution (VectaShield mounting medium, Vector Labs, Burlingame, CA, USA) and observed using a Zeiss Axiophot II microscope (Carl Zeiss, Göttingen, Germany) equipped with a triple band pass filter.
A total of 2,739 spermatozoa were evaluated, from seven distinct samples. For localization purposes the sperm head was divided into three equal quadrants (anterior, equatorial, posterior), with the tail insertion functioning as a landmark for the posterior side, as described previously [Luetjens et al. 1999].
Statistical analysis
Statistical analysis was performed using the IBM SPSS® 20 software (Chicago, IL, USA). All variables were checked for normal distribution through the one-sample Kolmogorov-Smirnov test. One-way ANOVA was used to compare the results obtained in the three regions considered and the post-hoc analyses were done using Tukey's test. Statistical significance was considered when p < 0.05.
Acknowledgments
All lab members are thanked for helpful discussions. This work was done as part of the Masters Program in Cell and Molecular Biology at the Department of Life Sciences, University of Coimbra (SAM).
Declaration of interests: The authors have no interests to declare.
Author contributions: Defined the project, and finalized the manuscript: JRS; Processed sperm samples for analysis: APS, TAS; Performed all FISH experiments, as well as flow cytometry data collection and initial analysis: SAM; Performed flow cytometry analysis: AP; All authors analyzed the data; Wrote the paper: JRS, APS, SAM; All authors approved the paper.
References
- Almeida Santos, T., Dias, C., Brito, R., Henriques, P. and Almeida Santos, A. (2002) Analysis of human spermatozoa by fluorescence in situ hybridization with preservation of the head morphology is possible by avoiding a decondensation treatment. J Assist Reprod Genet 19:291–294.
- Amaral, A., Ramalho-Santos, J. and St John, J.C. (2007) The expression of polymerase gamma and mitochondrial transcription factor A and the regulation of mitochondrial DNA content in mature human sperm. Hum Reprod 22:1585–1596.
- Cran, D.G. and Johnson, L.A. (1996) The predetermination of embryonic sex using flow cytometrically separated X and Y spermatozoa. Hum Reprod Update 2:355–363.
- Finch, K.A., Fonseka, K.G., Abogrein, A., Ioannou, D., Handyside, A.H., Thornhill, A.R. (2008) Nuclear organization in human sperm: preliminary evidence for altered sex chromosome centromere position in infertile males. Hum Reprod 23:1263–1270.
- Foster, H.A., Abeydeera, L.R., Griffin, D.K. and Bridger, J.M. (2005) Non-random chromosome positioning in mammalian sperm nuclei, with migration of the sex chromosomes during late spermatogenesis. J Cell Sci 118:1811–1820.
- Fugger, E.F., Black, S.H., Keyvanfar, K. and Schulman, J.D. (1998) Births of normal daughters after MicroSort sperm separation and intrauterine insemination, in-vitro fertilization, or intracytoplasmic sperm injection. Hum Reprod 13:2367–2370.
- Garner, D.L. (2006) Flow cytometric sexing of mammalian sperm. Theriogenology 65:943–957.
- Garner, D.L. (2009) Hoechst 33342: the dye that enabled differentiation of living X-and Y-chromosome bearing mammalian sperm. Theriogenology 71:11–21.
- Haaf, T. and Ward, D.C. (1995) Higher order nuclear structure in mammalian sperm revealed by in situ hybridization and extended chromatin fibers. Exp Cell Res 219:604–611.
- Hazzouri, M., Rousseaux, S., Mongelard, F., Usson, Y., Pelletier, R., Faure, A.K. (2000) Genome organization in the human sperm nucleus studied by FISH and confocal microscopy. Mol Reprod Dev 55:307–315.
- Hewitson, L., Dominko, T., Takahashi, D., Martinovich, C., Ramalho-Santos, J., Sutovsky, P. (1999) Unique checkpoints during the first cell cycle of fertilization after intracytoplasmic sperm injection in rhesus monkeys. Nat Med 5:431–433.
- Ioannou, D. and Griffin, D.K. (2011) Male fertility, chromosome abnormalities, and nuclear organization. Cytogenet Genome Res 133:269–279.
- Johnson, L.A., Welch, G.R., Keyvanfar, K., Dorfmann, A., Fugger, E.F. and Schulman, J.D. (1993) Gender preselection in humans? Flow cytometric separation of X and Y spermatozoa for the prevention of X-linked diseases. Hum Reprod 8:1733–1739.
- Luetjens, C.M., Payne, C. and Schatten, G. (1999) Non-random chromosome positioning in human sperm and sex chromosome anomalies following intracytoplasmic sperm injection. Lancet 353:1240.
- Manvelyan, M., Hunstig, F., Bhatt, S., Mrasek, K., Pellestor, F., Weise, A. (2008) Chromosome distribution in human sperm - a 3D multicolor banding-study. Mol Cytogenet 1:25.
- Mari, P.O., Verbiest, V., Sabbioneda, S., Gourdin, A.M., Wijgers, N., Dinant, C. (2010) Influence of the live cell DNA marker DRAQ5 on chromatin-associated processes. DNA Repair (Amst) 9:848–855.
- Millan, N.M., Lau, P., Hann, M., Ioannou, D., Hoffman, D., Barrionuevo, M. (2012) Hierarchical radial and polar organisation of chromosomes in human sperm. Chromosome Res 20:875–887.
- Mudrak, O., Tomilin, N. and Zalensky, A. (2005) Chromosome architecture in the decondensing human sperm nucleus. J Cell Sci 118:4541–4550.
- Olszewska, M., Wiland, E. and Kurpisz, M. (2008) Positioning of chromosome 15, 18, X and Y centromeres in sperm cells of fertile individuals and infertile patients with increased level of aneuploidy. Chromosome Res 16:875–890.
- Ramalho-Santos, J., Amaral, A., Brito, R., Freitas, M. and Almeida Santos, T. (2004) Simultaneous analysis of cytoskeletal patterns and chromosome positioning in human fertilization failures. Fertil Steril 82:1654–1659.
- Ramalho-Santos, J., Sutovsky, P., Simerly, C., Oko, R., Wessel, G.M., Hewitson, L. (2000) ICSI choreography: fate of sperm structures after monospermic rhesus ICSI and first cell cycle implications. Hum Reprod 15:2610–2620.
- Seidel, G.E. (2012) Sexing Mammalian sperm - where do we go from here? J Reprod Dev 58:505–509.
- Sills, E.S., Kirman, I., Thatcher, S.S., 3rd and Palermo, G.D. (1998) Sex-selection of human spermatozoa: evolution of current techniques and applications. Arch Gynecol Obstet 261:109–115.
- Sousa, A.P., Amaral, A., Baptista, M., Tavares, R., Caballero Campo, P., Caballero Peregrin, P. (2011) Not all sperm are equal: functional mitochondria characterize a subpopulation of human sperm with better fertilization potential. PLoS One 6:e18112.
- Tilgen, N., Guttenbach, M. and Schmid, M. (2001) Heterochromatin is not an adequate explanation for close proximity of interphase chromosomes 1–Y, 9–Y, and 16–Y in human spermatozoa. Exp Cell Res 265:283–287.
- Vidal, F., Fugger, E.F., Blanco, J., Keyvanfar, K., Catala, V., Norton, M. (1998) Efficiency of MicroSort flow cytometry for producing sperm populations enriched in X- or Y-chromosome haplotypes: a blind trial assessed by double and triple colour fluorescent in-situ hybridization. Hum Reprod 13:308–312.
- WHO (2010) WHO laboratory manual for the examination and processing of human semen, fifth edition, World Health Organization, Switzerland.
- Zalenskaya, I.A. and Zalensky, A.O. (2004) Non-random positioning of chromosomes in human sperm nuclei. Chromosome Res 12:163–173.
- Zalensky, A. and Zalenskaya, I. (2007) Organization of chromosomes in spermatozoa: an additional layer of epigenetic information? Biochem Soc Trans 35:609–611.
- Zhao, H., Traganos, F., Dobrucki, J., Wlodkowic, D. and Darzynkiewicz, Z. (2009) Induction of DNA damage response by the supravital probes of nucleic acids. Cytometry A 75:510–519