Abstract
Spermatozoa contain a complex population of RNAs including messenger RNAs (mRNAs) and small RNAs such as microRNAs (miRNA). It has been reported that these RNAs can be used to understand the mechanisms by which toxicological exposure affects spermatogenesis. The aim of our study was to compare mRNA and miRNA profiles in spermatozoa from eight smokers and eight non-smokers, and search for potential relationships between mRNA and miRNA variation. All men were selected based on their answers to a standard toxic exposure questionnaire, and sperm parameters. Using mRNA and miRNA microarrays, we showed that mRNAs from 15 genes were differentially represented between smokers and non-smokers (p < 0.01): five had higher levels and 10 lower levels in the smokers. For the microRNAs, 23 were differentially represented: 16 were higher and seven lower in the smokers (0.004 ≤ p < 0.01). Quantitative RT-PCR confirmed the lower levels in smokers compared to non-smokers for hsa-miR-296-5p, hsa-miR-3940, and hsa-miR-520d-3p. Moreover, we observed an inverse relationship between the levels of microRNAs and six potential target mRNAs (B3GAT3, HNRNPL, OASL, ODZ3, CNGB1, and PKD2). Our results indicate that alterations in the level of a small number of microRNAs in response to smoking may contribute to changes in mRNA expression in smokers. We conclude that large-scale analysis of spermatozoa RNAs can be used to help understand the mechanisms by which human spermatogenesis responds to toxic substances including those in tobacco smoke.
Introduction
It is now well established that spermatozoa contain messenger RNAs (mRNAs), which reflect germ cell transcriptional activity during different stages of spermatogenesis [Miller et al. Citation1999; Ostermeier et al. Citation2002]. In spite of the heterogeneity of the sperm RNA population, some transcripts seem to be strictly regulated [Lalancette et al. Citation2009]. Thus, the analysis of transcripts present in men’s spermatozoa represents a non-invasive method to study the processes involved during their spermatogenesis. More recently, it has also been shown that human spermatozoa bear small RNAs such as microRNAs (miRNAs) [Krawetz et al. Citation2011; Ostermeier et al. Citation2005]. MicroRNAs are short non-coding molecules that bind specifically, in conjunction with a protein complex, to several target messenger RNAs by complementary binding to the 3′-untranslated region [Eriksson et al. Citation2003] to trigger their degradation or transcriptional repression [Guo et al. Citation2010; Thomas et al. Citation2010]. Several studies have shown that many miRNAs are involved in the regulation of gene expression through the targeting of mRNAs during several processes such as cell proliferation, apoptosis, development, or cancer [Huang et al. Citation2011], including testis cancer [Voorhoeve et al. Citation2006]. One study has reported that spermatozoa mRNA profiles can be used to study the interaction between the environment and the germinal cells [Linschooten et al. Citation2009]. Moreover, microRNA expression levels have been associated with environmental exposures, especially cigarette smoke, making them potential biomarkers for better understanding the mechanisms of environmental diseases [Hou et al. Citation2011; Izzotti et al. Citation2009; Izzotti and Pulliero Citation2014].
Among the environmental agents to which humans can be exposed, cigarette smoke is one of the most common. Cigarette smoking releases more than 4,000 components into the human body. Dozens of them are classified as carcinogenic, mutagenic, and/or reproductive toxicants under European regulations [INRS 2008], and are thus likely to impact the quality of the gametes. Several studies have shown deleterious effects of tobacco smoking on semen quality [Rubes et al. Citation1998; Shrivastava et al. Citation2014]. Moreover, a link between paternal tobacco addiction and an increased risk of development of some childhood cancers has been reported [Ji et al. Citation1997]. The mechanisms by which cigarette smoking can impact spermatogenesis are multiple. Benzo(a)pyrene is formed during the combustion of tobacco, and one of its strongly reactive metabolites, the benzo(a)pyrene-diol-epoxide (BPDE), has the capacity to bind DNA covalently, leading to the formation of stable, potentially mutagenic, adducts. Indeed, the presence of such adducts can modify the three-dimensional conformation of the DNA. DNA adducts are recognized and repaired by the nucleotide excision-repair system, but repairs are error prone. Moreover, the large size of adducts and the resulting DNA distortion can cause DNA double-strand breaks, whose repair is likely to involve a loss of genetic material. These DNA adducts have been detected in the spermatozoa of smokers [Perrin et al. Citation2011a,Citationb; Zenzes et al. Citation1999a]. In addition, these adducts were also found in human embryos conceived by couples where the man but not the woman smoke, [Zenzes et al. Citation1999b], suggesting that the transmission of DNA adducts to the human embryos occurs via the spermatozoon. Moreover, a recent study of paternal exposure to benzo(a)pyrene showed that it might affect the expression of several microRNAs in mouse embryo. This suggests that smoking induced miRNA variations in human spermatozoa may also affect human embryo development and may have a potential trans generational effect [Brevik et al. Citation2012]. It must however be noted that the specific nature of spermatogenesis impairment in smokers has never been investigated. The metabolic pathways affected are probably multiple, and these may interact with each other. Large-scale molecular techniques are now available to explore these mechanisms, which will allow the development of assays that can be used to define the molecular basis of spermatogenic dysfunction [Anton and Krawetz Citation2012].
Our aim was to conduct a preliminary innovative study on spermatozoa RNAs, to compare mRNA and miRNA profiles in spermatozoa of smokers and non-smokers, and search for potential relationships between mRNA and miRNA variation. Our results show that this is a promising approach for beginning to understand how spermatogenesis is impacted in men exposed to potential reprotoxic molecules.
Results
Study group, semen parameters, and RNA extracts
The two study groups included eight smokers and eight non-smokers of similar ages (respective mean age 40.25 vs. 37.87, p = 0.393). The experimental design and the techniques used on each sperm sample are illustrated in (see details in the Materials and Methods section). Semen parameters for all men are available in . The quantity of spermatozoa available for this study varied from 25 to 145 million. RNA recovery ranged from 6.48 to 81.71 ng/106 spermatozoa (), a yield variability already described for RNA extraction from spermatozoa [Lalancette et al. Citation2009]. Comparison of semen parameters between smokers and non-smokers showed no statistical difference in sperm concentration (74.6+/−25.58 vs. 155.9+/−104.56, p = 0.065), or percentage of progressive sperm (motility a+b) (35.62+/−11.48 vs. 42.50+/−11.95, p = 0.260), respectively, but the percentage of morphologically normal sperm was lower in smokers (25.75+/−7.83 and 43.50+/−17.28 (p = 0.024), respectively.
Figure 1. Illustration of the experimental design of the study. (A) Methodology of participant selection. (B) Diagram illustrating the techniques used on each participant’s sperm sample. The eight smokers and the eight non-smokers are identified using letters A to H as indicated in . S: smoker; NS: non-smoker; miRNA: microRNAs; qPCR: quantitative PCR.
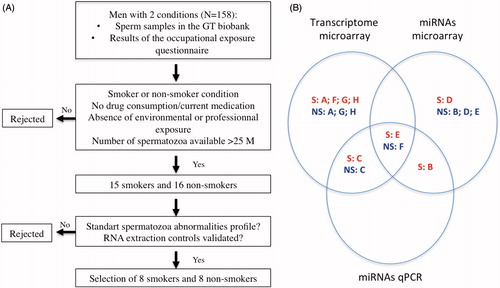
Table 1. Sperm parameters and profiles of smoker and non-smoker patients used for transcriptome study, miRNA chips, and miRNA quantitative PCR.
Table 2. RNA quantity obtained from each sperm sample.
Population selection and mRNA levels in spermatozoa in smokers and non-smokers
Samples from six smokers and five non-smokers were used on mRNA microarrays (). After microarray data normalization, we selected 18,836 genes that were detected in more than five out of the 11 samples. We further performed a t test to look for gene expression differences between smokers and non-smokers. We selected 15 genes having p < 0.01 (), and we performed unsupervised hierarchical clustering of samples based on the expression of the selected genes. As shown in , this procedure successfully classified six out of six smoker samples, and five out of five non-smoker samples. Moreover, we performed a two-way ANOVA to take into account a moderate level of alcohol as a factor (see Materials and Methods). Gene expression differences between smokers and non-smokers were not affected by alcohol for BAX, S100A13, B3GAT3, and PKD2 (p < 0.01).
Figure 2. Unsupervised hierarchical clustering of 15 differentially represented transcripts in spermatozoa after p value filtering of t test (p < 0.01) on six smokers and five non-smokers. Smoker samples are identified by the yellow bar and are located on the left of the cluster. Heatmap representation of over represented (red) and under represented (green) genes. The color scale bar indicates Log2 ratio of intensities.
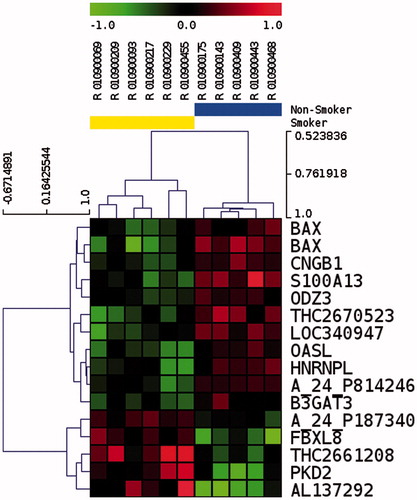
Table 3. List of transcripts with differential level of detection in spermatozoa between smokers and non-smokers after t test (p < 0.01).
miRNA levels in spermatozoa from smokers and non-smokers
We performed miRNA microarray analysis with spermatozoa RNA from four smokers and four non-smokers (), three of which were included in the mRNA microarray analysis. We first identified the miRNAs that showed the highest level of expression in the eight samples. To this end, we selected the miRNAs with an average level of detection corresponding to the 10% most represented miRNAs, and a variance corresponding to the 20% least variable. The 19 most represented miRNAs in spermatozoa of the eight patients are listed in .
Table 4. List of miRNAs present in human spermatozoa with an average level of expression included in the 10% of most represented miRNAs.
Thus, we selected 522 microRNAs with an expression above background in four out of eight samples (the background being defined by the average expression of all microRNAs), and identified 23 microRNAs differentially represented between smokers and non-smokers with a p value between 0.004 and 0.094 (median 0.0042) (), seven of them being down regulated, and 16 up regulated, in smokers. Among these 23 miRNAS, 12 had a p value < 0.05. In spite of the relatively low level of global expression in spermatozoa, the unsupervised hierarchical clustering of samples based on the 23 selected microRNAs allowed us to discriminate samples by smoking status (). As for mRNA analysis,we performed a two-way ANOVA to take into account moderate alcohol consumption. Differences between smokers and non-smokers were not affected by amoderate level of alcohol for hsa-miR-126*, hsa-miR-92a-2*, hsa-miR-26a-1, hsa-miR-517c, and hsa-miR-340* (p < 0.01).
Figure 3. Unsupervised hierarchical clustering of 23 differentially represented miRNAs in spermatozoa after p value filtering of t test (p < 0.01) on four smokers and four non-smokers. Smoker samples are identified by the yellow bar and are located on the left of the cluster. Heatmap representation of over represented (red) and under represented (green) genes. The color scale bar indicates Log2 ratio of intensities.
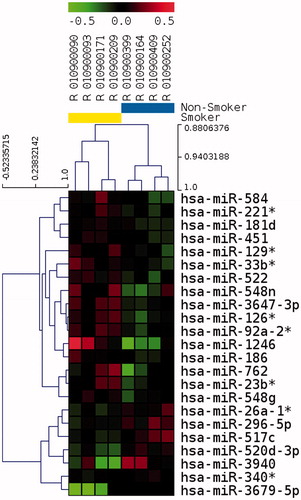
Table 5. List of miRNAs with differential level of detection in spermatozoa between smokers and non-smokers (p < 0.1).
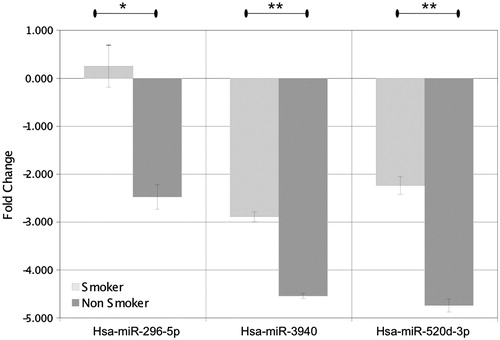
Quantitative real-time RT-PCR (RT-qPCR) analysis of smoker (R01090090, R010900209) and non-smoker samples (R010900175, R0109409) was performed for miRNAs differentially represented on the microarray. We choose two miRNAs up regulated in smokers (hsa-miR-129*, hsa-miR-1246), and three miRNAs down regulated in smokers (hsa-miR-296-5p, hsa-miR-3940, hsa-miR-520d-3p). Quantitative PCR (qPCR) confirmed that hsa-miR-296-5p, hsa-miR-3940 (miRBase mature sequence hsa-miR-3940-5p), hsa-miR-520d-3p were down regulated in smokers (). The qPCR did not confirm the up regulation for hsa-miR-129* and hsa-miR-1246 in smokers. The three down regulated miRNAs are at a level that is two to four times less than that present in smokers.
Putative miRNA target gene expression in human spermatozoa
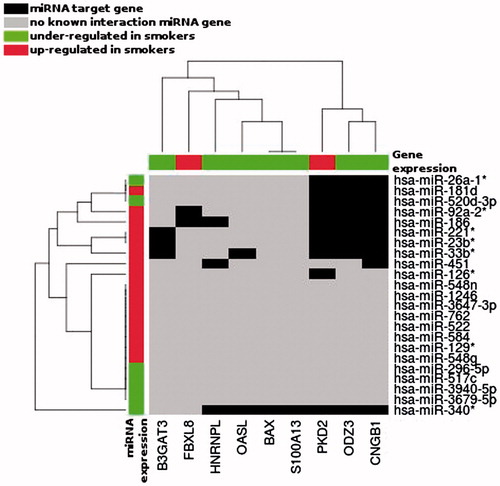
Putative targets for the 23 miRNAs differentially represented on the microarray according to smoker status, and for hsa-miR-638 (highly represented) used as control were determined using TargetScan (version 6.2, June 2012) [Lewis et al. Citation2005]. To determine whether miRNAs might be modulators of mRNAs that were differentially detected in smokers, we examined whether our selected mRNAs identified as affected by smoking included predicted targets of each of the miRNAs differentially represented between smokers and non-smokers. We found that nine out of the 15 are predicted to be targets of 11 miRNAs out of the 23 differentially detected in smokers (). This suggests that alterations in the levels of a relatively small number of miRNAs in response to smoking could potentially contribute to smoking-associated changes in mRNA expression. Interestingly, the microarray data show that the direction of variation of mRNAs compared to microRNAs is in agreement with a potential interaction during human spermatogenesis for five down-regulated genes (B3GAT3, HNRNPL, OASL, ODZ3, and CNGB1) and one up-regulated gene, PKD2. Indeed, PKD2 is a putative target of hsa-miR-520d-3p that we found to be down regulated in smokers by microarray and qPCR analysis.
Figure 5. Unsupervised hierarchical clustering of the differentially represented miRNAs (Y axis) and mRNAs (X axis) during spermatogenesis of studied patients, according to the smoking status. Only the miRNAs having target gene, in the TargetScan database v 6.2, corresponding to our 15 differentially represented mRNAs are represented. Black boxes correspond to miRNA target genes; red boxes: gene/miRNA over represented in Smokers; green boxes: gene/miRNA under represented in Smokers.
Discussion
In the present study, we examined the possibility that sperm mRNAs and miRNAs could be used as candidate markers to study the impact of toxicants such as cigarette smoke on human spermatogenesis. To the best of our knowledge, this is the first study of mRNA and miRNA profiles during spermatogenesis in men exposed to environmental toxicants, and let us highlight two interesting points.
First, we show that by using total RNA extracts from spermatozoa of smokers and non-smokers, we were able to select transcripts and miRNAs differentially represented in human spermatozoa based on smoking status. Among the 15 transcripts shown to be differential, PKD2, BAX, and HNRNPL are known to be involved in spermatogenesis regulation. Evidence from online resources indicate that the proteins they encode are present in human adult germ cells (www.proteinatlas.org, www.humanproteomemap.org [Fagerberg et al. Citation2014; Kim et al. Citation2014]). Polycystic Kidney Disease 2 (PKD2) overrepresented in smokers, encodes, the polycystin-2 (PC-2) involved in mesonephric tubule morphogenesis. PKD isoforms located in cilia have been demonstrated to play a role in the transduction of mechanosensation via calcium signalling [Nauli et al. Citation2003], and have been shown to operate in sperm as a determinant of directional movement inside the drosophila female reproductive tract [Gao et al. Citation2003; Yang et al. Citation2011]. The calcium channel PKD2 mediates sensory perception on cilia and flagella, and these defects can contribute to ciliopathic diseases. In human, asthenozoospermia and necrozoospermia have been described in patients bearing autosomal dominant polycystic kidney diseases [Vora et al. Citation2008]. If PKD2 may be involved in the modulation of flagellar properties, and given that there is no difference in motility between smokers and non-smokers in our study, one could hypothesize that an increased level of PKD2 transcripts in smokers may be a part in a compensatory mechanism that maintains normal motility despite a negative impact of smoking. The two other transcripts BAX and HNRNPL appear under represented in smokers. BAX encodes a well-known pro-apoptotic factor described as involved in male germ cell apoptosis [Carreau et al. Citation2011; Saygin et al. Citation2011; Sinha Hikim et al. Citation2003]. HNRNPL encodes an mRNA processing factor shown to be expressed in spermatids and involved in post-transcriptional expression during testicular spermatozoa maturation [Chapman et al. Citation2013; Li et al. Citation2012]. HNRNPL is supposed to be a key factor involved in the bioprocess of apoptosis during spermatogenesis. Interestingly, a recent study of spermatozoal RNA profile in smokers reported that apoptosis appeared to be the most influenced process, which means that damaged cells may not be eliminated as efficiently during the germ cell cycle in smokers [Linschooten et al. Citation2009]. Nevertheless, although differential transcripts between smokers and non-smokers in our study are not the same as those reported in this previous study, both support the hypothesis that tobacco smoking impacts the germ cell apoptotic process. Indeed, it has been shown that although discordances may be found between data sets for the same biological process, common patterns of expression and outcome predictions can be identified in data sets generated by independent labs [Hu et al. Citation2006]. However, it should be stressed that further studies with independent samples are required to clarify how smoking affects spermatozoa mRNA profiles.
Regarding miRNAs, we were able to distinguish spermatozoal miRNA profiles according to the smoking status despite the small amount of miRNAs that are present in our total human spermatozoa extracts. We show that 23 miRNAs were differentially represented between smokers and non-smokers. Among them, hsa-miR-1246 was previously shown to be up regulated in spermatozoa of smokers [Marczylo et al. Citation2012]. It is interesting to note that this previous study and ours find that miRNAs in spermatozoa of smokers seem to be more frequently up regulated than down regulated. This pattern is different than the one described in cancer or in airway epithelium of smokers where miRNAs are preferentially down regulated [Schembri et al. Citation2009]. This suggests that cigarette smoking could impact human spermatogenesis via mechanisms distinct from those acting during tumorigenesis. Using RTqPCR, three miRNAs were confirmed to be down regulated in smokers. Their roles in human spermatogenesis are still unclear, but two of them are known to be involved in human spermatogenesis or in tobacco smoke induced cell modifications. Hsa-miR-296-5p was shown to be involved in tumour angiogenesis and was present in embryonic stem cells, suggesting it may have a role in regulation of early embryonic development. Hsa-miR-520d-3p was shown to be present in human testis and was down regulated in the testis of patients with non-obstructive azoospermia. Altogether there are presumptions for a potential role of those miRNAs in the spermatogenetic modifications induced by cigarette smoking.
Second, we show that it is possible to find a relationship between the dysregulation of mRNAs and miRNAs in human spermatozoa according to smoking status. Among miRNAs that were down regulated in smokers, it is interesting to note that some of their theoretical targets are present among transcripts that appear up regulated. Indeed, hsa-miR-520d-3p potentially regulates PKD2, found to be up regulated in the same group and reported to be involved in regulating spermatogenesis [Gao et al. Citation2003; Yang et al. Citation2011]. Among miRNAs that were up regulated in smokers, hsa-miR-23b is predicted to target ODZ3, B3GAT3, and CNGB1, which we found to be down regulated. Hsa-miR-23b was reported to be up regulated in spermatozoa of asthenozoospermic patients [Abu-Halima et al. Citation2013]. ODZ3 encodes a protein expressed in neurons that may function as a cellular signal transducer [Beckmann et al. Citation2011; Nunes et al. Citation2005]. B3GAT3 encodes a member of the glucuronyltransferase gene family [Gulberti et al. Citation2003; Ouzzine et al. Citation2000], and CNGB1 a cyclic nucleotide-gated cation channel [Becirovic et al. Citation2010]. Although a biological relationship could not be demonstrated, our results suggest that alterations in the levels of a small number of miRNAs in response to smoking could potentially contribute to smoking–associated changes in mRNA expression, that potentially impact cellular transducers and possible consequences for apoptosis or spermatozoa motility.
Our study of mRNAs and miRNAs from human spermatozoa that contain very low levels of RNAs has nevertheless some limitations. In particular, the technique we used for RNAs extraction was not modified to fully recover the miRNAs and thus may explain our small yield of miRNAs and differences compared to miRNAs previously reported in spermatozoa [Krawetz et al. Citation2011; Hammoud et al. 2014]. Indeed, it has been reported that the concentration of total RNA in the starting material is critical in determining the yield of small RNA [Kim et al. Citation2012]. As a very low level of RNA is present in human spermatozoa, a technical adaptation may improve the significance of the results.
In conclusion, this pilot study shows that large-scale analysis of sperm RNAs can be used to help understand the mechanisms that mediate the response to tobacco smoke exposure during human spermatogenesis. However, the identification of the complete molecular pathways affected is a real challenge because of the low RNA levels present in human spermatozoa. While it has been shown that smoking-induced differences in spermatozoal mRNA can be detected with few sperm samples [Linschooten et al. Citation2009], the extension to a larger number of samples will improve the significance of our results. Moreover, new tools such as RNA-seq may soon provide the means to more fully characterize the RNAs present in human spermatozoa and thus to elucidate the fine regulatory mechanisms that mediate responses to environmental agent exposure during human spermatogenesis.
Materials and Methods
The Germetheque Biobank provided all samples to the authors. Germetheque obtained informed consent from each patient for inclusion of samples in the biobank and for their use for studies on human fertility in accordance with the Helsinki Declaration of 1975 on human experimentation. The Germetheque Scientific Committee approved the present study design. Consequently, no IRB approval was sought.
Participants and tobacco exposure
Smoker and non-smoker men consulted for primary infertility in the infertility clinic of the University Hospital of La Conception (Marseilles, France). A standardized questionnaire was used to evaluate cigarette smoking (with details about smoking levels, duration, and contribution of passive smoke), alcohol consumption, and occupational exposure [De Fleurian et al. Citation2009].
None of the participants had any known heritable condition that could favour an abnormal semen profile, no current medication known to impair spermatogenesis, no drug consumption (except cigarette), which can influence spermatogenesis. They had not taken medication for at least six months and their Body Mass Index was consistent with a normal body mass. Moreover, they were not occupationally exposed to chemical or physical agents likely to have effects on the spermatogenesis. Alcohol consumption of more than three drinks/w was a factor of exclusion. Patients smoking at least 10 cigarettes/d for at least six months defined the smokers group. Patients who had never smoked or had stopped cigarette smoking for at least 10 y were considered the non-smokers.
Sperm analysis and preparation
Semen was collected via masturbation after a period of sexual abstinence of 2-6 d. All patients gave an informed consent for the conservation of the remnant sperm after semen analysis in the Germetheque biobank. After 30 min of liquefaction, semen analysis was performed according to the WHO [Citation2009] criteria and according to the French David classification for the morphology analysis [Auger and Eustache Citation2000]. Within 30 min, a sample of one to two ml of sperm was then centrifuged at 3,000 rpm for 15 min. Seminal plasma was removed; the pellet was snap-frozen and stored in the Germetheque biobank in liquid nitrogen until use.
Experimental design
Sperm samples were selected from those stored in the Germetheque Biobank (n = 158), and were selected according to the result of the occupational exposures questionnaire, the number of spermatozoa available, and the sperm parameters. Therefore, eight smokers and eight non-smokers were selected, with a standard profile for morphological abnormalities. Whole mRNA microarray was performed on RNA extracts from six smokers and five non-smokers, miRNA microarray on RNA from four smokers and four non-smokers, and miRNA RT-qPCR analysis on RNA from two smokers and two non-smokers.
RNA extraction
Subsequent to storage, samples were thawed and washed twice in 2 ml of PBS, then suspended in round cell lysis buffer (0.1% SDS, 0.5% Triton X-100 in RNase free H2O). RNA was extracted using the RNeasy mini kit (Qiagen GmbH, Germany) with minor modifications. Sperm cells were suspended in 600 µl of lysis buffer for 107 cells. This cell suspension was lysed using a Politron as previously described [Saade et al. Citation2007], to maximize the recovery of spermatozoa RNA. The lysates were then processed according to the manufacturer’s instructions using the animal cell protocol. After RNase-free DNase treatment (Qiagen GmbH), the RNA was eluted with 30 µl of RNase free H2O. RNA concentration was determined using a nanodrop ND-1000 spectrophotometer (NanoDrop Technologies, Wilmington, DE, USA). RNA quality was assessed on an Agilent 2100 Bioanalyzer with the Agilent RNA 6000 nano kit (Agilent Technologies, Santa Clara, CA, USA), showing that the RNA is highly fragmented and that the 18S and 28S ribosomal RNAs are absent (see supplementary Figure S2).
Quality control of RNA extracts
One aliquot of 400 -500 ng of each sample was subjected to reverse transcriptase PCR using oligo-dT 25 primers and SuperScript II (Invitrogen, Carlsbad, CA, USA). The DNA was amplified by polymerase chain reactions using selected oligonucleotides in a final volume of 20 µl. To assure the purity of RNA extracts and be sure that there were no round cell extracts, we choose primers such as CD45, c-kit, or E-cadherin which are positive markers for leukocytes, testicular germ cells, and epithelial cells, respectively [Lambard et al. Citation2004]. Genomic DNA contamination was tested under conditions described above using a set of primers specific to PRM2 that span an intron [Miller et al. Citation1999] (see supplementary Figure S1). The positive controls for leukocytes and immature germ cell contamination were as follow: total RNA extracted from blood sample and total RNA extracted from human testis (636533 Clontech, purchased from Ozyme, Saint Quentin en Yvelines, France). Samples were also submitted to PCR.
Whole human genome Agilent microarray gene expression
Cy3 labelled cRNA was generated from 500 ng total RNA of patient’s spermatozoa, using the Low RNA Input Fluorescent Linear Amplification Kit (Agilent Technologies) following manufacturer’s instructions. Equal amounts of purified Cy3 labelled cRNA were hybridized for 17 h at 65°C to the Agilent whole human genome 4x44k microarray (G4112F). Each Agilent slide was hybridized with two smoker samples and two non-smoker samples according to the manufacturer’s instructions, taking care to change the order between smokers and non-smokers for each slide. The microarray was washed using the Gene Expression Wash Buffer Kit (Agilent Technologies) and scanned as for the standard Agilent protocol. The data were processed using Feature Extraction software. Data have been deposited in NCBI’s Gene Expression Omnibus and are accessible through GEO Series accession number GSE44133.
Microarray data normalization
Whole genome microarray quantification files derived from the Agilent Feature Extraction Software were analyzed using the Agilent R package (ftp://tagc.univ-mrs.fr/public/AgiND/) for quality control and normalization. The quantile method was used for normalization. Among probes obtained from Feature Extraction files, those for which at least 60% of samples are above background using the “gIsWellAboveBg” feature were conserved for statistical analysis (n = 18,836).
To reduce the effect of noise in the miRNA microarray data, we aggregated probe signals to have a more robust measure of expression, by computing the median of the intensity values “gProcessedSignal” of the set of probes corresponding to each miRNA. The samples were then normalized by the quantile method allowing a comparison of experimental samples between each other.
Statistical analysis
Statistical analyses were performed using the TIGR MeV (MultiExperiment Viewer) v4.5 and the GeneANOVA programs [Didier et al. Citation2002]. A student t test was applied to identify gene and miRNA whose expression differed between the smoker and the non-smoker groups. In addition, a two-way ANOVA was used to take simultaneously into account the effect of smoking and alcohol consumption on gene expression, each factor being binary: smokers/non-smokers and alcohol/non-alcohol.
MicroRNAs microarray preparation and hybridization
Total spermatozoa RNA extracts (100 ng) were labeled using the Agilent miRNA complete labeling and hybridization kit (5190-0456). Hybridization was performed on Agilent human miRNA microarray 8x60K (G-4872A-031181) sourced from the Sanger miRBASE. All procedures were performed according to the manufacturer’s instructions. Microarrays with labeled samples were hybridized 20 h at 55°C and washed using the Gene Expression Wash Buffer Kit (5188-5327). Arrays were scanned using a High-Resolution Microarray Scanner and images were analyzed using Feature extraction software. Data were corrected as described above using AgiND R library with the same parameters and filters. Data are accessible through GEO Series accession number GSE44134.
Real time miRNA qPCR
To validate miRNA microarray results, a subset of miRNAs identified as more concentrated or less concentrated in smokers by microarray analysis were analyzed for smoker-related differential expression. RT-qPCR was performed using a standard TaqMan MicroRNA assay according to the manufacturer’s standard protocol (Applied Biosystems, Carlsbad, CA, USA) [Chen et al. Citation2005]. Twenty nanograms of the isolated total RNA was reverse transcribed into cDNA using the TaqMan Multiscribe Reverse Transcription kit (4366596 Applied Biosystems). The MiRNA sequences and assay IDs are listed in . Each reaction was performed in triplicate in two smokers and two non-smokers. Quantification was performed using validated TaqMan probes, the TaqMan MicroRNA assay and the TaqMan Gene Expression Master Mix (Applied Biosystems). The fold change in miRNA level was calculated using has-miR-548q, as endogenous control.
Table 6. MicroRNAs (miRNA) sequences and assay IDs used for miRNA quantitative real-time PCR.
Supplementary material available online Supplementary Figures
UAAN_1022835_Supplementary_Information.zip
Download Zip (650 KB)Acknowledgments
We are grateful to the patients who gave their informed consent for the use of their samples for research. We thank C. Metton and M.J. Fays-Bernardin for technical assistance and Germetheque support. We also thank M. Mitchell for the text revision, and M. Guichaoua for her precious advise.
Declaration of interest
This work was supported by grants from INSERM: Institut National de la Santé Et de la Recherche Médicale and APHM: Assistance Publique – Hôpitaux de Marseille. The Germetheque biobank was supported by grants from the ANR (Agence Nationale de la Recherche), the Agence de la biomedicine, the Centre Hospitalier Universitaire de Toulouse, and APHM (Assistance Publique Hôpitaux de Marseille). All authors declare no declarations of interest and no competing financial interest.
Author contributions
Played a role in the sample collection, the biological experiments, the study design and the manuscript writing: CMG. Had a major role in the bioinformatics data analysis: CL, AB, PR. In charge of microarray and quantitative PCR experiments: GV, MY, NB. Played a role in the patient recruitment: JP, ISM. Direct responsibility for the manuscript: CN, PR. All authors contributed to the writing and revision of the manuscript.
References
- Abu-Halima, M., Hammadeh, M., Schmitt, J., Leidinger, P., Keller, A., Meese, E., et al. (2013) Altered microRNA expression profiles of human spermatozoa in patients with different spermatogenic impairments. Fertil Steril 99:1249–1255 e1216
- Anton, E. and Krawetz, S.A. (2012) Spermatozoa as biomarkers for the assessment of human male infertility and genotoxicity. Syst Biol Reprod Med 58:41–50
- Auger J. and Eustache F. (2000) Standardisation de la classification morphologique des spermatozoïdes humains selon la méthode de David modifiée. Andrologie 4:358–373
- Becirovic, E., Nakova, K., Hammelmann, V., Hennel, R., Biel, M. and Michalakis, S. (2010) The retinitis pigmentosa mutation c.3444+1G>A in CNGB1 results in skipping of exon 32. PLoS One 5:e8969
- Beckmann, J., Vitobello, A., Ferralli, J., Kenzelmann Broz, D., Rijli, F. M. and Chiquet-Ehrismann, R. (2011) Human teneurin-1 is a direct target of the homeobox transcription factor EMX2 at a novel alternate promoter. BMC Dev Biol 11:35
- Brevik, A., Lindeman, B., Brunborg, G. and Duale, N. (2012) Paternal Benzo[a]pyrene Exposure Modulates MicroRNA Expression Patterns in the Developing Mouse Embryo. Int J Cell Biol 2012:407431
- Carreau, S., Bouraima-Lelong, H. and Delalande, C. (2011) Estrogens: new players in spermatogenesis. Reprod Biol 11:174–193
- Chapman, K.M., Powell, H.M., Chaudhary, J., Shelton, J.M., Richardson, J.A., Richardson, T.E., et al. (2013) Linking spermatid ribonucleic acid (RNA) binding protein and retrogene diversity to reproductive success. Mol Cell Proteomics 12:3221–3236
- Chen, C., Ridzon, D.A., Broomer, A.J., Zhou, Z., Lee, D.H., Nguyen, J.T., et al. (2005) Real-time quantification of microRNAs by stem-loop RT-PCR. Nucleic Acids Res 33:e179
- De Fleurian, G., Perrin, J., Ecochard, R., Dantony, E., Lanteaume, A., Achard, V., et al. (2009) Occupational exposures obtained by questionnaire in clinical practice and their association with semen quality. J Androl 30:566–579
- Didier, G., Brezellec, P., Remy, E. and Henaut, A. (2002) GeneANOVA–gene expression analysis of variance. Bioinformatics 18:490–491
- Eriksson, M., Brown, W.T., Gordon, L.B., Glynn, M.W., Singer, J., Scott, L., et al. (2003) Recurrent de novo point mutations in lamin A cause Hutchinson-Gilford progeria syndrome. Nature 423:293–298
- Fagerberg, L., Hallstrom, B.M., Oksvold, P., Kampf, C., Djureinovic, D., Odeberg, J., et al. (2014) Analysis of the human tissue-specific expression by genome-wide integration of transcriptomics and antibody-based proteomics. Mol Cell Proteomics 13:397–406
- Gao, Z., Ruden, D.M. and Lu, X. (2003) PKD2 cation channel is required for directional sperm movement and male fertility. Curr Biol 13:2175–2178
- Gulberti, S., Fournel-Gigleux, S., Mulliert, G., Aubry, A., Netter, P., Magdalou, J., et al. (2003) The functional glycosyltransferase signature sequence of the human beta 1,3-glucuronosyltransferase is a XDD motif. J Biol Chem 278:32219–32226
- Guo, H., Ingolia, N.T., Weissman, J.S. and Bartel, D.P. (2010) Mammalian microRNAs predominantly act to decrease target mRNA levels. Nature 466:835–840
- Hammoud, S.S., Low, D.H., Yi, C., Carrell, D.T., Guccione, E. and Cairns, B.R. (2014) Chromatin and transcription transitions of mammalian adult germline stem cells and spermatogenesis. Cell Stem Cell 15:239--253
- Hou, L., Wang, D. and Baccarelli, A. (2011) Environmental chemicals and microRNAs. Mutat Res 714:105–112
- Hu, Z., Fan, C., Oh, D.S., Marron, J.S., He, X., Qaqish, B.F., et al. (2006) The molecular portraits of breast tumors are conserved across microarray platforms. BMC Genomics 7:96
- Huang, Y., Shen, X.J., Zou, Q., Wang, S.P., Tang, S.M. and Zhang, G.Z. (2011) Biological functions of microRNAs: a review. J Physiol Biochem 67:129–139
- Institut National de la Recherche et de Sécurité (INRS) pour la prévention des accidents du travail et des maladies professionnelles. (2008) Classification et étiquetage des produits chimiques
- Izzotti, A. and Pulliero, A. (2014) The effects of environmental chemical carcinogens on the microRNA machinery. Int J Hyg Environ Health 217:601–627
- Izzotti, A., Calin, G.A., Arrigo, P., Steele, V.E., Croce, C.M. and De Flora, S. (2009) Downregulation of microRNA expression in the lungs of rats exposed to cigarette smoke. Faseb J 23:806–812
- Ji, B.T., Shu, X.O., Linet, M.S., Zheng, W., Wacholder, S., Gao, Y.T., et al. (1997) Paternal cigarette smoking and the risk of childhood cancer among offspring of nonsmoking mothers. J Natl Cancer Inst 89:238–244
- Kim, M.S., Pinto, S.M., Getnet, D., Nirujogi, R.S., Manda, S.S., Chaerkady, R., et al. (2014) A draft map of the human proteome. Nature 509:575–581
- Kim, Y.K., Yeo, J., Kim, B., Ha, M. and Kim, V.N. (2012) Short structured RNAs with low GC content are selectively lost during extraction from a small number of cells. Mol Cell 46:893–895
- Krawetz, S.A., Kruger, A., Lalancette, C., Tagett, R., Anton, E., Draghici, S., et al. (2011) A survey of small RNAs in human sperm. Hum Reprod 26:3401–3412
- Lalancette, C., Platts, A.E., Johnson, G.D., Emery, B.R., Carrell, D.T. and Krawetz, S.A. (2009) Identification of human sperm transcripts as candidate markers of male fertility. J Mol Med (Berl) 87:735–748
- Lambard, S., Galeraud-Denis, I., Saunders, P.T. and Carreau, S. (2004) Human immature germ cells and ejaculated spermatozoa contain aromatase and oestrogen receptors. J Mol Endocrinol 32:279–289
- Lewis, B.P., Burge, C.B. and Bartel, D.P. (2005) Conserved seed pairing, often flanked by adenosines, indicates that thousands of human genes are microRNA targets. Cell 120:15–20
- Li, J., Guo, W., Li, F., He, J., Yu, Q., Wu, X., et al. (2012) HnRNPL as a key factor in spermatogenesis: Lesson from functional proteomic studies of azoospermia patients with sertoli cell only syndrome. J Proteomics 75:2879–2891
- Linschooten, J.O., Van Schooten, F.J., Baumgartner, A., Cemeli, E., Van Delft, J., Anderson, D., et al. (2009) Use of spermatozoal mRNA profiles to study gene-environment interactions in human germ cells. Mutat Res 667:70–76
- Marczylo, E.L., Amoako, A.A., Konje, J.C., Gant, T.W. and Marczylo, T.H. (2012) Smoking induces differential miRNA expression in human spermatozoa: A potential transgenerational epigenetic concern? Epigenetics 7:432–439
- Miller, D., Briggs, D., Snowden, H., Hamlington, J., Rollinson, S., Lilford, R., et al. (1999) A complex population of RNAs exists in human ejaculate spermatozoa: implications for understanding molecular aspects of spermiogenesis. Gene 237:385–392
- Nauli, S.M., Alenghat, F.J., Luo, Y., Williams, E., Vassilev, P., Li, X., et al. (2003) Polycystins 1 and 2 mediate mechanosensation in the primary cilium of kidney cells. Nat Genet 33:129–137
- Nunes, S.M., Ferralli, J., Choi, K., Brown–Luedi, M., Minet, A.D. and Chiquet-Ehrismann, R. (2005) The intracellular domain of teneurin-1 interacts with MBD1 and CAP/ponsin resulting in subcellular codistribution and translocation to the nuclear matrix. Exp Cell Res 305:122–132
- Ostermeier, G.C., Dix, D.J., Miller, D., Khatri, P. and Krawetz, S.A. (2002) Spermatozoal RNA profiles of normal fertile men. Lancet 360:772–777
- Ostermeier, G.C., Goodrich, R.J., Moldenhauer, J.S., Diamond, M.P. and Krawetz, S.A. (2005) A suite of novel human spermatozoal RNAs. J Androl 26:70–74
- Ouzzine, M., Gulberti, S., Netter, P., Magdalou, J. and Fournel-Gigleux, S. (2000) Structure/function of the human Ga1beta1,3-glucuronosyltransferase. Dimerization and functional activity are mediated by two crucial cysteine residues. J Biol Chem 275:28254–28260
- Perrin, J., Tassistro, V., Mandon, M., Grillo, J. M., Botta, A. and Sari-Minodier, I. (2011a) Tobacco consumption and benzo(a)pyrene-diol-epoxide-DNA adducts in spermatozoa: in smokers, swim-up procedure selects spermatozoa with decreased DNA damage. Fertil Steril 95:2013–2017
- Perrin, J., Tassistro, V., Paulmyer-Lacroix, O., Courbiere, B., Botta, A. and Sari-Minodier, I. (2011b) In smokers, swim-up and discontinuous gradient centrifugation recover spermatozoa with equally lower amounts of DNA damage than spermatozoa obtained from neat semen. Fertil Steril 95:2680–2682
- Rubes, J., Lowe, X., Moore, D., II Perreault, S., Slott, V., Evenson, D., et al. (1998) Smoking cigarettes is associated with increased sperm disomy in teenage men. Fertil Steril 70:715–723
- Saade, M., Irla, M., Govin, J., Victorero, G., Samson, M. and Nguyen, C. (2007) Dynamic distribution of Spatial during mouse spermatogenesis and its interaction with the kinesin KIF17b. Exp Cell Res 313:614–626
- Saygin, M., Caliskan, S., Karahan, N., Koyu, A., Gumral, N. and Uguz, A. (2011) Testicular apoptosis and histopathological changes induced by a 2.45 GHz electromagnetic field. Toxicol Ind Health 27:455–463
- Schembri, F., Sridhar, S., Perdomo, C., Gustafson, A. M., Zhang, X., Ergun, A., et al. (2009) MicroRNAs as modulators of smoking-induced gene expression changes in human airway epithelium. Proc Natl Acad Sci U S A 106:2319–2324
- Shrivastava, V., Marmor, H., Chernyak, S., Goldstein, M., Feliciano, M. and Vigodner, M. (2014) Cigarette smoke affects posttranslational modifications and inhibits capacitation-induced changes in human sperm proteins. Reprod Toxicol 43:125–129
- Sinha Hikim, A.P., Lue, Y., Diaz-Romero, M., Yen, P.H., Wang, C. and Swerdloff, R.S. (2003) Deciphering the pathways of germ cell apoptosis in the testis. J Steroid Biochem Mol Biol 85:175–182
- Thomas, M., Lieberman, J. and Lal, A. (2010) Desperately seeking microRNA targets. Nat Struct Mol Biol 17:1169–1174
- Voorhoeve, P.M., le Sage, C., Schrier, M., Gillis, A.J., Stoop, H., Nagel, R., et al. (2006) A genetic screen implicates miRNA-372 and miRNA-373 as oncogenes in testicular germ cell tumors. Cell 124:1169–1181
- Vora, N., Perrone, R. and Bianchi, D.W. (2008) Reproductive issues for adults with autosomal dominant polycystic kidney disease. Am J Kidney Dis 51:307–318
- WHO (2009) WHO laboratory manual for the examination of human semen and sperm-cervical mucus interaction, World Health Organization, Cambridge University Press, Cambridge
- Yang, Y., Cochran, D.A., Gargano, M.D., King, I., Samhat, N.K., Burger, B.P., et al. (2011) Regulation of flagellar motility by the conserved flagellar protein CG34110/Ccdc135/FAP50. Mol Biol Cell 22:976–987
- Zenzes, M.T., Bielecki, R. and Reed, T.E. (1999a) Detection of benzo(a)pyrene diol epoxide-DNA adducts in sperm of men exposed to cigarette smoke. Fertil Steril 72:330–335
- Zenzes, M.T., Puy, L.A., Bielecki, R. and Reed, T.E. (1999b) Detection of benzo[a]pyrene diol epoxide-DNA adducts in embryos from smoking couples: evidence for transmission by spermatozoa. Mol Hum Reprod 5:125–131