Abstract
Genetic factors are believe y a major role in the variation of treatment response and the incidence of adverse effects to medication. The aim of pharmacogenetics is to elucidate this variability according to hereditary differences. Considering current hypotheses for the mechanisms of action of antidepressants, most investigations to date have concentrated on mutations in genes coding either for the pathways in the serotonergic and noradrenergic systems or for drug-metabolizing enzymes. Recent studies shifted the emphasis on the mains mechanism of drug action from changes in neurotransmitter concentration or receptor function toward long-lasting adaptive processes within the neurons. Although the results are controversial, many studies support the hypothesis that psychopharmacogenetics will help predict an individual's drug response, while minimizing the side effects. The inclusion of functional genomics, investigate the complex gene and/or protein expression in response to a given drug, may lead to the development of novel and safer drugs.
Se cree que los factores genéticos juegan un papel principal en la variación de la respuesta terapéutica y en la incidencia de los efectos adversos a la medicación. El objetivo de la farmacogenética es aclarar esta variabilidad de acuerdo con las diferencias hereditarias. Considerando las hipótesis actuales para los mecanismos de acción de los antidepresivos, a la fecha, la mayoría de las investigaciones se han concentrado en las mutaciones de los genes que codifican tanto para las vías de los sistemas serotoninérgico y noradrenérgico como para las enzimas que metabolizan fármacos. El énfasis puesto en el principal mecanismo de acción del fármaco a partir de modificaciones en la concentración del neurotransmisor o en la función del receptor ha sido cambiado en estudios recientes hacía procesos de adaptación a largo plazo dentro de la neurona. Aunque los resultados son controvertidos, muchos estudios sustentan la hipótesis que la psicofarmacogenéiíca ayudará a predecir la respuesta de un individuo al fármaco y a reducir los efectos laterales. La inclusión de la genómica funcional, la cual investiga el complejo gen ylo expresión proteica en respuesta a un fármaco dado, podrá conducir al desarrollo de fármacos nuevos y más seguros.
On suppose que les facteurs génétiques jouent un rôle important dans la variation de la réponse au traitement et l'incidence des effets indésirables des médicaments. Le bus de la pharmacogénétique est d'expliquer cette variabilité d'après des différences héréditaires. Si l'on se réfère aux hypothèses actuelles sur le mécanisme d'action des antidépresseurs, la plupart des recherches à ce jour sont concentrées sur les mutations des gènes codant soit les voies des systèmes sérotoninergiques et noradrénergiques, soit les enzymes intervenant dans le métabolisme des médicaments. Des études récentes sur le principal mécanisme d'action médicamenteuse ont transféré leur intérêt partant des modifications des concentrations en neurotransmetteurs ou de la fonction des récepteurs vers les processus d'adaptation à long terme au sein des neurones. Malgré des résultats controversés, beaucoup d'études sont en faveur de l'hypothèse selon laquelle la pharmacogénétique pourra aider à prévoir une réponse individuelle au médicament tout en réduisant les effets indésirables. L'intervention de la génomique fonctionnelle qui explore le gène complexe et/ou l'expression protéique dans la réponse à un médicament prescrit, peut conduire au développement de nouvelles molécules mieux tolérées.
The first antidepressants (AD) were discovered by chance almost 50 years ago. Despite recent advances in the discovery and design of ADs, interindividual variability to treatment remains a serious problem in clinical psychiatry. It is well known that there are large differences in dosage requirements and that, with a standard dose of a given drug, a significant proportion of patients do not respond satisfactorily while others suffer from serious adverse effects. In both cases, patients do not benefit from the full therapeutic efficacy and a switch between different treatment regimens is often necessary to find a more suitable alternative.
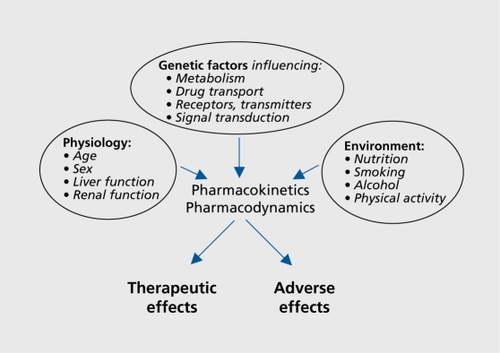
The variability in drug response is highly complex and can be attributed to several physiological and environmental factors, such as the patient's age, renal and liver function, nutritional status, smoking, alcohol consumption, and physical activity (). However, it has been recognized for almost 50 years now that genetic factors also influence both the efficacy of a drug and the likelihood of adverse reactions.Citation1 The concept of pharmacogenetics originated from clinical observations of patients with very high or very low plasma drug concentrations when given a fixed dose, and from the discovery that variations in the DNA sequences of genes coding for metabolizing enzymes are associated with these discrepancies.
The terms pharmacogenetics and pharmacogenomics are closely related and often used interchangeably. However, the terms do have distinct meanings. Pharmacogenetics represents the variability in drug response and metabolism due to genetic variants, while pharmacogenomics involves the systematic investigation of the human genome and alterations in complex gene and protein expression over time in response to a given drug. There are, however, many interactions between the two approaches and they complement each other at many levels; therefore, the distinction is easily blurred.Citation2
Polymorphisms are investigated in genes coding for either the pharmacokinetic pathways (encompassing the processes that influence bioavailability) or pharmacodynamic pathways (targets of drug action). The majority of studies to date involve single genes or single nucleotide polymorphisms (SNPs), but haplotype analyses of several variants within one or more genes are gain_ ing importance.Citation3 The overall aim of pharmacogenetics is to contribute to drug choice and dosage according to the individual genetic makeup, thus leading to a personalized, more efficacious, and less harmful therapy. This review will give a brief summary of the progress in the field and assess the prospects for future success in this area.
Polymorphic drug-metabolizing enzymes
All ADs are highly lipophilic compounds and, as such, subject to extensive metabolism by a number of enzymes, including those of the cytochrome P-450 (CYP) family. The CYPs were recognized quite as major source of pharmacokinetic variability, as they typically show large interlude vidual and sometimes intraindividual differences in activities due to genetic variants. More than 50 CYP genes have been described in the human genome to date, but less than 10 of them are of major significance in psychiatry Among those are CYP 3 A, which metabolizes about 50% of all psychotropic drugs, followed by CYP 2D6, CYP 2C19, CYP 1A2, and CYP 2C9.Citation4 CYPs show distinct but overlapping substrate specificities; their activities may be induced or inhibited by certain drugs or foodstuffs, such as grapefruit juice (an overview of the major CYPs, their AD substrates, inhibitors, and inducers is given in Table I).Citation5 Thus, swallowing the drugs with juice or combining them with other drugs (which is rather common in clinical psychiatry) might have uncontrollable, interactive effects on their bioavailabilityCitation6
Table I. Major cytochrome P450 isoenyzmes (CYP), their antidpressant (AD) substrates, enzyme inhibitors, and inducers.Citation5 TCA, tricyclic AD; SSRI, selective serotonin reuptake inhibitor.
The presence of allelic variants in CYP enzymes with varying degrees of functional significance may result in three main phenotypes, poor metabolizers (PMs), normal metabolizers (NMs), and extensive metabolizers (EMs). The PMs lack an active form of the expressed enzyme due to an inactivating allelic variant; NMs have at least one copy of an active gene; and EMs contain duplicated or amplified gene copies, thus leading to either increased (maybe toxic) or decreased (maybe ineffective) concentrations of the drug.Citation7
CYP 2D6 is the most extensively studied P-450 isoenzyme in psychiatry. More than 70 allelic variants have been identified so far, but only a few are clinically relevant, eg, CYP 2D6*3A, CYP 2D6*4B, and CYP 2D6*5, which all lead to the PM phenotype. Moreover, there are considerable ethnic variations in the frequencies of CYP 2D6 mutations, which are more common in Caucasians (7%) anci Africans (7% to 8%) than in the Asian population (1%).Citation8 In contrast, the incidence of PMs of CYP 2C19 substrates is much higher in Asians (15% to 30%) than in Caucasians.Citation5 Several studies have shown a significant contribution of the CYP 2D6 genotype on plasma concentrations of different ADs, and PMs had a higher incidence of side effects.Citation9-Citation11 Nevertheless, we should keep in mind that the involvement of several different enzymes in the various metabolic pathways may prevent large alterations in in vivo clearance.Citation11 Thus, CYP genotyping can be recommended as a complement to plasma concentration determination when aberrant metabolic capacity is suspected.
Pharmacodynamic drug targets
ADs have a wide variety of targets within the neurotransmitter systems, ranging from neurotransmitter synthesis, degrading enzymes, storage, receptors, and specific transport proteins (). Variations in DNA sequences of these genes can alter the function or levels of expression of neurotransmitters and enzymes and the binding properties of receptors and transport proteins. Newer concepts address signal transduction proteins and other downstream protein polymorphisms. Most notably, the superf amily of G-proteins, which have a key function in signal transduction and are target proteins for more than 50% of available drugs, is becoming a major goal of investigation. Other downstream proteins, such as the kinases or phosphatases, and proteins downstream to transcription factors, and the expression of proteins are target systems in pharmacogenetics and pharmacogenomics.Citation12 The proteins, which are related to synaptic and neuronal plasticity have become special goals of interest in terms of drug response.Citation13
Pharmacogemetic studies of ADs
According to the pathophysiological mechanisms of affective disorders, which mainly postulate deficiency in monoaminergic neurotransmission, ADs of various classes affect the serotonin, norepinephrine, and dopamine pathways (Table II).
Table II. Pharmacogenetics of antidepressant drugs and candidate genes. SERT, serotonin transporter; 5-HT2a, serotonin receptor 2A; TPH1, tryptophan hydroxylase 1; Gβ3, G-protein β3 subunit; NET, norepinephrine transporter; MAO-A, monoamine oxidase A; DRD2, dopamine D2 receptor; DRD3 D3 receptor; DRD4, dopamine D4 receptor; ACE, angiotensin-converting enzyme; BDNF, brain-derived neurotrophic factor; FKBP5: FK506 binding protein 5.
The serotonin transporter (5-HTT) is the initial target of most ADs, especially the widely used selective serotonin reuptake inhibitors (SSRIs). A functional variant was identified in the promoter region of the 5-HTT gene with an insertion/deletion of 44 bp, resulting in short (S) and long (L) alleles. The S allele reduces the transcriptional activity of the 5-HTT gene promoter, leading to reduced 5-HTT expression and 5-HT uptake.Citation14 A number of casecontrol association studies have outlined that individuals carrying at least one L allele, respond more favorably and rapidly to SSRIs, such as fluvoxamine and paroxetine,Citation15 and the S/S genotype had been associated with nonremission in citalopram and fluvoxamine treatment.Citation15 Taking all the findings together, the emerging picture suggests a marked influence of the 5-HTT promoter polymorphism on response to SSRIs in Caucasian population.Citation16 However, opposite findings were reported in the Asian population concerning the frequency of the L allele and the response to treatment. The L allele frequency in Asians was about one-third that in Caucasians, and persons with the S/S genotype responded more favorably to treatment.
These conflicting results are puzzling, but suggest that differential interactions in different populations exist, maybe via interaction with other functional gene variants (for a review, see reference 16). Interestingly, the 5-HTT variants are not only important for treatment with the SSRIs, but also for those with other compounds, eg, lithium, which is widely used as mood stabilizer. Serretti and colleagues have shown that patients with the L/L or L/S genotype have a better outcome than those with the S/S variant.Citation17
Current concepts: signal transduction pathways, neuronal plasticity, ami stress response
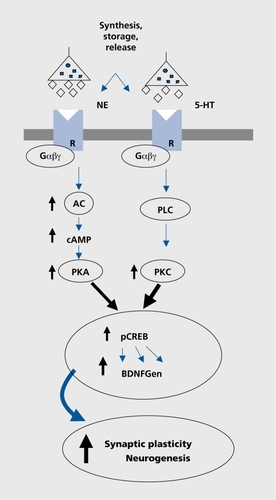
Our present knowledge about the pathophysiological basis of depression and the mechanisms of psychotropic drug action has increased during the last few years. There has therefore been some shift in emphasis from the focus on metabolizing enzymes, neurotransmitter receptors, and transporters toward long4asting adaptive processes, which are related to alterations in signal transduction pathways and mechanisms involving neuronal plasticity, or result from a response to environmental stressors (Figure 2).
The majority of studies concerning signal transduction pathways were carried out on polymorphisms within G-proteins, the key elements of intracellular pathway regulation by transmission of signals from receptors to effector proteins. We have investigated a functional polymorphism on the β3 subunit of the G-protein (C825T), which was shown to increase signal transduction,Citation18 and found an association of the T/T homozygosity to treatment response of ADs.Citation19 Our results were replicated in two further studies, which showed that patients with the Gβ3 T/T variant responded better to AD treatment and that this effect was independent of the analyzed demographic and clinical variables.Citation20,Citation21 This find_ ing highlights the possible influence of downstream messenger systems in treatment-mediated responses and may facilitate the identification of novel key targets that may also be relevant in the etiology of depression.
Secondary signaling mechanisms downstream from the receptors and G-proteins, such as cyclic adenosine monophosphate (cAMP), phosphodiesterase, cAMP response element, and cAMP binding proteins are currently under investigation, but only a marginal association has been found so far.Citation22
In recent years, it has become obvious that chronic treatment with ADs influences the gene expression of potential target genes, eg, neurotrophic factors (brain-derived neurotrophic factor [BDNF]), its receptor (trkB), and vesicle proteins (synapsin I-IIl, synaptophysin). These proteins are involved in neuronal or synaptic plasticity mechanisms and the transcription factor CREB (cAMP response element binding protein) represents the link between the observed short- and long-term treatment effects (Figure 2).Citation23 In postmortem studies, an increase of BDNF and trkB levels were found in depressive patients who were receiving AD treatment at the time of death.Citation24 Moreover, the serum levels of BDNF were also decreased in untreated patients and showed a correlation with the severity of symptoms.Citation25 BDNF has not only been associated with affective disorders; it also seems to be essential in mediating the neuroprotective effect of lithium and has been implicated in the mode of action of antipsychotics.Citation26 However, pharmacogenetic studies with polymorphisms in the BDNF gene were thus far inconclusive.
It has been postulated that decreased BDNF seen in depressed patients may be secondary to increased Cortisol levels, a phenomenon which has been repeatedly described in alterations of stress-hormone regulation in affective disorders. Hyperactivity of the hypothalamopituitary adrenal (HPA) axis with elevated secretion of corticotrophin-releasing factor (CRF), and subsequently Cortisol, as well as decreased glucocorticoid receptor sensitivity and disturbed feedback mechanisms are well known.Citation27 In this context, our own results from two independent clinical studies from a cooperation with the Max Planck Institute for Psychiatry in Munich are of importance. To investigate a possible association between genes regulating HPA axis and response to ADs and susceptibility for depression, we genotyped SNPs in genes regulating the HPA axis activity in depressed patients and matched controls. We found significant associations between the response to ADs and SNPs in the FKBP5 gene, a glucocorticoid receptor (GR)-regulating cochaperone of hsp-90 in two independent samples. Patients homozygous for the minor allele of the associated SNPs responded almost 2 weeks faster to AD drug treatment than patients with the other genotypes.Citation28
Disturbances of the HPA axis are also mirrored by genetic findings in the angiotensin-converting-enzyme (ACE) gene. ACE is not only involved in blood pressure regulation, but is also highly expressed within the central nervous system (CNS), where its primary function comprises degradation of neuropeptides, including bradykinin and substance P.ACE is further supposed to modulate the regulation of the HPA axis, thereby interacting with synthesis and production of neuroactive steroids.Citation29
Within our own studies, we could show that the D allele of a functional insertion/deletion (I/D) polymorphism (the D allele is associated with higher ACE levels and higher neuropeptide degradation capabilities) was associated with several methods of AD treatment, including pharmacological treatment, electroconvulsive treatment (ECT), transcranial magnetic stimulation (TMS), and sleep deprivation.Citation30,Citation31 Moreover, we observed a relationship between the D allele and the hyperactivity of the HPA axis, determined by the combined dexamethasone/corticotropin-releasing hormone test.Citation32 Our findings concerning the ACE gene may have a great impact, not only as a missing link between affective disorders and cardiovascular diseases,Citation33 but also due to a probable function of the ACE gene as a signal transduction component. A recent study has demonstrated that the expression of ACE and other target genes is enhanced by ACE-associated kinases and, therefore, ACE might have an influence on signal transduction mechanisms in the CNS and the periphery.Citation34
Future aspects: from pharmacogenetics to pharmacogenomics
Although the above findings in the field of pharmacogenetics are important, none of the positive results can fully account for the heterogeneity in response to AD treatment. Moreover, due to the complexity of drug response, single mutations in one gene are unlikely to cause the continuous variability in response.
The pharmacogenomic approach uses recent advances in experimental genomics and proteomics (the investigation of all the proteins in a cell or organism), together with the available sequence information of the Human Genome Project. These developments will not only enable genome-wide screens of several millions of SNPs without the use of specific hypotheses and a candidate gene strategy but also functional investigations of gene and/or protein expression over the whole genome or proteome.Citation35 Although most of the data available so far derived from animal studies, the finding of Landgrebe et al,Citation36 who performed a gene expression analysis in mice treated with paroxetine and mirtazapine, is remarkable. The authors found that both drugs led to a downregulation of four common genes, thus suggesting that ADs with different pharmacological principles of action can share the same molecular targets even through the primary pathways on which they act are different. This finding underlines the recent hypothesis that the initial mechanisms of action trigger subsequent events in the signal transduction cascade and, finally, protein expression.
Although all the above results have to be replicated and validated in further experiments and also analyzed in vivo, there is no doubt that large-scale gene and/or protein expression analysis will be performed in the near future in many laboratories by using all these rapidly growing technologies of functional genomics. The expected results will provide new insights in the pathophysiology of psychiatric disorders. Such detailed knowledge will have profound effects on the diagnosis, prevention, and treatment of these diseases.
Conclusion
Although many investigations have shown that genetic variations in target proteins influence their interaction with ADs, the results are still not conclusive and far from the original concept of tailoring the drug regimen to an individual's predisposition and predicting a patient's response to therapeutic agents. We have to be aware that the phenotype of drug response is highly complex, representing a classical example of the outcome of gene-gene or gene-environment interactions. Thus, single mutations are unlikely to cause the continuous variability observed in response to psychiatric treatment.
Moreover, clinical factors can be problematic, as they include the confounding variable of placebo response, the issue of inadequate sample sizes, the study of different drug-response phenotypes and patient populations, as well as the use of continual versus categorical outcome measures. Two key elements are needed to perform valid pharmacogenomic studies: explicit and consistent definition of drug-response phenotype and knowledge of polymorphic candidate genes with relevance to the mechanism of drug action. Nevertheless, the field of pharmacogenetics and pharmacogenomics is expanding rapidly, the development of new, fast, and cost-effective methods for the investigation of the individual genetic/genomic profile is under way, and the incorporation of novel research areas, such as pharmacoproteomics, will lead to better insights into disease and treatment processes. The identification of novel drug targets and the determination of patient subpopulations are ambitious methods that may help individualize pharmacological therapy in psychiatry.
Selected abbreviations and acronyms
| ACE | = | angiotensin-converting enzyme |
| AD | = | antidepressant |
| BDNF | = | brain-derived neurotrophic factor |
| cAMP | = | cyclic adenosine monophosphate |
| CYP | = | cytochrome P-450 |
| HPA | = | hypothalamo-pituitary-adrenal (axis) |
| 5-HTT | = | serotonin transporter |
| PM | = | poor metabolizer |
| SNP | = | single nucleotide polymorphism |
| SSRI | = | selective serotonin reuptake inhibitor |
Some of the work reviewed in this article was supported by the German Federal Research Ministry within the promotional emphasis “Competence Nets in Medicine.”
REFERENCES
- WeinshilboumR.Inheritance and drug response.N Engl J Med.200334852953712571261
- LererB.MacciardiF.Pharmacogenetics of antidepressant and mood-stabilizing drugs: a review of candidate-gene studies and future research directions.Int J Neuropsychopharmacol.2002525527512366879
- JohnsonJA.LimaJJ.Drug receptor/effector polymorphisms and pharmacogenetics: current status and challenges.Pharmacogenetics.20031352553412972951
- LindpaintnerK.Pharmacogenetics and pharmacogenomics in drug discovery and development: an overview.Clin Chem Lab Med.20034139841012747581
- PoolsupN.Li WanPA.KnightTL.Pharmacogenetics and psychopharmacotherapy.J Clin Pharm Ther.20002519722010886465
- DahlML.Cytochrome p450 phenotyping/genotyping in patients receiving antipsychotics: useful aid to prescribing?Clin Pharmacokinet.200241770
- OscarsonM.Pharmacogenetics of drug metabolising enzymes: importance for personalised medicine.Clin Chem Lab Med.20034157358012747605
- CichonS.NothenMM.RietschelM.ProppingP.Pharmacogenetics of schizophrenia.Am J Med Genet2000979810610813809
- GrasmaderK.VerwohltPL.RietschelM.et al.Impact of polymorphisms of cytochrome-P450 isoenzymes 2C9, 2C19 and 2D6 on plasma concentrations and clinical effects of antidepressants in a naturalistic clinical setting.Eur J Clin Pharmacol.20046032933615168101
- CharlierC.BrolyF.LhermitteM.PintoE.AnsseauM.PlomteuxG.Polymorphisms in the CYP 2D6 gene: association with plasma concentrations of fluoxetine and paroxetine.Ther Drug Monit.20032573874214639062
- StormerE.von MoltkeLL.ShaderRI.GreenblattDJ.Metabolism of the antidepressant mirtazapine in vitro: contribution of cytochromes P-450 1A2, 2D6, and 3A4.Drug Metab Dispos.2000281168117510997935
- YoungLT.BakishD.BeaulieuS.The neurobiology of treatment response to antidepressants and mood-stabilizing medications.J Psychiatry Neurosci.20022726026512174735
- SchlossP.HennFA.New insights into the mechanisms of antidepressant therapy.Pharmacol Ther.2004102476015056498
- HeilsA.TeufelA.PetriS.et al.Allelic variation of human serotonin transporter gene expression.J Neurochem.199666262126248632190
- SerrettiA.LilliR.SmeraldiE.Pharmacogenetics in affective disorders.Eur J Pharmacol.20024383128
- MancamaD.KerwinRW.Role of pharmacogenomics in individualising treatment with SSRls.CNS Drugs.20031714315112617694
- SerrettiA.ArtioliP.Predicting response to lithium in mood disorders: role of genetic polymorphisms.Am J Pharmacogenomics.20033173012562213
- SiffertW.RosskopfD.SiffertG.et al.Association of a human G-protein beta3 subunit variant with hypertension [see comments].Nat Genet.19981845489425898
- ZillP.BaghaïTC.ZwanzgerP.et al.Evidence for an association between a G-protein beta3-gene variant with depression and response to antidepressant treatmentNeuroreport.2000111893189710884039
- SerrettiA.LorenziC.CusinC.et al.SSRls antidepressant activity is influenced by G beta 3 variants.Eur Neuropsychopharmacol.20031311712212650956
- LeeHJ.ChaJH.HamBJ.et al.Association between a G-protein beta 3 subunit gene polymorphism and the symptomatology and treatment responses of major depressive disorders.Pharmacogenomics J.20044293314647404
- SerrettiA.ArtioliP.From molecular biology to pharmacogenetics: a review of the literature on antidepressant treatment and suggestions of possible candidate genes.Psychopharmacology (Berl).200417449050314997279
- DumanRS.Role of neurotrophic factors in the etiology and treatment of mood disorders.Neuromol Med.200451125
- ChenB.DowlatshahiD.MacQueenGM.WangJF.YoungLT.Increased hippocampal BDNF immunoreactivity in subjects treated with antidepressant medication.Biol Psychiatry20015026026511522260
- ShimizuE.HashimotoK.OkamuraN.et al.Alterations of serum levels of brain-derived neurotrophic factor (BDNF) in depressed patients with or without antidepressants.Biol Psychiatry.200354707512842310
- HashimotoK.ShimizuE.lyoM.Critical role of brain-derived neurotrophic factor in mood disorders.Brain Res Brain Res Rev.20044510411415145621
- HolsboerF.The corticosteroid receptor hypothesis of depression.Neuropsychopharmacology.200023477501 11027914
- BinderEB.SalyakinaD.LichtnerP.et al.Polymorphisms in FKBP5 are associated with increased recurrence of depressive episodes and rapid response to antidepressant treatment.Nat Genet.2004361319132515565110
- MoskowitzDW.Is angiotensin l-converting enzyme a “master” disease gene?Diabetes Technol Ther.2002468371112458570
- BaghaiT.SchüleC.ZwanzgerP.et al.Possible influence of the insertion/deletion polymorphism in the angiotensin I-converting enzyme gene on therapeutic outcome in affective disorders.Mol Psychiatry.2001625825911326291
- BaghaiTC.SchuleC.ZwanzgerP.et al.Influence of a functional polymorphism within the angiotensin I-converting enzyme gene on partial sleep deprivation in patients with major depression.Neurosci Lett.200333922322612633893
- BaghaiTC.SchuleC.ZwanzgerP.et al.Hypothalamic-pituitary-adrenocortical axis dysregulation in patients with major depression is influenced by the insertion/deletion polymorphism in the angiotensin l-converting enzyme gene.Neurosci Lett.200232829930312147330
- BondyB.BaghaiTC.ZillP.et al.Combined action of the ACE D- and the G-protein β3-allele in major depression: a possible link to cardiovascular disorder?Mol Psychiatry.200271120112612476328
- KohlstedtK.BrandesRP.Muller-EsterlW.BusseR.FlemingI.Angiotensin-converting enzyme is involved in outside-in signaling in endothelial cells.Circ Res.200494606714615289
- GoldsteinDB.TateSK.SisodiyaSM.Pharmacogenetics goes genomic.Nat Rev Genet.2003493794714631354
- LandgrebeJ.WelzlG.MetzT.et al.Molecular characterisation of antidepressant effects in the mouse brain using gene expression profiling.J Psychiatr Res.20023611912911886689