Abstract
Background
Vermicomposting is a mesophilic process using earthworms to efficiently and at low cost process large volumes of organic waste. It has been suggested to not only increase soil fertility but also increase biomass of beneficial bacteria while reducing harmful bacteria. The aim of this study was to set up a strategy to investigate and characterise the viral as well as the bacterial composition of a vermicomposting system.
Material and methods
The vermicomposting unit used in this study was placed at the Makerere University Agricultural Research Institute Kabanyolo on the outskirts of Kampala, Uganda, and was fed with 80% cattle manure and 20% food waste. On Day 172, the compost was terminated and compost samples were collected from three layers of the unit: the top, the middle and the bottom layer. A metagenomic approach was then applied to characterise the viral and bacterial composition of the vermicomposting system.
Results and discussion
A high abundance and diversity of bacteria were identified. Proteobacteria was the largest phyla in the compost (mainly Alpha-, Gamma- and Betaproteobacteria), constituting almost 65% of the bacterial reads in the data sets. DNA samples from several possible pathogenic bacteria, such as Salmonella spp., Escherichia coli, Enterobacter spp., Enterococcus spp. and Clostridium spp, were detected in the vermicompost, suggesting that there might still be harmful bacteria in the vermicast. Phages constituted the main viral group; apart from phages, mainly insect viruses were identified. The only animal or human virus identified was kobuvirus. In summary, metagenomic analysis was shown to be an efficient technology to characterise the microbial composition of vermicast. The data from this study contribute to a better understanding of the microbes present in this kind of composting system and can help determine measures necessary for safe manure handling.
The production of organic waste from domestic and agricultural sources has led to the need for technologies to process and recycle nutrients and reduce the negative environmental impacts of insufficient organic waste management (Citation1). Release of unprocessed organic wastes such as animal manure, crop residues and household waste can pose a potential risk of spreading plant and/or animal pathogens. Hence, different composting techniques have been suggested not only as a strategy to recycle nutrients back to nature or society but also as a way to reduce the risk of spreading pathogens (Citation2, Citation3).
Vermicomposting is an attractive approach for the treatment of organic waste (Citation4), particularly in areas where there is no functioning of organic waste management (Citation5). Vermicomposting is the biooxidation of organic matter facilitated by worms (Citation6). The most commonly used earthworm in vermicomposting is Eisenia feotida. Eudrilus eugeniae, known as the African night crawler, is another common earthworm that is native to the African continent (Citation7). It is a large worm with a rapid reproduction cycle (Citation8). Its preferred temperature range is somewhat lower compared to that of E. feotida, but under ideal conditions (25°C and 80% moisture) it is capable of decomposing large quantities of organic material (Citation9, Citation10). The worms fragmentise the material increasing the contact area for microorganisms, while also aerating the material, which increase aerobic microbial activity (Citation11, Citation12). The passage through the worms is believed to change the biochemical property of the passed material, the so-called worm casting or vermicast (Citation13, Citation14). This process has been shown to increase the soil fertility and the biomass of beneficial bacteria while reducing the rate of harmful ones (Citation15–Citation17).
Most microbial investigations of compost material focus mainly on bacteria; however, many viruses are potent pathogens of crops, animals and/or humans that may be spread when the compost is used on arable land. The aim of this study was to set up a strategy to investigate and characterise the viral as well as the bacterial composition in a vermicomposting system, using shotgun metagenomics. This technique allows for an unbiased and deep characterisation of the complete microbiota, which contributes to a better understanding of the measures necessary for safe manure handling.
Materials and methods
Unit handling and process
The vermicomposting unit was placed at the Makerere University Agricultural Research Institute Kabanyolo (MUARIK) on the outskirts of Kampala, Uganda, and was run by research technicians at the site. In Lalander et al. (Citation18), a detailed description of the unit setup and performance is given. The unit was fed with 80% cattle manure (collected from the farm at MUARIK) and 20% food waste (obtained from the household of the principal technician at MUARIK maintaining the vermicomposting unit). The contents of Salmonella spp. and thermotolerant coliforms were investigated in the fresh material fed into the vermicomposting unit (Citation18), and the concentrations were found to be around 150 CFU g−1 and 1.4×105 CFU g−1, respectively. The feeding rate was adjusted to the number of worms present in the unit. On Day 67, 87% of the worms and a large part of the material were harvested from the unit. The process continued for another 105 days and was finally terminated on Day 172. The material reduction was 45.9% on a dry matter basis over the course of the experiment (Citation18).
Sampling
Samples were collected on Day 172 from three layers in the unit – the top (C1), the middle (C2) and the bottom (C3) layer – in order to verify the effect of the vermicomposting process on the microbial and viral community. From each level, around 50 g of sample was collected in 50 mL sterile centrifuge tubes. The samples were placed in an envelope with cooling pads and sent to Sweden via express delivery (3 days) for further analysis.
Pre-treatment of compost material
To reduce the background of non-microbial nucleic acids, the samples were pre-treated prior to extraction of nucleic acid. From each of the three layers, one compost material sample was prepared to investigate the microbial flora. A total of 800 mg compost material per sample was handled using the Meta-G-Nome DNA isolation kit (Epicentre, Madison, WI, USA) according to the instructions of the manufacturers to isolate the bacterial DNA from the material. However, some additional steps were included in order to also isolate genomic material from possible viruses. After the 0.45-µm filtration step, which was used to capture the bacteria on the filter, the filtered liquid was subjected to ultracentrifugation at 32,000 rpm (175,000×g) for 2 h in order to pellet the virus. The pellet was resuspended in 1x DNase buffer (Roche, Mannheim, Germany) and subjected to DNase (100 U) (Roche, Mannheim, Germany) treatment.
Nucleic acid extraction
The bacterial DNA was extracted using the Meta-G-Nome kit (Epicentre, Madison, WI, USA). For the pelleted virus, each pre-prepared sample was divided into two aliquots and viral RNA and DNA were extracted, respectively. The viral DNA was extracted using the DNA mini kit (Qiagen, Hilden, Germany), according to the manufacturer's instructions, and eluted in 50 µL EB. The RNA from the pelleted virus was extracted using a combination of QIAzol and RNeasy kit (Qiagen, Hilden, Germany). In brief, the samples were lysed using QIAzol Lysis Reagent, and phase separation was performed by the addition of chloroform. The RNA phase was mixed with ethanol and transferred to RNeasy columns for further washing and elution. The RNA was eluted in 30 µL EB.
cDNA synthesis and labelling of the viral nucleic acid
To amplify all viral nucleic acid in the different samples, the nucleic acid was labelled with tag sequence at both 5′and 3′ end. For the RNA, this was done through double-stranded cDNA synthesis, using the primer FR-20N (GCC GGA GCT CTG CAG ATA TCN NNN NN) (Citation19). The first strand synthesis was performed using Superscript III (Invitrogen, Carlsbad, CA, USA) according to the manufacturer's instructions. After the inactivation of superscript at 70°C for 10 and 2 min on ice, the second strand synthesis was performed with the addition of Klenow Fragment (3′→ 5′ exo-) (NEB, Ipswich, MA, USA). The reaction was run for 1 h at 37°C before a 10 min termination step at 75°C. The DNA was labelled during a Klenow Fragment (3′→ 5′ exo-) reaction using the same primer and temperature as for the cDNA reaction. After the labelling reaction, the samples were treated in an identical way.
Random amplification of labelled viral cDNA/DNA
PCR amplification was performed on all viral tag-labelled samples using the primer FR-26RV (GCC GGA GCT CTG CAG ATA TC) (Citation19) according to the following procedure: 1x PCR buffer, 2.5 mM MgCl2, 2.5 mM dNTP, 0.4 mM F primer, 0.4 mM R primer and 1.25 U AmpliTaq Gold DNA Polymerase (Applied Biosystems, Foster City, CA, USA). The amplification was initiated with a 10 min heating step at 95°C, followed by 40 cycles of 30 s at 95°C, 30 s at 58°C and 90 s at 72°C. The reaction was ended with an extra elongation step at 72°C for 10 min. The primer sequence was cleaved off with EcoRV (NEB, Ipswich, USA), and the product was purified using PCR purification kit (Qiagen, Hilden, Germany) before large-scale sequencing.
Large-scale sequencing
The samples were sequenced using the Ion Torrent platform with three samples per a 316-chip (barcoded fragment libraries) at the Uppsala Genome Center, SciLifeLab. The sequence data from this study have been deposited in the Sequence Read Archive (SRA) and are available at www.ncbi.nlm.nih.gov/sra/?term=SRP071135.
Sequence analysis
CLC Genomic Workbench (Version 7) was used to perform the quality trimming of the sequences as well as to perform the de novo assembly of each sequence set. After assembly, the singletons as well as the contigs were uploaded into the CAMERA portal (Citation20), where BLASTn and BLASTx analyses were performed using default settings and an e-value threshold of 10−4. The generated XML files were uploaded into the MEGAN software (Citation21) in order to visualise and compare the blast results. It should be noted that in this study only one sample per layer was sequenced, making it hard to make conclusions about the differences in the amount of specific microbes in the different layers. Thus, this article focuses on the total microbial composition and the type of microbes that are present in the compost.
Results and discussion
Sequencing output
The output of the Ion Torrent sequencing and the initial bioinformatics analysis (trimming and assembly) is summarised in . In total, 1,885,978 reads (9,419 contigs and 1,825,412 singletons) from the bacteria-enriched samples were further analysed (CB1-3). The corresponding number for the virus-enriched samples (CV1-3) was 1,932,253 reads (4,761 contigs and 496,076 singletons).
Table 1 Summary of the sequence data obtained from the Ion Proton for each individual sample
Composition of viruses
Almost 90% of the viral sequences originated from DNA viruses, of which almost 78% came from different phages within the Microviridae, Siphoviridae, Podoviridae and Myoviridae families, reflecting the abundance of bacteria present in the investigated samples. Also, viruses infecting insects from the Iridoviridae family constituted a large proportion, around 20%, of the DNA viruses identified in the samples. Almost no human and/or animal viruses were identified in the sequences. The only animal virus identified was kobuvirus belonging to the Picornaviridae family, a virus associated with enteric disorders. However, it should be noted that only four reads were annotated to this virus. Around 9% of the virus sequences identified were homologous to RNA viruses, and the majority of these were RNA viruses of insects, such as members of the Dicistroviridae, Partitiviridae and Leviviridae families. Also, plant viruses within the sobemovirus were identified.
Composition of bacteria
There was a high abundance of bacteria in the vermicompost sample. The phylum that most bacteria fell within was Proteobacteria, with over 60% of all the bacterial hits, while all other phyla contained around 10% or less of the total bacterial sequences identified (a). The abundance of Proteobacteria was also reflected in the major orders (b), classes (c) and genera (d), as many of these belong to this phylum. All the results from this study are presented as the top phyla, classes, orders and genera identified in the total compost (combining all three layers).
Fig. 1 Bacterial composition of the compost. Top-10 most abundant phyla (a), classes (b), orders (c) and genera (d) in the compost. The figures display the percentage of the total bacteria from the three layers falling in each respective category.
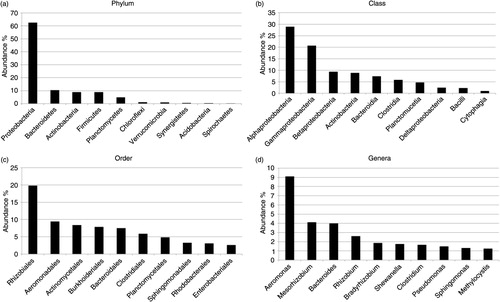
Proteobacteria is a phylum which contains many bacteria responsible for nitrogen fixation as well as a number of pathogens. The abundance of these bacteria is in accordance with the findings of Hill et al. (Citation22) who found a significant increase in nitrogen-fixing bacteria during vermicomposting of faecal matter using coir as bedding material. Out of the six classes of Proteobacteria, very few sequences belonged to Zetaproteobacteria, while Alphaproteobacteria and Gammaproteobacteria were the most abundant classes, 47 and 34%, respectively. Rhizobiales, Aeromonades and Burkholderiales were the most highly represented orders within this phylum. Specifically looking for potential pathogens, sequences from Brucella spp., Yersinia spp. and the enterobacteria Salmonella spp. and Escherichia coli were identified. The diverse phylum Bacteroidetes containing bacteria often found in the environment, such as soil and water, constituted a little over 10% of the identified bacteria. The major class identified was the anaerobic class Bacteroidia, constituting 71% of the identified bacteria within this phylum. Actinobacteria was the third largest phylum, constituting 8.7% of the identified bacteria, and 93% of these falls within the diverse Actinomycetales order. The largest genera were Microbacterium, Mycobacterium, Corynebacterium, Streptomyces and Cellulomonas. The Firmicutes phylum constituted 8.8%, of which two-thirds belonged to the Clostridia class. The results correspond to that of Romero-Tepal et al. (Citation23), who, using 16S rRNA gene pyrosequencing, identified Proteobacteria, Actinobacteria, Tenericutes, Bacteroidetes, Chloroflexi, Firmicutes and Planctomycetes as the most important phylum in wormbed leachate obtained during vermicomposting of cow manure. Similarly, Alpha-, Beta- and Gammaproteobacteria were the main classes, and they also had a high abundance of the Rhizobiales order (Citation23). Yasir et al. (Citation24) identified Proteobacteria, Bacteroidetes, Verrucomicrobia, Actinobacteria and Firmicutes as the major phyla in vermicompost material with the classes Bacteroidia, Gammaproteobacteria and Actinobacteria increased in vermicompost compared to the starting material. Many of these bacteria have been highly associated with the earthworm gut. Comparing the major microbial communities between the vermicomposting and thermophilic composting reveals differences, most likely reflecting their very different approaches to degrade the material. In thermophilic composting, the major bacteria genera under the thermophilic phase are Bacillus and Thermus, and during the mature phase, Bacillus, Flavobacterium, Pseudomonas and Cellulomonas (Citation25) are the major genera. Although these genera were identified in vermicompost, they were not among the top genera identified, with the exception of Pseudomonas.
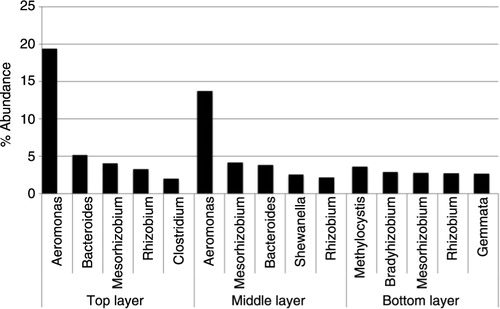
As mentioned before, the results from this article are presented as the top phyla, classes, orders and genera identified in the total compost (combining all three layers). However, similar results were obtained when investigating the layers individually. The main difference found between the different layers was that the lowest layer, unlike the middle and bottom layers, did not have a high abundance of bacteria within the genus Aeromonas (order Aeromonadales) (). The top and middle layers had 19 and 14% of bacteria, respectively, while in the bottom layer only 0.7% of bacteria belong to this genus.
Fig. 2 Comparison of top genera identified in the individual layers. The top-five most abundant genera detected in layer one (top), two (middle) and three (bottom), respectively. The abundance is displayed in percentage.
As vermicomposting is a mesophilic process, it has been argued that a smaller reduction of pathogens can be expected compared to that achieved in thermophilic composting. However, many studies have demonstrated the vermicomposting process capacity for pathogen reduction. Yadav et al. (Citation26) detected very low concentrations or no total and faecal coliforms, Salmonella spp. and viable helminth ova after 6 months vermicomposting of faecal slurry. The bacterial reduction has been confirmed in several other studies in different materials (Citation27, Citation28). Monroy et al. (Citation17) came to a different conclusion; they conducted a number of trials on vermicomposting of organic waste and found that the concentration of total coliforms was reduced greatly when applied in low dosage, but there was no reduction found when applied in high dosage. They drew the conclusion that the reduction of coliforms was due to passage through the worms and not due to interaction with the earthworm-shaped microbial community (Citation17). In 2001, Eastman et al. published a study about the effectiveness of vermiculture to reduce pathogens. They reported a significant improvement in the removal of Salmonella spp., faecal coliforms, enteric virus and helminth ova, in comparison to a system with no worms (Citation29). The removal or inactivation of helminth ova by vermicomposting has been questioned by other researchers, who found no inactivation or destruction of helminth ova (Citation22, Citation30). Salmonella spp. sequences were detected in all the layers of the investigated vermicompost, although only a limited number of sequences were observed. However, Lalander et al. (Citation18) showed (in the same vermicomposting unit as investigated in this paper) that no active Salmonella spp. was found in the vermicompost material, although it was found in the untreated material, suggesting that salmonella was inactivated during vermicomposting. It is thus possible that the salmonella sequences identified come from bacteria that are not viable. Aira et al. (Citation31) studied the pathogen reduction in cow manure in a continuous feed system and found that the levels of faecal enterococci, faecal coliforms and E. coli were reduced to acceptable levels, which is in agreement with previous studies. However, the concentrations of Clostridium spp., total coliforms and enterobacteria were not changed during the vermicomposting process (Citation31). Lalander et al. (Citation18) observed no inactivation of total coliforms or Enterococcus spp. in this vermicomposting unit, which was believed to be due to recontamination of already processed material by untreated material that was continuously placed on top of the treatment unit in this vertical system. In this metagenomic investigation, E. coli, Enterobacter spp., Enterococcus spp. and Clostridium spp. were detected, and out of these four Clostridium spp. was detected in a higher abundance. Clostridia are anaerobic sulphite-reducing bacteria commonly found in the environment, in particular in soil. In compost, they are involved in degradation of cellulose (Citation32).
Hygienic aspects of the reuse of vermicompost
DNA from many possible pathogenic bacteria was detected in the vermicompost. It is not known whether the bacteria are viable; they could be viable but not cultivable. It should also be noted that no ‘day zero’ sample was analysed. Therefore, it is not possible to know from these metagenomic data whether there was a reduction of pathogenic bacteria in the final vermicompost material compared to the input. The findings of Lalander et al. (Citation18), on the reduction in pathogenic Salmonella spp. in the studied vermicompost, however, suggest that the vermicomposting process studied here reduced the load of pathogenic bacteria. It can thus be deduced that, in this case, it is hygienically safer to vermicompost and handle the vermicompost than it is to leave untreated material in the environment or use it untreated in agriculture. Although the risk is decreased with vermicomposting, a post-sanitisation step should be included if the handling and the use of vermicomposted material needs to be completely safe. Since the amount of easily available carbon is decreased with vermicomposting, it is not likely that very high temperatures to ensure sanitisation would be achieved (Citation33). A viable post-treatment strategy is ammonia sanitisation (Citation34); it is an inexpensive, robust sanitisation technique that would have the added benefit of increasing the fertiliser value of the sanitised vermicompost.
Conclusion
Vermicomposting, at a low cost, can process large amounts of organic waste, which can be used to improve soil structure and fertility and hence plant growth and yield (Citation35). However, it is important to minimise the risk of spreading human or animal or plant pathogens when recycling organic matter back into the food production chain. In this study, the microbial composition (virus and bacteria) of a vermicomposting unit was investigated using metagenomics. The only human or animal virus identified was a picornavirus (kobuvirus), while DNA samples from many possible pathogenic bacteria were detected in the vermicompost. Removing the material from the environment and preventing the use of untreated material in agriculture reduce the risk of disease transmission. A post-sanitisation step is required to ensure the production of hygienically safe organic fertiliser if it has to be used in agriculture.
Conflict of interest and funding
The authors wish to acknowledge The SLU Global Food Security Research and Education Program 2010–2013 (UD40), the Ministry for Foreign Affairs, Sweden, for financial support. The authors would like to acknowledge support of the National Genomics Infrastructure (NGI)/Uppsala Genome Center and UPPMAX for providing assistance in massive parallel sequencing and computational infrastructure. The work performed at NGI/Uppsala Genome Center has been funded by RFI/VR and Science for Life Laboratory, Sweden. The authors also thank research technicians Charles Azizi and Ronald Nsobya at MUARIK for maintenance of the system.
References
- Hoornweg D, Bhada-Tata P. What a waste – a global review of solid waste management. Urban development series. 2012; Washington, DC: World Bank.
- Senesi N, Plaza C. Role of humification processes in recycling organic wastes of various nature and sources as soil amendments. Clean (Weinh). 2007; 35: 26–41.
- Vinnerås B. Comparison of composting, storage and urea treatment for sanitising of faecal matter and manure. Bioresour Technol. 2007; 98: 3317–21.
- Dominguez J, Gomez-Brandon M. Kumar S, Bharti A. Vermicomposting: composting with earthworms to recycle organic wastes. Mangagemnet of organic waste . 2012; Rijeka, Croatia: In Tech. 29–48.
- Aalok A, Tripathi AK, Soni P. Vermicomposting: a better option for organic solid waste management. J Hum Ecol. 2008; 24: 59–64.
- Dominguez J, Edwards CA. Edwards CA, Arancon NQ, Sherman R. Relationships between composting and vermicomposting. Vermiculture technology: earthworms, organic wastes, and environmental management. 2010; Boca Raton, FL: CRC Press. 12–24.
- Dominguez J, Edwards CA. Edwards CA, Arancon NQ, Sherman R. Biology and ecology of earthworm species used for vermicomposting. Vermiculture technology: earthworms, organic wastes, and environmental management. 2010; Boca Raton, FL: CRC Press. 28–38.
- Reinecke AJ, Viljoen SA. Reproduction of the African earthworm, Eudrilus eugeniae (Oligochaeta) – cocoons. Biol Fertil Soils. 1988; 7: 23–7.
- Dominguez J, Edwards CA, Dominguez J. The biology and population dynamics of Eudrilus eugeniae (Kinberg) (Oligochaeta) in cattle waste solids. Pedobiologia (Jena). 2001; 45: 341–53.
- Reinecke AJ, Viljoen SA, Saayman RJ. The suitability of Eudrilus eugeniae, perionyx excavatus and Eisenia fetida (Oligochaeta) for vermicomposting in southern Africa in terms of their temperature requirements. Soil Biol Biochem. 1992; 24: 1295–307.
- Lavelle P. Earthworm activities and the soil system. Biol Fertil Soils. 1988; 6: 237–51.
- Dominguez J, Edwards CA. Hanna SH, Mikhail WZA. Vermicomposting organic wastes: a review. Soil zoology for sustain development in the 21st century. 2004; Egypte, Cairo: Geocities. 369–95. In: .
- Loehr RC, Neuhauser EF, Malecki MR. Factor affecting the vermistabilization process – temperature, moisture-content and polyculture. Water Res. 1985; 19: 1311–17.
- Aira M, Monroy F, Domínguez J. Changes in microbial biomass and microbial activity of pig slurry after the transit through the gut of the earthworm Eudrilus eugeniae (Kinberg, 1867). Biol Fertil Soils. 2006; 42: 371–76.
- Arancon NQ, Edwards CA, Bierman P. Influences of vermicomposts on field strawberries: part 2. Effects on soil microbiological and chemical properties. Bioresour Technol. 2006; 97: 831–40.
- Atiyeh RM, Subler S, Edwards CA, Bachman G, Metzger JD, Shuster W. Effects of vermicomposts and composts on plant growth in horticultural container media and soil. Pedobiologia (Jena). 2000; 44: 579–90.
- Monroy F, Aira M, Domínguez J. Reduction of total coliform numbers during vermicomposting is caused by short-term direct effects of earthworms on microorganisms and depends on the dose of application of pig slurry. Sci Total Environ. 2009; 407: 5411–16.
- Lalander C, Komakech AJ, Vinnerås B. Vermicomposting as manure management strategy for urban small-holder animal farms – Kampala case study. Waste Manag. 2015; 39: 96–103.
- Allander T, Tammi MT, Eriksson M, Bjerkner A, Tiveljung-Lindell A, Andersson B. Cloning of a human parvovirus by molecular screening of respiratory tract samples. Proc Natl Acad Sci USA. 2005; 102: 12891–6.
- Sun S, Chen J, Li W, Altintas I, Lin A, Peltier S, etal. Community cyberinfrastructure for advanced microbial ecology research and analysis: the CAMERA resource. Nucleic Acids Res. 2011; 39: D546–51.
- Huson DH, Weber N. Microbial community analysis using MEGAN. Methods Enzymol. 2013; 531: 465–85.
- Hill GB, Lalander CH, Baldwin SA. The effectiveness and safety of vermi-versus conventional composting of human feces with Ascaris suum Ova as model Helminthic parasites. J Sustain Dev. 2013; 6: 1–10.
- Romero-Tepal EM, Contreras-Blancas E, Navarro-Noya YE, Ruiz-Valdiviezo VM, Luna-Guido M, Gutierrez-Miceli FA, etal. Changes in the bacterial community structure in stored wormbed leachate. J Mol Microbiol Biotechnol. 2014; 24: 105–13.
- Yasir M, Aslam Z, Kim SW, Lee SW, Jeon CO, Chung YR. Bacterial community composition and chitinase gene diversity of vermicompost with antifungal activity. Bioresour Technol. 2009; 100: 4396–403.
- Mehta CM, Palni U, Franke-Whittle IH, Sharma AK. Compost: its role, mechanism and impact on reducing soil-borne plant diseases. Waste Manag. 2014; 34: 607–22.
- Yadav KD, Tare V, Ahammed MM. Integrated composting – vermicomposting process for stabilization of human faecal slurry. Ecol Eng. 2012; 47: 24–9.
- Contreras-Ramos SM, Escamilla-Silva EM, Dendooven L. Vermicomposting of biosolids with cow manure and oat straw. Biol Fertil Soils. 2005; 41: 190–8.
- Kumar R, Yadav S. Removal of pathogens during vermi-stabilization. J Environ Sci Tech. 2011; 4: 621–9.
- Eastman BR, Kane PN, Edwards CA, Trytek L, Gunadi B, Stermer AL, etal. The effectiveness of vermiculture in human pathogen reduction for USEPA biosolids stabilization. Compost Sci Util. 2001; 9: 38–49.
- Bowman DD, Liotta JL, McIntosh M, Lucio-Forster A. Ascaris suum egg inactivation and destruction by the vermicomposting worm, Eisenia foetida . Proc Water Environ Fed. 2006; 2006: 11–18.
- Aira M, Gomez-Brandon M, Gonzalez-Porto P, Dominguez J. Selective reduction of the pathogenic load of cow manure in an industrial-scale continuous-feeding vermireactor. Bioresour Technol. 2011; 102: 9633–7.
- Haagsma J. Pathogenic anaerobic bacteria and the environment. Rev Sci Tech. 1991; 10: 749–64.
- Niwagaba C, Nalubega M, Vinnerås B, Sundberg C, Jönsson H. Substrate composition and moisture in composting source-separated human faeces and food waste. Environ Tech. 2009; 30: 487–97.
- Nordin A, Nyberg K, Vinnerås B. Inactivation of Ascaris eggs in source-separated urine and feces by ammonia at ambient temperatures. Appl Environ Microbiol. 2009; 75: 662–7.
- Arancon NQ, Edwards CA, Bierman P, Welch C, Metzger JD. Influences of vermicomposts on field strawberries: 1. Effects on growth and yields. Bioresour Technol. 2004; 93: 145–53.
