Abstract
Background: In all Streptococcus mutans strains, 5–13% carry a 5.6-kb plasmid. Despite its frequency, little is known about its mediated functions with most of the information coming from a single study focussing on plasmid pUA140.
Objective: Here, we describe the sequence and genetic organization of two S. mutans 5.6-kb plasmids, pDC09 and pNC101.
Results: Based on PicoGreen dsDNA quantification and Real-Time quantitative PCR (RTQ-PCR), the plasmid copy number was found to range between 10 and 74, depending on the strain tested. In contrast to literature, we identified six instead of five open reading frames (ORFs). While the putative gene products of ORF1 (as a Rep-protein) and ORF2 (as a Mob-protein) could be confirmed as being identical to those from pUA140, the functions of ORF3 (unknown) and ORF 4 (possibly AtpE homologue) could not be further revealed. However, the product of ORF5 showed a fairly high identity (38–50%) and structural similarity (58–74%) to RelE of Streptococcus pneumoniae, Streptococcus equi, and Streptococcus downei. In addition, we identified a functionally corresponding ORF6 encoding a protein with 61–68% identity (81–86% similarity) to the S. equi and S. downei antitoxin of the RelB family. RelE and RelB together form a plasmid-encoded toxin-antitoxin (TA) system, RelBEplas. Despite its rather limited sequence similarity with chromosomal TA systems in S. mutans (RelBEchro, MazEF, HicBA), we found similar tertiary structures applying I-Tasser protein prediction analysis.
Conclusion: Type II-toxins, as the plasmid-encoded RelE, are RNA endonucleases. Depending on their mRNA cleavage activity, they might 1) kill every plasmid-free progeny, thereby stabilizing plasmid transfer at the expense of the host and/or 2) help S. mutans enter a dormant state and survive unfavourable environmental conditions. Whilst a function in plasmid stabilization has been confirmed, a function in persistence under nutritional stress, tested here by inducing amino acid starvation, could not be demonstrated so far.
Plasmids present in many gram-positive cocci usually encode for a variety of biological activities, including resistance to antibiotics, heavy metals, production of and/or resistance to bacteriocins, metabolic properties, immunity, and factors of pathogenesis. However, a 5.6-kb (corresponding to 3.6 megadalton) plasmid with a G + C content between 31 and 34 mol% in Streptococcus mutans has unknown functions and is thus still designated as ‘cryptic’. Interestingly, in clinical populations, it shows a relatively constant prevalence ranging from 5 to 13% Citation1 Citation2. Almost all plasmid-containing strains of S. mutans produce distinct mutacins which are not directly plasmid-encoded Citation3 Citation4. Furthermore, S. mutans plasmids can be divided into two types classified by bacteriocin profile and restriction enzyme digest: Group I plasmids isolated from strains of individuals from African/Asian descent and Group II plasmids from Caucasians Citation5 Citation6. Efforts to transform plasmid-positive into plasmid-negative strains have all been unsuccessful Citation2 Citation4. This makes studies regarding biological functions difficult even though they give evidence of an inherent plasmid-stabilizing system. By sequence analysis, Zou et al. Citation7 previously found five open reading frames (ORFs) on the representative pUA140 Group I plasmid. Based on cloning experiments ‘stability determinants’ could be assigned to ORF 1 and 5. However, toxin–antitoxin loci, which are frequently present in plasmids to confer stability and maintenance, were not known at that time.
The primary aim of our study was to get a better insight into the frequency, heterogeneity, and function of S. mutans 5.6-kb plasmids. By testing 40 S. mutans strains applying a PCR with primers directed to conserved regions, we found four plasmid positive strains. The sequence analysis revealed a toxin-antitoxin (TA) system of the RelBE family with RelE (toxin) known for cleaving mRNA positioned at the ribosomal A site and drastically reducing translation Citation8. We further tested the hypothesis that plasmid-encoded TA systems (TAplas) in S. mutans could 1) compensate function of mutated and defect chromosomal TA modules (we further abbreviate TAchro) and/or 2) generate a plethora of TA genes facilitating dormancy and persistence of the host under (amino acid) starvation.
Methods
Bacterial strains, plasmids, and media
Forty strains of S. mutans were cultured under standard conditions on Columbia blood agar aerobically, with 10% CO2 at 37°C, and screened for the 5.6-kb (3.6 MDa) plasmid by PCR using the primer pair pF6/pR3 (). The four positive strains were DC09, DD09, NC101, and NC102 (all originated from the collection of D. Beighton, KCL Dental Institute, London, UK). A selection of plasmid-free strains, namely UA159, ATCC25175, NCTC11060, OMZ918 (Oral Microbiology, Zürich, Switzerland), KK21, R187 (both from the Biological Laboratory, University Hospital, Jena, Germany), 5DC8 (D. Beighton), and AC4446 (National Reference Centre for Streptococci, Aachen, Germany) were used for contrast.
Table 1. Amplification and sequencing primers for S. mutans 5.6-kb plasmids
Growth conditions for persistence experiments
The basal synthetic medium (BM) consisting of buffering salts, 0.8% glucose, 0.2% casaminoacid, 2 mM MgSO4, vitamins, and four essential amino acids (see below) were used to induce starvation. Amino acid starvation causes guanosine tetraphosphates (ppGpp and pppGpp, known as alarmones) and, secondarily, PolyP to accumulate Citation9. PolyP in turn is a signalling molecule that activates the Lon protease to digest ribosomal proteins and readjust and adapt translation rate to starving conditions Citation10. Lon also degrades antitoxin of the RelB-type so that the toxin (RelE), an endonuclease, clears ribosomes from mRNAs further supporting readjustment of translation rate Citation8.
Amino acid starvation was induced by two different strategies: 1) Naturally induced amino acid starvation: The amount of casaminoacid (from 0.2 to 0.02%) as well as the amount of essential amino acids (L-glutamic acid from 4 to 0.4 mM, L-arginine from 1 to 0.1 mM, L-cysteine from 1.3 to 0.1 mM, and L-tryptophan from 0.1 to 0.01 mM) was reduced. The BM-broths (10 ml) were inoculated with 100 µl of an overnight S. mutans (strains as above) preculture, incubated for 48 hours at 37°C in a 10% CO2 atmosphere, and then kept at room temperature (RT). On a daily basis, 10–100 µl samples were struck on Columbia agar to stop dormancy and test for viability. When strains missed growth on agar, the suspensions were concentrated in 1 ml by centrifugation and again samples were struck on agar. If the final culture failed to grow a single S. mutans colony, the strain was reported as ‘dead’. This way differences in starvation tolerance and persistence could be assessed for all strains.
2) Chemically induced amino acid starvation: S. mutans strains (UA159, ATCC25175, DC09, DD09) were grown in standard BM broth. At A540=0.2, serine hydroxamate (SHX), an analogue of serine, was added (0.5 mg/ml) for the induction of amino acid starvation by inhibiting the seryl-tRNA synthase. After further incubation for 4 hours at 37°C in a 10% CO2 atmosphere, the cultures were kept at RT for the rest of the experiment. Again daily, 10–100 µl samples were struck on Columbia agar to test for viability. The protocol was finished as described above.
DNA preparation and sequence analyses
Plasmid DNA preparation, restriction enzyme digestion, and sequencing were performed by standard methods and as previously described Citation11. DNA sequencing was accomplished via an automatic sequence facility (Applied Biosystems 310 DNA sequencer, Applied Biosystems, Foster City, CA). The complete base composition of both strands of plasmids pDC09 and pNC101 was determined by sequencing and assembling PCR amplified stretches of the plasmid using a newly designed primer system () and the Vector NTI advance 9.0 program (Informax, Invitrogen life science software, Darmstadt, Germany). For PCR amplification, the following conditions were used: 94°C for 2 min; followed by 35 cycles of 94°C for 45 s, annealing temperature (calculation based on ) for 30 s, and 72°C for 45 s (amplicon <800 bp) or 90 s (amplicon >800 bp), respectively; with a final extension at 72°C for 5 min. DNA and protein sequences were analyzed with the software programs GeneDoc (www.psc.edu/biomed/genedoc), Pfam (Sanger Institute, Cambridge, England), MOTIFsearch (GenomeNet, Kyoto, Japan), Softberry (MolQuest, Mount Kisco, USA), EMBOSS (European Molecular Biology Open Software Suite, Cambridge, England), and I-Tasser Citation12 Citation13. Database searches were carried out through NCBI and EMBL with the BLAST and ORF Finder search programs.
Determination of plasmid copy number
To determine the plasmid copy number of all four strains, pNC101 was selected for the preparation of standard reactions. Amplification products generated using the primer pFChyper/pRChyper and pNC101 () were purified using the Qiagen Purification Kit (Qiagen, Hilden, Germany) according to the manufacturer's instructions. Purified products were subsequently quantified with the PicoGreen dsDNA quantification kit (Quant-iT PicoGreen, Invitrogen, Darmstadt, Germany). Knowing the exact size of the amplicons and using the average molecular weight of a single DNA base pair, the measured DNA value could then be converted to target molecule numbers per microlitre. Dilution series of these PCR products were further used as calibration standards to measure DNA obtained from all S. mutans cultures with an unknown content of plasmids via Real-Time Quantitative PCR (RTQ-PCR) on a LightCycler 2.0 using the assay-primers pF5/pR2 (). For determination of total S. mutans cells, the glucosyltransferase-gene gtf was quantified by RTQ-PCR in parallel as previously described by Horz et al. Citation14. The calculated plasmid/gtf(B/C)–gene ratio per S. mutans strain then gave an estimate of the average number of plasmids per cell. Various dilutions were tested independently to increase the accuracy of the measurement ().
Table 2. Determination of plasmid number per cell in four S. mutans strains
Analysis of chromosomally encoded TA systems
All S. mutans strains possess at least two chromosomal TA systems, referred to as mazEF and hicBA, and additionally in some strains like UA159, DC09, NC101-relBE Citation15 Citation16. We designed the following primers to amplify chromosomal TA cassettes in plasmid positive and negative strains for sequence and mutation analysis: mazEF-F 5’ AATAGACACAGAAATAGGAGGT 3’, mazEF-R 5’ GATTTCAGTAGAAGTTTAACTG 3’; relBEchro-F 5’ GGTATAATAGATATAAAAGGAGTT 3’, relBEchro-R 5’ AACAGGAAGAACTACCTTTGAG 3’; Hic-F 5’ CGCTAGAAATCAATCTTTCTA 3’, Hic-R 5’ TAGTTTCTAAATCAGGGGCT 3’. For PCR amplification, the following conditions were used: 94°C for 2 min; followed by 32 cycles of 94°C for 30 s, 48°C for 30 s, and 72°C for 45 s; with a final extension of 72°C for 5 min. DNA and protein sequences were analyzed using the software programs mentioned above and the TADB as well as the GenBank database.
Results
A total of 40 different S. mutans strains were initially tested for the presence of a plasmid by PCR-amplification of a conserved 198 bp fragment using primer pair pF6/pR3 (). Four strains (10%) tested positive (confirmed by sequence analysis of the PCR-products) of which DC09 and NC101 were selected for an entire sequence analysis of their plasmids pDC09 and pNC101 and the resulting sequences deposed in GenBank under accession numbers HQ156229 and HQ156230. However, all four plasmid carriers were subjected to plasmid copy number determination.
Physical map of plasmids pDC09 and pNC101
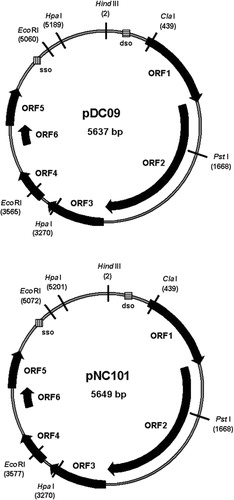
Both plasmids, pDC09 and pNC101, were found to contain identical restriction sites to the reference plasmid pUA140. They exhibited both single (e.g. HindIIII [position 2], ClaI [439], PstI [1668], NdeI [2,247], AfllI [3,082], XbaI [4,621], EspI [4,473], and XcmI [3,750]) as well as multiple restriction sites (e.g. HpaI [3,270; 5,189], AfllII [2,542; 5,415], and EcoRI [3,565; 5,060]) ( with selected restriction sites). With the exception of HindIII and EcoRI, which were tested experimentally by restriction analysis, the other sites were deduced from in silico analysis. The position of each restriction site is given according to pDC09 positions (in brackets). The exact size of pDC09 was 5,637 bp and 5,649 bp for pNC101, which is consistent with earlier estimations of related S. mutans plasmids Citation1 Citation2 Citation17 Citation18 and especially consistent with the size of pUA140 in the study of Zou et al. Citation7. The G + C content of pDC09 and pNC101 was not homogenous over the entire plasmid, approximately 32.6% (67.4% A + T) and 32.9% (67.1% A + T), respectively.
We found six ORFs encoding peptides larger than 90 amino acids (aa). All ORFs are located on the same strand with the following lengths of corresponding peptides: ORF1 (258 aa), ORF2 (555 aa), ORF3 (188 aa), ORF4 (105 aa), ORF5 (120 aa), and ORF6 (92 aa).
Coding regions
ORF1
In silico analysis revealed that ORF1 in both plasmids was located between position 397 and 1,170 (according to the pUA140 reference sequence numbering Citation7), thus 774 bp in length and encoding for a 258 aa Rep-protein. A Softberry search for promoter sequences revealed the -35-Box (5′TTCACA3′), a -10-Box (5′TAGTTATAAC3′), and a Shine-Dalgarno (SD) sequence starting with position -4 (5'AGGA3’) upstream from the ORF1 start codon ATG. Analysis of the amino acid sequence deduced from ORF1 of pDC09 and pNC101 gave results identical to the corresponding pUA140 sequence and showed similarity with the Streptococcus thermophilus Rep-protein. It demonstrated two motifs (II and III) universally associated with Rep proteins involved in rolling circle replication (RCR). Motif II may be involved in the binding of essential cofactor metal ions, such as Mg2 + and Mn2 +. Motif III is thought to be involved in DNA binding.
ORF2
The ORF2 in both plasmids was located between position 1,158 and 2,822, thus 1,665 bp in length, and encoding for a 555 aa Mob-protein. The search for promoter sequences revealed the -35-Box (5’ TTTCAA 3’), a -10-Box (5’ ATATAGAAT 3’) and a potential SD-sequence starting with position -14 (5'AGGA3’) upstream from ATG. The deduced amino acid Mob-sequence (at its N-terminus) was clearly related to TraD proteins and to the c109099 superfamily represented by P-loop-NTPases. The TraD protein performs an essential coupling function in conjugative secretion systems. This protein connects the inner membrane of the cell–cell contact site with the relaxosome-plasmid DNA complex. Members of this superfamily are characterized by conserved nucleotide phosphate-binding motifs, also referred to as the Walker A motif and the Walker B motif.
The 3’ end of ORF1 and the 5’ start of ORF2 were found to share 16 nucleotides (5’ ATGGACAACAATTTAG 3’). Within ORF1, a 38 nucleotide sequence (5’ TTATATCTCTTGGATAAATTTAGGGTATTAGATGCTAA 3’) was found to be an almost inverted repeat structure of an ORF2-motif (5’ TTAGCAACTGTTACACTAAAATTAGCTAAAAAATATAA 3’). Its function could not be revealed but may be associated with the DSO repeats (see below).
ORF3
The ORF3 in both plasmids was located between position 2,838 and 3,401 (564 bp in length) and encoding for a 188 aa protein with an unknown function. The Softberry search revealed the -35-Box (5’ TAGTAA 3’), a -10-Box (5’ TTTTAAAAT 3’), and a potential SD-sequence starting with position -8 (5'AGGA3’) upstream ATG. However, even extensive search for homology in GenBank and other databases could not reveal any potential function of protein 3 or homology except the pUA140 homologue.
ORF4
The ORF4 was located between position 3,465 and 3,779 in pDC09, and between position 3,477 and 3,791 in pNC101, because of a 12 bp insert in the promoter region. Both fragments are 315 bp in length and encode for a 105 aa protein. The search for promoter sequences revealed the -35-Box (5'TATCTA 3’), a -10-Box (5’ TGATATCAT 3’), and a potential SD-sequence starting with position -6 (5'AGGA3’) upstream from the ORF4 start. In accordance with Zou et al. Citation7, we found that the putative protein 4 had a certain degree of similarity to the putative H+ transporting ATPase subunit E (atpE) in Archaeoglobus fulgidus (GenBank Accession No. B69395, 23% identity, 57% similarity). Since the proton translocation complex confers tolerance to low pH aciduricity Citation19, a plasmid-borne version of one of the ATPases may complement or replace the intrinsic protein thus altering or even enhancing ATPase function.
ORF5
The ORF5 was located between position 4,230 and 4,589 in pDC09 and between position 4,242 and 4,601 in pNC101, respectively in both cases, 360 bp in length and encoding for a 120 aa protein. No promoter sequences were revealed. Search for homology at the protein level revealed protein 5 to be involved in plasmid stabilization (super family cl11422). It shows a high identity (38–50%) and similarity (58–74%) to proteins of Streptococcus pneumoniae (hypothetical protein, GenBank No. ZP_01832116.1), Streptococcus equi (plasmid stabilization system, No. YP_002747188), and Streptococcus downei (addiction module toxin No. ZP_07726556.1, plasmid stabilization system No. ZP_07725301.1). This protein is involved in stabilization of plasmid transfer as it functions as a stable toxin of the RelE family.
ORF6
Finally, ORF6 was located between position 3,977 and 4,264 in pDC09 and between position 3,989 and 4,273 in pNC101, respectively, in both cases, 288 bp in length and encoding for a 92 aa protein. The search for promoter sequences revealed the -35-Box (5’ TTGCAC 3’), a -10-Box (5’ TTATATAAT 3’), and a potential SD-sequence starting with position -22 (5'AGGA3’) upstream from the start. Search for homology at the protein level revealed (putative) protein 6 to belong to the RelB superfamily cl01171 which was also involved in plasmid stabilization, with 68% identity to the corresponding protein of S. equi 4047 (GenBank No. YP_002747189) and 61% identity to S. downei F0415 (GenBank No. ZP_07725256), respectively.
Rolling circle replication of pDC09 and pNC101
Rolling circle replication (RCR) plasmids typically share a number of common features. This includes a double- and single-stranded origin (DSO and SSO), respectively, a gene encoding for a replication (Rep) protein, and a single-stranded intermediate. Furthermore, the nic region is highly conserved among replicons of the same family while the bind loci usually vary Citation20 Citation21. The sequence 5’-CAAACGAACTACTAATAGCAGTAA-3’ on the coding strand from 296 to 319 bp of both plasmids pDC09 and pNC101 was identical or at least remarkably similar to the nic region of pUA140 Citation7, pT181 Citation22, and that of other plasmids or phages from the same plasmid family Citation20 Citation21. In particular, the nucleotide sequence 5’ CT/AATAGC 3’ between bases 307 and 314, which includes the nic site (T/A), was identical to the consensus sequence within the origins of pT181-like replicons. The putative DSO region contains several inverted repeats capable of generating hairpin structures, including one large hairpin with the nic site downstream. As the DSO usually lies upstream of or within Rep Citation23 Citation24 and the location of the nic site (T/A) in pDC09 and pNC101 was 89 bp upstream of the ORF1 start codon, further evidence exists that ORF1 encodes for the Rep protein.
Analysis of DNA sequence structure revealed a region between 4,998 and 5,174 on the encoding strand of pDC09 (5,011 and 5,192 on the encoding strand of pNC101, respectively), with a high level of secondary structure upstream of the Rep gene and close to the high G + C region. This agrees with the fact that the SSO region, although less conserved than that of the DSO region, usually resides within a region of strong secondary structure located upstream of the coding sequences for the Rep gene. Within this region, the hexanucleotide sequence 5’-TAGCGT-3’ was located as part of the stem of a large hairpin structure presumably acting as the site of second-strand initiation (ssi) and transcriptional terminator for the synthesis of an RNA primer Citation23.
Determination of plasmid copy numbers
Purified and standardized plasmid PCR products were quantified with the PicoGreen dsDNA quantification kit as described above using different dilutions. A gtf B- and gtfC-targeting RTQ-PCR was applied to assess the number of S. mutans cells. The results are given in calculated from different dilutions of pDC09, pDD09, pNC101, and pNC102. Whereas strains NC101 and NC102 exhibited medium copy numbers of 10 and 15, the copy numbers of strains DC09 and DD09 were 23 and 74. The latter might be very high which could be due to a high multiplication rate of host cells at the time tested. Regardless, the associated number of TA systems significantly increases the RNA endonuclease potential of the plasmid carriers.
Sequence analysis of chromosomal TA systems in plasmid positive and negative strains
As shown in , a number of differences in the chromosomal TA systems (hicBA, mazEF, and relBEchro) were found between strains. However, none of the resulting differences in amino acid sequence was found exclusively in plasmid carriers (DC09 & NC101) or non-carriers (KK23, UA159) and were essential for toxin or antitoxin function. Unexpectedly, the sequence homology between relEplas and relEchro and also between relBplas and relBchro was relatively low. However, as mentioned by Guglielmini and van Melderen Citation25, it is known that some toxin-superfamilies exhibit dramatic sequence diversities but have a similar tertiary structure. By running the I–Tasser protein prediction analysis, we were able to confirm this assumption for both RelE and RelB ().
Fig. 2. Amino acid sequence comparison of three chromosomally encoded TA systems (HicBA, MazEF, RelBE) between plasmid carrier strains DC09/NC101 and plasmid-free strains UA159/KK23. None of the alterations are typical for plasmid carriers or non-carriers or were found to be essential for the principal toxin or antitoxin function. In case of RelB, the plasmid-encoded version (RelBplas of DC09 and NC101) has little similarity with RelBchro (of UA159, DC09, NC101, all identical). In case of RelE, the plasmid-encoded version (RelEplas of DC09 and NC101) has little similarity with the RelEchro of UA159, DC09, and NC101. However, we found similar tertiary structures applying I–Tasser protein prediction analysis ().
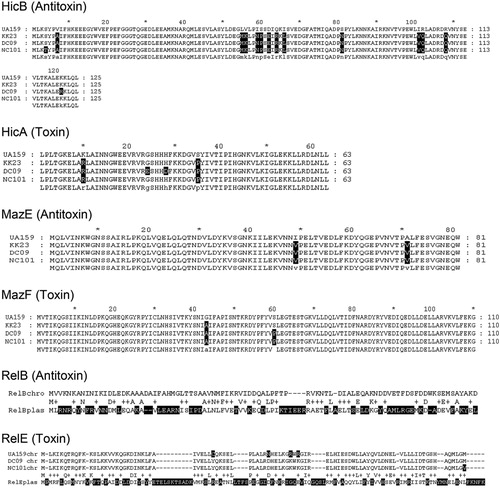
Fig. 3. Protein prediction model based on I-Tasser, A) left: RelEchro UA159, right: RelEplas DC09/NC101; B) left: RelBchro UA159, right: RelBplas DC09/NC101.
Source: http://zhanglab.ccmb.med.umich.edu/I-TASSER/ Citation12 Citation13
Survival under amino acids starvation
After incubation of S. mutans strains in BM-medium with 1) reduced casaminoacid and essential amino acids concentrations (i.e. 10% of standard concentration) or 2) with SHX as a starvation stress inductor, the cultures were monitored for viability daily.
Cleary, neither the medium copy number strains NC101 and NC102 (with 10–14 days) nor the high copy number strains DC09 and DD09 (with 10–14 days) persisted significantly longer than the plasmid-free S. mutans representatives (7–24 days).
Discussion
We analyzed the genetic architecture of two new S. mutans 5.6-kb plasmids, pDC09 and pNC101. Both were highly similar to pUA140 and fall into what Zou et al. designated as the ‘Group I plasmids’ Citation7. They are usually found in S. mutans strains of humans of African or Asian descent. DC09 and NC101 were isolated in London making such an origin – even if not documented – plausible. With this evolutionary significance together with the inherent stability, these plasmids might be a good marker for S. mutans (and for its human host) evolutionary history.
Complementing the pUA140 data of Zou et al. Citation7, we found six ORFs which are located on the same DNA strand and encoded six proteins with a length between 92 (ORF6, product = antitoxin) and 555 (ORF2, product = Mob protein) amino acids. No meaningful sequence homologue was found for ORF3. As shown for pUA140, the plasmids replicate by RCR and belong to the pT181 family based on similarities in the Rep protein sequence (ORF1), the DSO, and the secondary structure-rich SSO Citation7. It is also noteworthy that DSO and SSO flank a high G + C region with many secondary structures, which supports replication initiation and termination. The Mob protein encoded by ORF2 plays a role in plasmid stability.
RelE (encoded by ORF5) and RelB (encoded by ORF6) form a toxin–antitoxin system (type II loci). In addition, the terms ‘poison/antidote system’ and ‘plasmid addiction system’ are used in literature. The corresponding genes, relE and relB, are usually under the control of the same promoter that we found located upstream of relB. RelE, an RNA endonuclease, is stable and terminates global translation through digestion of mRNA positioned at the ribosomal A site. RelB, as a counterpart, is unstable. It forms complexes with RelE, blocking the endonuclease activity and repressing its own promoter. Progenies that inherit a plasmid copy at cell division grow normally. In contrast, cells that do not inherit a plasmid copy still carry the stable toxin in the cytoplasm while the antitoxin is degraded. This leads to the selective post-segregational killing (PSK) of the plasmid-free progenies, but a dormant state of these cells should also be considered. In this way, TA loci prevent the proliferation of plasmid-free cells in growing bacterial cultures, leading to what is called ‘plasmid addiction’. In addition and according to the ‘selfish theory’, TA systems allow a conjugative plasmid to outcompete a second and TA-negative plasmid belonging to the same incompatibility group Citation26. Mathematical models demonstrate that the PSK phenomenon allows the propagation of TA systems in bacterial populations, independent of their original frequencies. This might provide a rational explanation for the evolutionary success of TA systems Citation27 and lead to the assumption that TA + plasmids might have an advantage over TA-plasmids under certain conditions Citation28–Citation30.
A question yet to answer is if there are any links or interactions (cross reactivity, dosage effects) between TAplas and TAchro, the latter involved in persistence under unfavourable conditions. Leung and Lévesque demonstrated that ectopic expression of RelBEchro or of MazEF caused cell growth arrest and forms persisters under stress, in their case antibiotic treatment Citation31. From other studies, it is known that under amino acid starvation RelBEchro reduces the global level of translation to save amino acids for the most essential housekeeping processes. As this is a mechanism present across bacterial phyla, cross reactivity must be considered as feasible. In any case, RelE is a global regulator of translation and is activated by stress via alarmones and via Lon proteases (digest of antitoxin), finally inducing mRNA cleavage Citation8 Citation9 Citation32. According to Gotfredsen and Gerdes Citation33, the chromosomal relBE locus stabilizes plasmids as efficiently as the plasmid-borne relBE loci confirming a kind of trans-activity. Recently, the same group showed that successive deletion of 10 different TA loci in E. coli progressively reduced the level of persisters, demonstrating a dosage effect Citation8.
We also tested the hypothesis that RelBEplas compensates a dysfunction of corresponding TAchro modules and is therefore beneficial or even indispensable for certain S. mutans strains. As shown by Lemos et al. Citation32, S. mutans UA159 has at least two TA loci on the chromosome (relBE and mazEF) plus a hicBA module as Song et al. recently found Citation16. In our study, we compared the amino acid sequences corresponding to all three loci between plasmid carrier (DC09, NC101) and non-carrier strains (UA159, KK23). Some alterations were found but none of them seem to be unique for plasmid carriers or non-carriers or accumulating in consensus regions essential for toxin or antitoxin function (). Altogether, in the case of RelB and RelE, the plasmid-encoded proteins show limited homology to their chromosomal pendants in their primary structure. However, when comparing the tertiary structures (), similarities become obvious and a grouping into the same family seems justified. Based on the three-dimensional structure, trans-activity of Antitoxinplas inhibiting Toxinchro and of Antitoxinchro inhibiting Toxinplas is conceivable.
The present study provides complete sequence information of two additional 5.6-kb S. mutans plasmids and revises the function of ORFs. It provides evidence that the ORF5 product belongs to the RelE (toxin) and that the ORF6 product belongs to the RelB (antitoxin) family. After comparing chromosomal and plasmid TA modules and corresponding protein structures, we speculate, depending on their netted endonuclease activity, whether they might 1) kill every plasmid-free progeny and stabilize plasmid transfer at the expense of the host and/or 2) help S. mutans to enter a dormant state and survive unfavourable conditions, as is known for chromosomal systems of the same kind. A function in plasmid stabilization has been confirmed: the 5.6-kb plasmids appear to be extremely stable in their S. mutans host as indicated by the apparent difficulty in curing strains of the plasmid according to the literature Citation3 Citation18 Citation34 Citation35 and our observations. The stability of such plasmids makes them attractive as cloning/expression vectors.
A function in persistence, tested here by inducing amino acid starvation using two methods, could not be confirmed. Clearly, further experiments with modified environmental factors (pH, temperature, oxygen, substrates, etc.) are needed to clearly exclude or confirm any advantage in the persistence of plasmid-carriers.
Conflict of interest and funding
There is no conflict of interest in the present study for any of the authors. This work was funded as part of the BioInSys project No. 0315411D by the German Ministry of Education and Research (BMBF, call MedSys), and we thank all collaborative partners here.
Acknowledgements
For providing strains, we are grateful to Susanne Kneist (University Hospital Jena) and David Beighton (KCL Dental Institute, London).
Notes
GenBank accession No.: HQ156229 (pDC09), HQ156230 (pNC101).
References
- Caufield PW, Wannemuehler YM, Hansen JB. Familial clustering of the Streptococcus mutans cryptic plasmid strain in a dental clinic population. Infect Immun. 1982; 38: 785–7.
- Macrina FL, Reider JL, Virgili SS, Kopecko DJ. Survey of the extra chromosomal gene pool of Streptococcus mutans. Infect Immun. 1977; 17: 215–26.
- Caufield PW, Shah G, Hollingshead SK, Parrot M, Lavoie MC. Evidence that mutacin II production is not mediated by a 5.6-kb plasmid in Streptococcus mutans. Plasmid. 1990; 24: 110–8. 10.3402/jom.v5i0.19729.
- Noji S, Date S, Abiko Y, Takiguchi H, Taniguchi S. Cloning and characterization of Streptococcus mutans LM7 plasmid pAM7. Infect Immun. 1987; 55: 2538–40.
- Caufield PW, Childers NK, Allen DN, Hansen JB. Distinct bacteriocin groups correlate with different groups of Streptococcus mutans plasmids. Infect Immun. 1985; 48: 51–6.
- Caufield PW, Ratanapridakul K, Allen DN, Cutter GR. Plasmid-containing strains of Streptococcus mutans cluster within family and racial cohorts: implications for natural transmission. Infect Immun. 1988; 56: 3216–20.
- Zou X, Caufield PW, Li Y, Qi F. Complete nucleotide sequence and characterization of pUA140, a cryptic plasmid from Streptococcus mutans. Plasmid. 2001; 46: 77–85. 10.3402/jom.v5i0.19729.
- Maisonneuve E, Shakespeare LJ, Jorgensen MG, Gerdes K. Bacterial persistence by RNA endonucleases. Proc Natl Acad Sci USA. 2011; 108: 13206–11. 10.3402/jom.v5i0.19729.
- Kuroda A, Murphy H, Cashel M, Kornberg A. Guanosine tetra– and pentaphosphate promote accumulation of inorganic polyphosphate in Escherichia coli. J Biol Chem. 1997; 272: 21240–3. 10.3402/jom.v5i0.19729.
- Gerdes K, Christensen SK, Lobner-Olesen A. Prokaryotic toxin–antitoxin stress response loci. Nat Rev Microbiol. 2005; 3: 371–82. 10.3402/jom.v5i0.19729.
- Shah GR, Caufield PW. Enhanced transformation of Streptococcus mutans by modifications in culture conditions. Anal Biochem. 1993; 214: 343–6. 10.3402/jom.v5i0.19729.
- Roy A, Kucukural A, Zhang Y. I-TASSER: a unified platform for automated protein structure and function prediction. Nat Protoc. 2010; 5: 725–38. 10.3402/jom.v5i0.19729.
- Zhang Y. I-TASSER server for protein 3D structure prediction. BMC Bioinformatics. 2008; 9: 40. 10.3402/jom.v5i0.19729.
- Horz HP, Scheer S, Huenger F, Vianna ME, Conrads G. Selective isolation of bacterial DNA from human clinical specimens. J Microbiol Methods. 2008; 72: 98–102. 10.3402/jom.v5i0.19729.
- Shao Y, Harrison EM, Bi D, Tai C, He X, Ou HY, et al.. TADB: a web-based resource for Type 2 toxin–antitoxin loci in bacteria and archaea. Nucleic Acids Res. 2011; 39: D606–11. 10.3402/jom.v5i0.19729.
- Song L, Sudhakar P, Wang W, Conrads G, Brock A, Sun J, et al.. A genome-wide study of two-component signal transduction systems in eight newly sequenced mutans streptococci strains. BMC Genomics. 2012; 13: 128. 10.3402/jom.v5i0.19729.
- Dunny GM, Birch N, Hascall G, Clewell DB. Isolation and characterization of plasmid deoxyribonucleic acid from Streptococcus mutans. J Bacteriol. 1973; 114: 1362–4.
- Hansen JB, Abiko Y, Curtiss R3rd. Characterization of the Streptococcus mutans plasmid pva318 cloned into Escherichia coli. Infect Immun. 1981; 31: 1034–43.
- Smith AJ, Quivey RGJr, Faustoferri RC. Cloning and nucleotide sequence analysis of the Streptococcus mutans membrane-bound, proton-translocating ATPase operon. Gene. 1996; 183: 87–96. 10.3402/jom.v5i0.19729.
- del Solar G, Moscoso M, Espinosa M. In vivo definition of the functional origin of replication (ori(+)) of the promiscuous plasmid pLS1. Mol Gen Genet. 1993; 237: 65–72.
- del Solar G, Moscoso M, Espinosa M. Rolling circle-replicating plasmids from gram-positive and gram-negative bacteria: a wall falls. Mol Microbiol. 1993; 8: 789–96. 10.3402/jom.v5i0.19729.
- Rasooly A, Novick RP. Replication-specific inactivation of the pT181 plasmid initiator protein. Science. 1993; 262: 1048–50. 10.3402/jom.v5i0.19729.
- del Solar G, Giraldo R, Ruiz-Echevarria MJ, Espinosa M, Diaz-Orejas R. Replication and control of circular bacterial plasmids. Microbiol Mol Biol Rev. 1998; 62: 434–64.
- Gruss A, Ehrlich SD. The family of highly interrelated single-stranded deoxyribonucleic acid plasmids. Microbiol Rev. 1989; 53: 231–41.
- Guglielmini J, Van Melderen L. Bacterial toxin–antitoxin systems: translation inhibitors everywhere. Mob Genet Elements. 2011; 1: 283–90. 10.3402/jom.v5i0.19729.
- Mochizuki A, Yahara K, Kobayashi I, Iwasa Y. Genetic addiction: selfish gene's strategy for symbiosis in the genome. Genetics. 2006; 172: 1309–23. 10.3402/jom.v5i0.19729.
- Van Melderen L, Saavedra De Bast M. Bacterial toxin–antitoxin systems: more than selfish entities?. PLoS Genet ; : e. 2009; 5: 1000437. 10.3402/jom.v5i0.19729.
- Cooper TF, Heinemann JA. Selection for plasmid post-segregational killing depends on multiple infection: evidence for the selection of more virulent parasites through parasite-level competition. Proc Biol Sci. 2005; 272: 403–10. 10.3402/jom.v5i0.19729.
- Cooper TF, Paixao T, Heinemann JA. Within-host competition selects for plasmid-encoded toxin–antitoxin systems. Proc Biol Sci. 2010; 277: 3149–55. 10.3402/jom.v5i0.19729.
- Nordstrom K, Austin SJ. Mechanisms that contribute to the stable segregation of plasmids. Annu Rev Genet. 1989; 23: 37–69. 10.3402/jom.v5i0.19729.
- Leung V, Levesque CM. A stress-inducible quorum-sensing peptide mediates the formation of persister cells with noninherited multidrug tolerance. J Bacteriol. 2012; 194: 2265–74. 10.3402/jom.v5i0.19729.
- Lemos JA, Brown TAJr, Abranches J, Burne RA. Characteristics of Streptococcus mutans strains lacking the MazEF and RelBE toxin–antitoxin modules. FEMS Microbiol Lett. 2005; 253: 251–7. 10.3402/jom.v5i0.19729.
- Gotfredsen M, Gerdes K. The Escherichia coli relBE genes belong to a new toxin–antitoxin gene family. Mol Microbiol. 1998; 29: 1065–76. 10.3402/jom.v5i0.19729.
- Clewell DB. Plasmids, drug resistance, and gene transfer in the genus Streptococcus. Microbiol Rev. 1981; 45: 409–36.
- Murchison HH, Barrett JF, Cardineau GA, Curtiss R3rd. Transformation of Streptococcus mutans with chromosomal and shuttle plasmid (pYA629) DNAs. Infect Immun. 1986; 54: 273–82.