Abstract
Background
The malQ and glgP genes, respectively, annotated as putative 4-α-glucanotransferase and putative glycogen phosphorylase are located with a 29 nucleotide overlap on the Streptococcus mutans genome. We found that the glgP gene of this organism was induced with maltose, and the gene likely constituted an operon with the upstream gene malQ. This putative operon was negatively regulated with the malR gene located upstream from the malQ gene and a MalR-binding consensus sequence was found upstream of the malQ gene. S. mutans is not able to catabolize starch. However, this organism utilizes maltose degraded from starch in the presence of saliva amylase. Therefore, we hypothesized that the MalQ/GlgP system may participate in the metabolism of starch-degradation products.
Methods
A DNA fragment amplified from the malQ or glgP gene overexpressed His-tagged proteins with the plasmid pBAD/HisA. S. mutans malQ and/or glgP mutants were also constructed. Purified proteins were assayed for glucose-releasing and phosphorylase activities with appropriate buffers containing maltose, maltotriose, maltodextrin, or amylodextrin as a substrate, and were photometrically assayed with a glucose-6-phosphate dehydrogenase–NADP system.
Results
Purified MalQ protein released glucose from maltose and maltotriose but did not from either maltodextrin or amylodextrin. The purified GlgP protein did not exhibit a phosphorylase reaction with maltose or maltotriose but generated glucose-1-phosphate from maltodextrin and amylodextrin. However, the GlgP protein released glucose-1-phosphate from maltose and maltotriose in the presence of the MalQ protein. In addition, the MalQ enzyme activity with maltose released not only glucose but also produced maltooligosaccharides as substrates for the GlgP protein.
Conclusion
These results suggest that the malQ gene encodes 4-α-glucanotransferase but not α-1,4-glucosidase activity. The malQ mutant could not grow in the presence of maltose as a carbon source, which suggests that the malQ gene is essential for the utilization of starch-degradation products.
Streptococcus mutans is a major etiologic agent of human dental caries (Citation1). Although some strains of this organism have been recently reported as agents of infective endocarditis (Citation2) as well as aggravating agents of hemorrhagic stroke (Citation3) and ulcerative colitis (Citation4) when they entered the blood streams of animals, the natural habitat of S. mutans was an oral biofilm called dental plaque. Here, S. mutans is subjected to continual cycles of abundance and depletion (so-called ‘feast’ and ‘famine’) (Citation5) with respect to carbohydrate energy sources. S. mutans accumulates intracellular polysaccharides as energy reserve materials similar to glycogen and starch in animals and plants, respectively (Citation6). Therefore, S. mutans harbors genes encoding intracellular polysaccharide synthesis and degradation enzymes in its genome. The malQ and glgP genes respectively annotated as putative 4-α-glucanotransferase and putative glycogen phosphorylase (Citation7) are located with a 29 nucleotide overlap, likely constituting an operon on its genome. Since a debranching enzyme that mediates glycogen degradation is a member of the 4-α-glucanotransferases, we initially speculated that the enzyme encoded by the malQ gene may be involved in glycogen degradation as a debranching enzyme together with the glycogen phosphorylase encoded by the glgP gene.
However, we recently reanalyzed the genome sequence around the glgP gene region and found a potential promoter-like sequence located in the 114-bp intergenic region between the malQ and upstream malR genes. In addition, a MalR-binding consensus sequence reported in Streptococcus pyogenes (Citation8) was detected between the putative malQ -35 and -10 sequences. We constructed a malR mutant in strain UA159. The phenotype of this mutant concerning GlgP expression was constitutive. Therefore, we concluded that the malR gene was the negative regulator of the putative malQ/glgP operon. In addition, this consensus sequence was also found in the promoter regions of the malT (symbolized as ptsG in the genome data) and malXFGK operons respectively encoding IIABCmaltose of the phosphoenolpyruvate-dependent maltose phosphotransferase system (PTS) (Citation9) and the MalXFGK-binding protein-dependent ABC transporter for maltooligosaccharides (Citation10, Citation11). Furthermore, the other glycogen phosphorylase gene phsG were located on another part of the chromosome (Citation7) as a cluster with the glycogen synthesis genes (glgBCDA), although a candidate for a debranching enzyme was not present in this gene cluster.
A major carbon source during a ‘feast’ period in the human oral environment is dietary starch. However, S. mutans could not grow with starch as a direct carbon source, but grew well with maltose or maltooligosaccharides derived from starch in the presence of a small amount of saliva. Therefore, we presumed that the malQ/glgP genes may be involved in the catabolism of maltose or maltooligosaccharides derived from the starch supplied in food in the oral environment rather than the degradation of intracellular glycogen-like polysaccharides. This suggested that this organism, as well as other oral streptococci (Citation12), indirectly utilizes starch in oral biofilms. However, the energy metabolism of starch-degradation products in S. mutans has not yet been fully elucidated. Therefore, we characterized the malQ and glgP genes in this study.
Materials and methods
Bacterial strains
S. mutans strains used were UA159 (Citation7) and its mutants fkU1 (malR), zJU1 (malQ), zKU1 (glgP), and aeU1 (malQ, glgP c ). Streptococci were maintained on Todd-Hewitt (TH) broth/agar plates with or without appropriate antibiotics. Escherichia coli strain TOP10 was used as a host with the vector pBAD/HisA for the expression of cloned genes as N-terminal histidine-tagged proteins.
PCR amplification of fragments to express or inactivate specific genes
The polymerase chain reaction (PCR) primers used in this study are listed in . All amplification reactions were carried out with high-fidelity DNA polymerase, KOD-Plus (Toyobo, Osaka, Japan) without terminal deoxynucleotidyl transferase activity. Regions corresponding to the malQ and glgP genes in strain UA159 were amplified with the primer sets exmalQ5/malQ32 and expglg51/expglg31, respectively. Amplified fragments were purified, digested with XhoI, and subcloned into XhoI/PvuII-double-digested pBAD/HisA. The fragment (fk) was constructed by inserting the kanamycin-resistant gene cassette into the BamHI site in the middle of the malR gene, which was subcloned into a pUC-vector () (Citation13). Splicing by the overlapping extension method (Citation14) was employed to construct the linear fragments (zJ), (zK) and (ae) used to transform S. mutans UA159 resulting in malQ- and glgP-mutants (zJU1, zKU1 and aeU1). Target regions containing primer-annealing sites that were used in this technique are noted in and the targeted fragments corresponded to those in , indicating spliced linear fragments for transforming S. mutans UA159.
Fig. 1 Construction of linear fragments for transforming S. mutans. Transformation was carried out as described previously (Citation13). Numbers depicted above and below the fragments (fk), (zJ), (zK), and (ae) correspond to the nucleotide positions on the complementary strand of the UA159 genome. Ovals in the figure indicate promoter regions, which are abbreviated as ‘prom’ containing a 114-bp intergenic region between the malR and malQ genes. A MalR-binding consensus sequence is located between the -35 and -10 regions of a putative promoter sequence. CTC in the fragment (ae) are added nucleotides for splicing the fragments and are located immediately upstream from the Shine–Dalgarno sequence of the glgP gene.
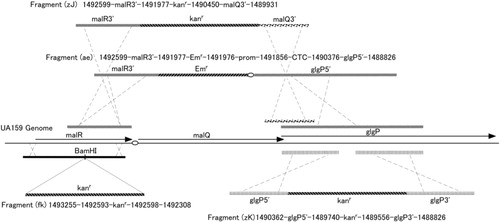
Table 1 Primers used in this study
Sample preparation, SDS-PAGE, and Western blot analysis
S. mutans strains and mutants were grown in 10-ml BTR-Sugar broth (Citation15) (1% Bacto Tryptone, 0.1% Bacto yeast extract, 0.05% sodium thioglycolate, 0.61% K2HPO4, 0.2% KH2PO4, 1 mM MgSO4, 0.1 mM MnSO4, 0.2% sugar). Cells were harvested, washed, disrupted with a Tissue Lyser (Qiagen, Hilden, Germany) with 0.2 mm diameter ceramic beads, and subjected to centrifugation (12,000×g, 5 min) with a microfuge to remove undisrupted cells. Four hundred microliters of supernatant fluid was obtained as a crude extract sample and its protein concentration was determined with DC protein assay reagents (BioRad, Hercules, CA, USA). SDS-PAGE was run with 2.5 µg of samples for Quick-CBB-PLUS (Wako Pure Chemical Industries, Osaka, Japan) staining and 0.4 µg of samples were used for Western blot analysis with previously prepared anti-GlgP serum (Operon Biotechnology, Tokyo, Japan) as described previously (Citation16).
E. coli clones designated as ZF27 and ZF32 expressed the GlgP and MalQ proteins, respectively. The cells of these strains grown with 100-ml LB broth supplemented with 4×10–3% arabinose as an inducer were collected, washed, and subjected to 20 cycles of 30-second ultrasonication and 60-second incubation periods in an iced water container to obtain crude cell-free extracts.
The GlgP and MalQ proteins were purified with Ni-Sepharose 6 Fast Flow resin (GE Healthcare KK, Tokyo, Japan) as described previously, and the protein concentrations of the purified proteins were determined with DC protein assay reagents. Purified samples were frozen at −20°C for later enzyme assays. The enzyme activities of these samples were stable for at least 8 months.
Enzyme assays for glucose-releasing and phosphorylase activities
Enzyme reactions both for the MalQ and GlgP proteins were performed in 50-µl reaction mixtures within a thin-wall-PCR tube with starch-degradation products as substrates. The standard enzyme reaction mixtures for glucose- and glucose-1-phosphate (G1P) assays were composed as recommended by the supplier (Oriental Yeast Co. Ltd, Tokyo, Japan) of enzymes used in the assay system. The enzyme reaction mixture contained 0.7–1.0 µg of purified MalQ protein or 2.5–3.2 µg of purified GlgP protein in addition to 50 mM potassium phosphate (pH 7), 0.5 mM MgCl2, and sugar substrate. The sugar substrates used in the enzyme reaction were 1% maltose, maltotriose, maltodextrin, and 0.8% amylodextrin (final concentrations). Both glucose-releasing and phosphorylase reactions were not linear as a function of time. Therefore, the incubation time was fixed at 10 min at 37°C followed by 5-min inactivation at 95°C. Glucose and G1P were formed in the reaction mixtures containing MalQ and GlgP proteins, respectively. Glucose or G1P in aliquots were spectrophotometrically determined by the end-point method of absorbance changes at 340 nm as a result of the amount of generated NADPH in the assay mixtures as recommended by the supplier (Oriental Yeast Co. Ltd) of the enzymes. Glucose and G1P assays were started with the addition of 2 IU of hexokinase and phosphoglucomutase, respectively.
Monitoring for growth of UA159 and its specific mutants
The growth of S. mutans strains and mutants in BTR-sugar broth was measured at an optical density (OD) of 660 nm with the Ultrospec 500 pro spectrophotometer (GE Healthcare Life Sciences, Uppsala, Sweden). Values of OD660 nm were recorded at 1-hour intervals following the inoculation of cultures into screw-capped glass tubes containing BTR-sugar broth. Sugars included maltose, maltotriose, maltodextrin, and amylodextrin as starch-degradation products, other disaccharides, as well as glucose as a control.
Thin-layer chromatography
The reaction products liberated in the MalQ enzyme reaction mixture described above incubated with 1% maltose as a substrate for 100 min were characterized with respect to molecular size by thin-layer ascending chromatography on a 0.25-mm silica-gel-coated glass plate (TLC Silica Gel 60; Merck, Darmstadt, Germany) employing a solvent system of 1 M lactic acid/acetone/2-propanol (2: 4: 4, by vol.) (Citation17). Carbohydrate spots were visualized by soaking the chromatogram into p-anisaldehyde solution containing ethanol/sulfuric acid/acetic acid/p-methoxybenzaldehyde (68: 2.5: 1.2: 1.9) and heated on a hotplate (Model 49SH, Fisher Scientific, Hampton, NH, USA) for 40 min at a dial setting of 4.5.
Results
Enzyme activities of the MalQ and GlgP proteins
The MalQ or GlgP protein was expressed as a His-tagged protein, purified, and the enzyme activities were determined as described in ‘Materials and methods’ section. The MalQ protein exhibited glucose-releasing activities with maltose and maltotriose and low activity with maltodextrin but not with amylodextrin. The GlgP protein did not exhibit phosphorylase activities with maltose and maltotriose but did with maltodextrin and amylodextrin (). Moreover, no glucose-releasing activity by the GlgP protein and no phosphorylase activity by the MalQ protein from any of these sugars were also confirmed.
Table 2 Enzyme activities of the MalQ and GlgP proteins
Expression of the GlgP protein and construction of the malQ/glgP mutants
The GlgP protein was not induced in wild-type strain UA159 when grown with glucose but was markedly induced with maltose and maltotriose, which are starch-degradation products, as indicated by Western blot results (). GlgP protein bands were also observed with CBB (Coomassie Brilliant Blue) staining of the SDS gel (arrow in ). Their expression levels were similar to those of glucose-grown malR-mutant fkU1 and aeU1 (), in contrast to no GlgP expression in glgP-mutant zKU1 and mutant zJU1 affected by the polar effect of the upstream malQ insertional inactivation. These results suggest that the MalR protein acted as a negative regulator of the malQ/glgP operon and that maltose and other starch-degradation products induced the transcription of this operon by inactivating the MalR protein. S. mutans was unable to ferment starch but able to grow with starch in the presence of saliva (a). Therefore, we hypothesized that the malQ and glgP genes are involved in the energy metabolism of starch-degradation products.
Fig. 2 Expression of the GlgP protein in S. mutans. SDSPAGE and Western blot analysis were performed as described in the text. An arrow indicates GlgP protein bands appeared in the SDSPAGE. Cells were grown in BTR-sugar broth.
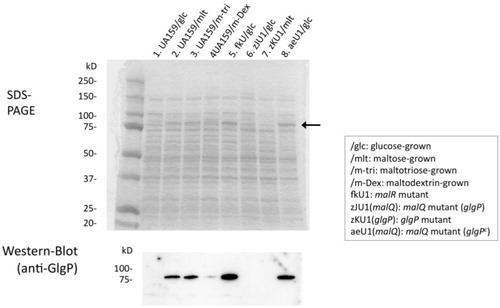
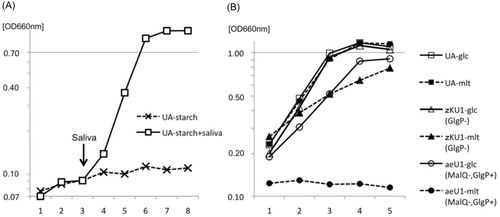
Fig. 3 Growth of UA159, malQ mutant aeU1, and glgP mutant zKU1. The growth medium used was BTR-broth, whose composition is described in the text. Ten microliters of filter-sterilized saliva were added at the arrow into 10-ml culture of strain UA159 in panel (a). Abbreviations: UA-, UA159. The mutant aeU1 did not grow with maltotriose or maltodextrin and the mutant zJU1 also did not utilize these maltooligosaccharides as well as maltose. These results were eliminated from panel (b) of this figure to avoid ambiguity but are presented with the results for aeU1-mlt.
We initially constructed the mutant zJU1 in which the malQ gene was inactivated by replacement of the chromosomal DNA fragment encompassing a malQ upstream region and 93% of the malQ 5′ gene region with the kanamycin-resistant gene kanr (). This mutant contained the intact glgP gene but did not express GlgP protein likely due to a polar effect on the malQ/glgP operon as suggested above. A characteristic phenotype of the mutant zJU1 was no growth with maltose as the sole carbon source. However, we could not determine whether the phenotype resulted from the absence of the MalQ or GlgP proteins in this mutant. Therefore, we constructed another malQ mutant in an attempt to be able to constitutively express the GlgP protein. The chromosomal DNA region around the malQ gene and the fragment (ae), which was used to transform S. mutans strain UA159, are also indicated in . The resultant MalQ-negative mutant was designated as aeU1. When we constructed the fragment (ae), we planned to use the segment malR3′ () by employing the corresponding region from the chromosome of a strain harboring a mutation in the malR gene. We confirmed the constitutive expression of the GlgP protein with glucose-grown cells in contrast to the absence of GlgP expression in glucose-grown UA159 or zJU1 cells as indicated in . The GlgP-negative mutant zKU1 was constructed by the transformation of strain UA159 with the fragment (zK), in which a glgP internal fragment was replaced with the streptococcal kanamycin-resistant gene kanr ().
Growth of UA159 and its specific mutants, aeU1 and zKU1
Parental strain UA159 grew well with either glucose or maltose as a carbon source (b). In contrast, the malQ mutant aeU1 exhibited no growth with maltose. This mutant still utilized glucose, although the growth rate and yield of this mutant were slightly lower than those of strain UA159. No clear differences in growth rate and yield were observed between UA159 and aeU1 when melibiose, raffinose, lactose, trehalose, or sucrose was supplied as sole carbon sources (data not shown). The finding that this mutant could not grow with maltotriose or maltodextrin (data not shown) in addition to maltose suggests that the malQ gene is very likely essential for the utilization of starch-degradation products, but does not participate in the energy metabolism of the other sugars mentioned above. The GlgP mutant zKU1 grew well with glucose similar to UA159. However, the growth of this mutant in the presence of maltose as the sole carbon source was slightly less than that when glucose was used as a carbon source. The reason for this phenomenon will be discussed below. These results together with the observation that the GlgP protein had no phosphorylase activities for maltose or maltotriose described above suggest that the glgP gene is not essential for, but somewhat influences the utilization of maltose and maltotriose.
Reciprocally additive effect on the enzyme activities of the MalQ and GlgP proteins
The malQ and glgP genes were likely co-transcribed because the malQ mutant zJU1 was GlgP negative as described above. Therefore, these two proteins may work together on their substrates or products in vivo. Glucose-releasing and phosphorylase activities on starch-degradation products were determined in the presence of both of the purified MalQ and GlgP proteins (). The most significant changes were the phosphorylase activities of GlgP proteins with maltose and maltotriose in the absence and presence of the MalQ protein. Glucose-releasing activities in the presence of the GlgP protein were almost unchanged from those without the GlgP protein.
Table 3 Reciprocally additive effect on the enzyme activities of the MalQ and GlgP proteins
The reaction products liberated with the MalQ enzyme reaction from maltose
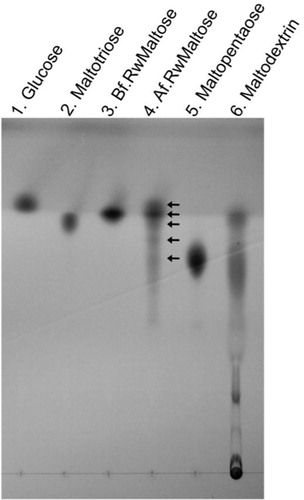
The MalQ enzyme assay to examine the reaction products released from maltose was performed as described above (Table and 3 ) except for the extended reaction time (100 min). Thin-layer chromatography (TLC) indicated that maltooligosaccharides including maltotriose, maltotetraose, maltopentaose and higher molecular oligomers as well as glucose were generated in the reactions (, lane 4). This suggests that the MalQ protein does not encode α-1,4-glucosidase activity but is a 4-α-glucanotransferase.
Fig. 4 Thin-layer chromatography (TLC) analysis. An aliquot (2.5 µl) of the reaction mixture incubated for 100 min containing products from the activity of the purified MalQ enzyme was spotted at lane 4 next to the control (lane 3) spotted with a 1 µl aliquot of enzyme reaction mixture before incubation. Lanes: 1, 0.5 µl of 1% glucose; 2, 1 µl of 1% maltotriose; 5, 1 µl of 1% maltopentaose; 6, 1 µl of 1% maltodextrin. Arrows indicate glucose, maltose, maltotriose, maltotetraose, and maltopentaose. Abbreviations: Bf.RwMaltose, before reaction with maltose; Af.RwMaltose, after reaction with maltose.
Discussion
4-α-Glucanotransferase was first reported in potato tubers in the 1950s as a disproportionating enzyme with distinct activity from the glycogen debranching enzyme, and genes encoding this enzyme activity were characterized later among Streptococcus pneumoniae (malM) (Citation18), Clostridium butyricum (malQ) (Citation17), and E. coli (malQ) (Citation19) as well as potato (malQ) (Citation20). 4-α-Glucanotransferase in the above organisms catalyzes a reaction in which single or multiple glucose units from the non-reducing ends of maltooligosaccharides are transferred to the 4-hydroxyl group of acceptor sugars. Glucose and maltose act only as acceptors, whereas maltotriose is the smallest donor substrate for these enzymes (Citation17, Citation19) (Citation20). Therefore, these enzymes do not release glucose from maltose, but release it from maltodextrin or amylodextrin. In contrast, the S. mutans MalQ enzyme released glucose from maltose (). How could the S. mutans MalQ enzyme release glucose from maltose? One possibility is that this enzyme may be able to use maltose as a donor molecule. Medda reported amylomaltase (4-α-glucanotransferase) activity in partially purified preparations obtained from S. mutans strain 6715-49 (this strain is currently classified as S. sobrinus) cells grown with maltose as the main carbon source, and indicated that maltose participated as a donor as well as an acceptor for the enzyme activity (Citation21). S. mutans MalQ reaction products were not only glucose but also maltotriose and higher molecular weight maltooligosaccharides as shown in , and these products were derived initially from maltose. Therefore, maltose participated as a donor of the reaction mediated by the MalQ enzyme likely to be a homolog of the S. sobrinus 4-α-glucanotransferase enzyme.
We demonstrated the glucose-releasing activity of the MalQ protein from maltose and maltotriose. However, the physiological substrate of this protein in S. mutans cells may be maltose-6-phosphate, which is taken up into cells predominantly through IIABCmaltose (the malT product) of the phosphoenolpyruvate-dependent maltose PTS in S. mutans (Citation9). In this respect, Mokhtari et al. recently reported a novel maltose-6-phosphate phosphatase (MapP) in Enterococcus faecalis (Citation22). The mapP gene was located downstream from the enterococcal malT gene encoding a maltose-specific EIICBA of the PTS, and was previously suggested to encode an endonuclease/exonuclease/phosphatase family protein of unknown function. The S. mutans malT gene was followed by a gene locus tagged as ‘SMU_2046c’, which encoded the same family protein suggested by the conserved domain search of the BLAST program. Therefore, SMU_2046c is very likely the mapP homolog, although this remains to be confirmed. Accordingly, the MalQ enzyme of S. mutans likely utilizes maltose as a substrate.
The malQ mutant aeU1 exhibited no growth with maltose but still utilized other disaccharides including sucrose, lactose, trehalose, and melibiose as described above, which is consistent with the finding that S. mutans possesses transporter and sugar (phosphate) hydrolase protein pairs specific for these disaccharides that induce the corresponding pairs (Citation23–Citation27). In contrast, the malT gene encoding the maltose transporter (IIABC) of PTS was not proximal to the corresponding sugar phosphate hydrolase gene, which may be compatible with a malT-mapP gene arrangement in E. faecalis.
Even if such a hydrolase for maltose is absent in S. mutans cells, starch-degradation products including maltose as the smallest molecule would be catabolized in concert with the malQ, glgP, and amy (Citation28) gene products. The GlgP protein did not phosphorylize maltotriose or maltose, but phosphorylized saccharides larger than maltodextrin as a substrate (). However, this enzyme apparently catabolized maltotriose or maltose () in the presence of the MalQ protein. This did not appear to result from changes in the substrate specificities of the GlgP protein but may have resulted from the generation of maltooligosaccharides and glucose from maltose and maltotriose mediated by MalQ enzyme reactions (). The growth of the glgP mutant zKU1 was suppressed when maltose was used as the sole carbon source (b). This may be explained by the intracellular over-accumulation of maltooligosaccharides mediated by the MalQ glucanotransferase activity in the absence of GlgP phosphorylase activities.
S. mutans alone was unable to ferment starch (a) similar to other oral streptococci (Citation29). However, oral biofilm bacteria as well as tooth hydroxyapatite actively bind salivary amylase (Citation30), which degrades starch to maltose, maltotriose, and maltodextrin fermentable by S. mutans.
We now report that the purified MalQ and GlgP proteins exhibited glucose release from maltose/maltotriose and phosphorylase activity from maltodextrin/amylodextrin, respectively. In addition, the malQ gene involved in glucose-releasing activity is essential for utilizing starch-degradation products similar to the malP maltose phosphorylase gene in E. faecalis (Citation22). Therefore, the malQ gene may be a target for controlling S. mutans in oral environments.
Conflict of interest and funding
There are no conflicts of interest in the present study for any of the authors.
Acknowledgements
The authors thank H. K. Kuramitsu (State University of New York at Buffalo, NY) for critically reading this article.
References
- Burne RA. Oral streptococci … products of their environment. J Dent Res. 1998; 77: 445–52.
- Nomura R, Nakano K, Nemoto H, Mukai T, Hata H, Toda K, etal. Molecular analyses of bacterial DNA in extirpated heart valves from patients with infective endocarditis. Oral Microbiol Immunol. 2009; 24: 43–9.
- Nakano K, Hokamura K, Taniguchi N, Wada K, Kudo C, Nomura R, etal. The collagen-binding protein of Streptococcus mutans is involved in haemorrhagic stroke. Nat Commun. 2011; 2: 485.
- Kojima A, Nakano K, Wada K, Takahashi H, Katayama K, Yoneda M, etal. Infection of specific strains of Streptococcus mutans, oral bacteria, confers a risk of ulcerative colitis. Sci Rep. 2012; 2: 332.
- Hamilton IR, Reizer J, Peterkofsky A. Effects of changing environment on sugar transport and metabolism by oral bacteria. Sugar transport and metabolism in Gram-positive bacteria. 1987; Chichester: Ellis Horwood. 94–133.
- Walker GJ, Jacques NA, Reizer J, Peterkofsky A. Polysaccharides of oral streptococci. Sugar transport and metabolism in Gram-positive bacteria. 1987; Chichester: Ellis Horwood. 39–68.
- Ajdic D, McShan WM, McLaughlin RE, Savic G, Chang J, Carson MB, etal. Genome sequence of Streptococcus mutans UA159, a cariogenic dental pathogen. Proc Natl Acad Sci USA. 2002; 99: 14434–9.
- Shelburne SA 3rd, Sahasrobhajane P, Suber B, Keith DB, Davenport MT, Horstmann N, etal. Niche-specific contribution to streptococcal virulence of a MalR-regulated carbohydrate binding protein. Mol Microbiol. 2011; 81: 500–14.
- Webb AJ, Homer KA, Hosie AH. A phosphoenolpyruvate-dependent phosphotransferase system is the principal maltose transporter in Streptococcus mutans. J Bacteriol. 2007; 189: 3322–7.
- Kilic AO, Honeyman AL, Tao L. Overlapping substrate specificity for sucrose and maltose of two binding protein-dependent sugar uptake systems in Streptococcus mutans. FEMS Microbiol Lett. 2007; 266: 218–23.
- Webb AJ, Homer KA, Hosie AH. Two closely related ABC transporters in Streptococcus mutans are involved in disaccharide and/or oligosaccharide uptake. J Bacteriol. 2008; 190: 168–78.
- Nikitkova AE, Haase EM, Scannapieco FA. Effect of starch and amylase on the expression of amylase-binding protein A in Streptococcus gordonii. Mol Oral Microbiol. 2012; 27: 284–94.
- Perry D, Kuramitsu HK. Genetic transformation of Streptococcus mutans. Infect Immun. 1981; 32: 1295–7.
- Heckman KL, Pease LR. Gene splicing and mutagenesis by PCR-driven overlap extension. Nat Protoc. 2007; 2: 924–32.
- Sato Y, Yamamoto Y, Suzuki R, Kizaki H, Kuramitsu HK. Construction of scrA::lacZ gene fusions to investigate regulation of the sucrose PTS of Streptococcus mutans. FEMS Microbiol Lett. 1991; 63: 339–45.
- Sato Y, Ishikawa A, Okamoto-Shibayama K, Takada K, Hirasawa M. Four gbpC gene homologues in Streptococcus sobrinus. J Oral Biosci. 2007; 49: 303–8.
- Goda SK, Eissa O, Akhtar M, Minton NP. Molecular analysis of a Clostridium butyricum NCIMB 7423 gene encoding 4-alpha-glucanotransferase and characterization of the recombinant enzyme produced in Escherichia coli. Microbiology. 1997; 143: 3287–94.
- Espinosa M, Lopez P, Lacks SA. Transfer and expression of recombinant plasmids carrying pneumococcal mal genes in Bacillus subtilis. Gene. 1984; 28: 301–10.
- Palmer TN, Ryman BE, Whelan WJ. The action pattern of amylomaltase from Escherichia coli. Eur J Biochem. 1976; 69: 105–15.
- Takaha T, Yanase M, Okada S, Smith SM. Disproportionating enzyme (4-alpha-glucanotransferase; EC 2.4.1.25) of potato. Purification, molecular cloning, and potential role in starch metabolism. J Biol Chem. 1993; 268: 1391–6.
- Medda S, Smith EE. A radioisotope method for assays of amylomaltase and D-enzyme. Anal Biochem. 1984; 138: 354–9.
- Mokhtari A, Blancato VS, Repizo GD, Henry C, Pikis A, Bourand A, etal. Enterococcus faecalis utilizes maltose by connecting two incompatible metabolic routes via a novel maltose 6’-phosphate phosphatase (MapP). Mol Microbiol. 2013; 88: 234–53.
- Hiratsuka K, Wang B, Sato Y, Kuramitsu H. Regulation of sucrose-6-phosphate hydrolase activity in Streptococcus mutans: characterization of the scrR gene. Infect Immun. 1998; 66: 3736–43.
- Wang B, Kuramitsu HK. Control of enzyme IIscr and sucrose-6-phosphate hydrolase activities in Streptococcus mutans by transcriptional repressor ScrR binding to the cis-active determinants of the scr regulon. J Bacteriol. 2003; 185: 5791–9.
- Honeyman AL, Curtiss R 3rd.. Isolation, characterization and nucleotide sequence of the Streptococcus mutans lactose-specific enzyme II (lacE) gene of the PTS and the phospho-beta-galactosidase (lacG) gene. J Gen Microbiol. 1993; 139: 2685–94.
- Ajdic D, Pham VT. Global transcriptional analysis of Streptococcus mutans sugar transporters using microarrays. J Bacteriol. 2007; 189: 5049–59.
- Russell RR, Aduse-Opoku J, Sutcliffe IC, Tao L, Ferretti JJ. A binding protein-dependent transport system in Streptococcus mutans responsible for multiple sugar metabolism. J Biol Chem. 1992; 267: 4631–7.
- Simpson CL, Russell RR. Intracellular alpha-amylase of Streptococcus mutans. J Bacteriol. 1998; 180: 4711–7.
- Nikitkova AE, Haase EM, Scannapieco FA. Taking the starch out of oral biofilm formation: molecular basis and functional significance of salivary alpha-amylase binding to oral streptococci. Appl Environ Microbiol. 2013; 79: 416–23.
- Scannapieco FA, Torres G, Levine MJ. Salivary alpha-amylase: role in dental plaque and caries formation. Crit Rev Oral Biol Med. 1993; 4: 301–7.