Abstract
The study aimed at determining the prevalence antibiotic resistance of species – identified enterococci and Escherichia (E.) coli isolated from typical fresh Slovak cheese, bryndza. Antibiotic resistance of enterococci was determined by disk diffusion method. Of isolated enterococci, 240 were obtained from bryndza cheese. The first two decimal dilutions from 24 bryndza cheese samples purchased at supermarkets in Košice (0.1 mL) were spread on the surface of Slanetz and Bartley agar and incubated for 48±2 h at 37±1°C. Species identification of enterococci and E. coli was detected by means of matrix-assisted laser desorption/ionization time-of-flight mass spectrometry (MALDI-TOF MS) based on bacterial protein profiling. The following species of enterococci were identified by MALDI-TOF MS: Enterococcus (Ent.) faecalis (22 strains), Ent. faecium (18 strains), Ent. sacharolyticus (6 strains), Ent. gilvus (4 strains), Ent. durans (9 strains), and Ent. casseliflavus (6 strains). All of the 45 E. coli strains and 74 strains of enterococci identified by MALDI-TOF MS were determined for occurrence of blaTEM, blaSHV and blaCTX-M genes. The results of our study suggest that the highest resistance of enterococci was on tetracycline (29.73%) and any resistance was recorded on vancomycin (0%). The highest multidrug-resistance was recorded on two antibiotics (32.43%). Neither one isolate of enterococci was resistant to all 6 antibiotics used in the experiment. In total, 19 (42.22%) E. coli were found to be producers of extended-spectrum β-lactamase.
Introduction
Enterococci and Escherichia (E.) coli are ubiquitous microorganisms and belong to the part of the normal gastrointestinal microbiota of mammals and other warm - blooded animals, as well as in soil, plants and water. By intestinal or environmental contamination, these microorganisms colonize raw food such as milk and meat, throughout any fermentation process (Ribeiro et al., Citation2011). As enterococci in food are not always due to faecal contamination, the legislation in force (European Commission, Citation2007) sets no limit for enterococcal presence in food. In fact in some kinds of food such as cheeses and fermented meats, enterococci are added during the production process, both to extend their shelf life and to improve their organoleptic properties (Cocolin et al., Citation2007). One of the most severe problems in human and veterinary medicine is microbial resistance to antibiotics. The main risk factor for the increase in the antibiotic resistance is an extensive use of antibiotic. High level of resistance is considered to be a good indicator for selection pressure by antibiotic use.
Monitoring the prevalence of resistance in indicator bacteria such as faecal E. coli and enterococci in food and different populations, animals and humans, makes it feasible to compare the prevalence of resistance and to detect transfer of resistant bacteria or resistance genes between animals and humans within food chain (Lukášová and Šustačková, Citation2003). Virulence of these bacteria is strongly enhanced by their frequent resistance to commonly used antibiotics. Antibiotic resistance, which can be both inward and acquired, makes enterococci and E. coli effective opportunists in nosocomial infections (Deshapande et al., Citation2007 Giraffa, Citation2002). Enterococci are naturally resistant to cephalosporins, low-level aminoglycozides, lincomycin, clindamycin, and often quinolones, and can acquire resistance to macrolides, tetracyclines, chloramphenicol and ampicilin (Barbosa et al., Citation2009). Over the last few years, enterococci resistance to β-lactams, glycopeptides, and aminoglycozides as well as to linezolid (Scheetz et al., Citation2008) has been increasing. E. coli is the most important microorganism associated with extended-spectrum beta-lactamases (ESBL)-mediated resistance. ESBLs have been classified into types, based on amino-acid sequences (i.e., the TEM, SHV, and CTX-M, PER, VEB, GES, TLA, BES and OXA types). TEM and SHV derivates have been the most prevalent types of ESBL, but the prevalence of CTX-M type has increased in most part of the world, including Europe (Romero et al., Citation2005).
Bryndza is a typical Slovak cheese made from raw milk with no special starter culture. The prevalence of antibiotic resistance raises the threat of the cheese ecosystem as a potential reservoir of resistance genes exchanges between bacteria.
The goal of this study was to determine the phenotypic antibiotic resistance of Enterococcus spp. and E. coli strains isolated and identified from retail bryndza cheese. In isolates of Enterococcus spp. and E. coli also the occurrence of blaTEM, blaSHV and blaCTX-M genes by PCR method was detected.
Materials and methods
Bryndza cheese sampling and isolation of enterococci and Escherichia coli
In order to research Enterococcus spp. and E. coli 24 samples of bryndza cheese purchased at supermarkets in Košice were analysed. All samples were obtained between May and October 2013. A 10 g portion of each sample was aseptically taken, added in 90 mL of sterile peptone/saline solution and homogenized trough a Stomacher for 2 min. The tenfold decimal dilution was prepared according to STN EN ISO 6887-3/03 (ISO, Citation2003).
The first two decimal dilutions were spread in an amount of 0.1 mL on the surface of Slanetz and Bartley Agar (HiMedia Laboratories, Mumbai, India) and Endo Agar (Thermo Scientific-Oxoid, Basingstoke, UK) in duplicate. The plates were incubated for 48±2 h at 37±1°C. Ten of the suspected colonies (pink or dark red, with a narrow whitish border) from Slanetz and Bartley Agar (n=240) and ten pink to rose red with metallic sheen of suspected colonies from Endo agar (n=240) were randomly selected and restreaked twice for purification and submitted for species identification.
Species identification of Enterooccus spp. and Escherichia coli
The species identification of enterococci and E. coli was subsequently provided with help of matrix-assisted laser desorption/ionization (MALDI) Biotyper. Matrix-assisted laser desorption/ionization time-of-flight mass spectrometry (MALDI-TOF MS) analysis was performed on a Microflex MALDI Biotyper (Bruker Daltonics, Fremont, CA, USA) according to a standard sample preparation protocol of Bruker Daltonics. Matrix-assisted laser desorption/ionization time-of-flight mass spectra were subjected to numerical analysis (BioTyper 3.1 software; Bruker Daltonics). The similarity between the MALDI-TOF mass spectra of the isolates and the reference MALDI-TOF mass spectra was indicated by BioTyper scoring, where the score value exceeding 2.000 indicated identification of the genus and probable identification of the species, and score value exceeding 2.300 indicated highly probable identification of the species. The MALDI-TOF mass spectra-based dendrogram was generated using the correlation distance measure with the average linkage algorithm (MALDI-TOF MS Biotyper; Bruker Daltonics).
Phenotypic and genotypic assessment of antibiotic resistance
All 74 strains of enterococci isolates and 45 strains of E. coli were tested by the standard disk diffusion method on Müeller-Hinton Agar (HiMedia Laboratories, India) incubated at 35±1°C for 18±2 h. The following antibiotic disks (all from Thermo Scientific-Oxoid) were used: ampicillin – 10 µg, chloramphenicol – 30 µg, erythromycin – 15 µg, penicillin – 10 µg, tetracycline – 30 µg, vancomycin – 30 µg. Inhibition zones were interpreted following the Clinical and Laboratory standards Institute guidelines (CLSI, Citation2011). The isolates with identical antibiotic resistance patterns isolated from the same sample were considered as the similar strain.
The total genomic DNA was isolated from all identified enterococci and E. coli strains and further tested by polymerase chain reaction (PCR) identification analysis for the detection of resistance genes: blaTEM, blaSHV and blaCTX-M. For amplification of blaTEM gene (516 bp) the following primers were used: forward: TEM-A (5 CCCCGAAGAACGTTTTC 3), reverse: TEM-B (5 ATCAGCAATAAACCAGC 3) (Mabilat and Courvalin, Citation1990). The reaction mixture in a volume of 25 µL contained 1 µL genomic DNA, 50 pmol.L-1 primer, 1.25 U Taq DNA polymerase, 50 mmol/L KCl, 30 mmol/L Tris-HCl pH 8.3, 0.1% Igepal CA360 and 0.2 mmol/L from each dNTP. The PCR protocol was as follows: initial denaturation at 95°C for 2 min, 35 cycles consisting of denaturation at 95°C for 1 min, annealing at 49°C for 1 min, extension at 72°C for 1 min and final extension 72°C for 7 min followed the last cycle.
Amplification of blaSHV gene (475 bp) was done by the primers of SHV-A (5 TCAGCGAAAAACACCTTG 3) (forward) and SHV-B (5 TCCCGCAGATAAATCACCA 3) as reverse, according to M’Zali et al. (Citation1997). The reaction mixture in a volume of 25 µL contained 1 µL genomic DNA, 30 pmol/L primer, 1.25 U Taq DNA polymerase, 50 mmol/L KCl, 30 mmol/L Tris-HCl pH 8.3, 0.1% Igepal CA360 and 0.2 mmol/L from each dNTP. The PCR protocol was as follows: initial denaturation at 95°C for 3 min, 35 cycles consisting of denaturation at 95°C for 1min, annealing at 53°C for 1 min, extension at 72°C for 1 min and final extension 72°C for 7 min followed the last cycle.
The forward primer CTX-M (5 TCAGCGAAAAACACCTTG 3), and reverse primer CTX-M (5 GATATCGTTGGTGGTGCCAT 3) were used for amplification of blaCTX-M gene (543 bp) (Edelstein et al., Citation2003). The reaction mixture in a volume of 25 µL contained 2.5 µL genomic DNA, 0,4 µmol/L primer, 1.25 U Taq DNA polymerase, 50 mmol/L KCl, 10 mmol/L Tris-HCl pH 9, 0.1% TritonxX-100, 200 µL dNTP. The PCR protocol was as follows: initial denaturation at 94°C for 2 min, 35 cycles consisting of denaturation at 94°C for 20 s, annealing at 51°C for 30 s, extension at 72°C for 30 s, and final extension 72°C for 3 min followed the last cycle. Polymerase chain reaction products were separated in 1.5% agarose gel stained with Goldview™ Nucleic acid stain (Beijing SBS Genetech Co. LTD, Beijing, China) and visualized with ethidium bromide staining under UV light.
Results and discussion
Contamination by Enterococcus spp. and Escherichia coli
It is generally accepted opinion that the food chain has been recognized as one of the main passage for the transfer of antibiotic resistant bacteria between the human and animal population. Enterococci can be present in many different kinds of ingredients for their high resistance and multiplication capability (Pesavento et al., Citation2014). The aim of this study was focused on enterococci and E. coli isolated from bryndza cheese – traditional Slovak sheep cheese with relatively short period of ripening, 8 days, and different ways of processing throughout the year (Jurkovič et al., Citation2006). The presence of enterococci in bryndza cheese samples ranged between 107 and 108 CFU/g. The concentration of enterococci in bryndza cheese is much higher than in Mediterranean-type cheese curds (104 to 106 CFU/g), and fully ripened cheeses (105 to 107 CFU/g) (Franz et al., Citation1999). According to Lauková et al. (Citation2004) due to the fact that, this count is higher, it doesn’t express low quality for product, because enterococci belong also to the technological microflora especially in cheese making. On the other hand, their higher count can be due to the technological sanitary aspect by process of bryndza production from unpasteurized milk as their source, respectively. In this study, 240 strains were isolated and identified by means of MALDI Biotyper. Enterococcus spp. was confirmed in 74 isolates from bryndza cheese samples. Six species were identified among 74 isolates of enterococci as follows: Enterococcus faecalis, Enterococcus faecium, Enterococcus durans, Enterococcus gilvus, Enterococcus sacharolyticus, and Enterococcus casseliflavus. Enterococcus faecalis and Enterococcus faecium were found as dominating species in all samples of bryndza cheese (). The origin of enterococci from bryndza cheese samples isolated in our study is from the environment of ewe’s farm and processing plant. It is because during the technological process any enterococci are not used as starter culture.
Table 1. Identification of enterococci from bryndza cheese by means of matrix-assisted laser desorption/ionization biotyper.
Escherichia coli in bryndza cheese samples ranged from 101 to 103 CFU/g. Little et al. (Citation2008) reported raw or thermized milk cheeses that were of unsatisfactory quality due to levels of E. coli at ≥105 CFU/g. On the other hand, Holko et al. (Citation2006) showed that the strains of E. coli isolated from unpasteurized sheep milk in Slovakia of the same pathotype are genetically similar and carry the same virulence determinants which are ideal targets for the determination of the pathogenic potential of any given E. coli isolate. Out of 240 suspected colonies of E. coli only 45 strains of E. coli were identified () from the same bryndza cheese samples.
Table 2. Identifications of Escherichia coli from bryndza cheese by means of matrix-assisted laser desorption/ionization biotyper.
Antibiotic resistance
A number of studies have attempted to compare the resistance spectra of different enterococci according to their human, animal or food origins. Although antibiotic resistant enterococci are isolated from foods, only few are resistant to the clinically important antibiotics (Ogier and Serror, Citation2008). ESBL-producing microorganisms are among the most problematic multiresistant factor worldwide and are being isolated with increased frequency (Bouchillon et al., Citation2004; Husičková et al., Citation2011). According to EFSA (Citation2011), the commensal bacteria present in animal intestine are considered a potential reservoir of resistance genes that can be horizontally transferred to other bacteria through food chain. reports the resistance profiles to 6 antibiotics for the enterococci and E. coli isolated from bryndza cheese. It was observed, that in enterococci, the highest percentage of resistance was to tetracycline (29.73%) and erythromycin (27.02%). The zero resistance to vancomycin was detected in our investigation. In a study of European cheeses, Teuber et al. (Citation1999) also described a low incidence of vancomycin-resistant enterococci. Vancomycin and teicoplanin seemed to be the most effective anti-microbial antibiotics, with a percentage of susceptible strains of about 90% (Mannu et al., Citation2003). No strains tested were resistant to all six antibiotics used in this study, while multidrug resistance to two or more antibiotics was observed (). Our results also showed that 14.87% () of the enterococci were resistant to the β-lactams ampicillin and penicillin. Enterococci are intrinsically more resistant to ampicillin and penicillin than to other streptococci. Some early studies suggested that higher levels of ampicillin resistance in enterococci were achieved by increasing levels of penicillin-binding protein 5 (PBP5) expressions (Ramos et al., Citation2009). Commonly, mutations that are presumed to lower the affinity for β-lactam antibiotics have been identified within pbp5 genes of highly resistant clinical isolates (Rice et al., Citation2004). The genes blaTEM, blaSHV and blaCTX-M were not detected in enterococci from our experiment. This fact suggests that entrococcal resistance to β-lactams is not related to genes blaTEM, blaSHV and blaCTX-M. It was observed in our study, that the highest resistance of E. coli was on β-lactams (68.88%) antibiotics (). Multidrug resistance of E. coli was detected on two antibiotics. Any of E. coli strains isolated from bryndza cheese was not resistant on five and six antibiotics used in this study ().
Table 3. Isolated strains of Enterococcus spp. resistant to antibiotics used in this study.
Table 4. Multidrug-resistance of Enterococcus spp. and Escherichia coli.
Data analysed in this study revealed a 42.22% prevalence of ESBL-positive E. coli isolates from the bryndza cheese samples. Genetic analysis performed in 45 E. coli isolates did not revealed the presence of the blaCTX-M gene encoding CTX-M broad-spectrum β-lactamases. The blaTEM and blaSHV genes were detected in 15 and 4 isolates, respectively (, and ). It is generally known, that production of broad-spectrum β-lactamases has a significant clinical impact, because resistance due to the production of these enzymes may result in failure of antibiotic therapy thus resulting in higher morbidity and mortality raise not only in human but also in veterinary medicine (D’Andrea et al., Citation2013; Doi et al., Citation2013).
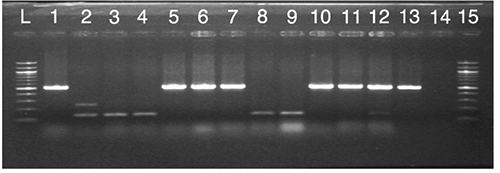
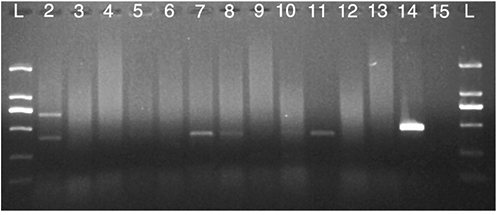
Table 5. Frequency of Escherichia coli strains resistant to antimicrobial determinants.
Conclusions
The results of present study confirmed that enterococci and E. coli belong to the common contaminants of bryndza cheese, and their antibiotic resistance can have a significant clinical impact if they are present in the final product. The presence of enterococci with significant antibiotic resistance that can be transmitted to humans after food consumption makes it necessary an increased microbiological examination of food of animal origin. The relationship between enterococci and E. coli contamination of the retail bryndza cheese tested suggests that the resistance determinant in entrococci should not be acquired through exchange of genetic material from β-lactam-antibiotic resistant E. coli also isolated from the same bryndza cheese samples.
Acknowledgments
The authors wish to acknowledge the financial support obtained from the grant KEGA 011 UVLF-4/2012 and KEGA 005UVLF-4/2015.
References
- BarbosaJ. FerreiraV. TeixeiraP., 2009. Antibiotic susceptibility of enterococci isolated from traditional fermented meat products. Food Microbiol. 26:527–532.
- BouchillonS.K. JonsonB.M. HobanD.J. JohnsonJ.L. DowzickyM.J. WuD.H. VisalliM.A. BradfordP.A., 2004. Determining incidence of extended spectrum beta-lactamase producing Enterobacteriaceae, vancomycin-resistant Enterococcus faecium and methicillin-resistant Staphylococcus aureus in 38 centres from 17 countries: the PEARLS Study 2001-2002. Int. J. Antimicrob. Ag. 24:119–124.
- CLSI, 2011. Performance standards for antimicrobial susceptibility testing, 21st informational supplement M100-S21. Clinical & Laboratory Standards Institute, Wayne, PA, USA.
- CocolinL. FoschinoR. ComiG. Grazia FortinaM., 2007. Description of the bacteriocins produced by two strains of Enterococcus faecium isolated from Italian goat milk. Food Microbiol. 24:752–758.
- D’AndreaM.M. ArenaF. PallecchiL. RossoliniG.M., 2013. CTX-M-typeβ-lactamases: a successful story of antibiotic resistance. Int. J. Med Microbiol. 303:305–317.
- DeshpandeL.M. FritscheT.R. MoetG.J. BiedenbachD.J. JonesR.N., 2007. Antimicrobial resistance and molecular epidemiology of vancomycin-resistant enterococci from North America and Europe: a report from the SENTRY antimicrobial surveillance program. Diagn. Micr. Infec. Dis. 58:163–170.
- DoiY. ParkY.S. RiveraJ.I. Adams-HaduchJ.M. HingweA. SordilloE.M. LewisJ.S.2nd HowardW.J. JohnsonL.E. PolskyB. JorgensenJ.H. RichterS.S. ShuttK.A. PatersonD.L., 2013. Community-associated extended-spectrum β-lacta-mase producing Escherichia coli infection in the United States. Clin. Infect. Dis. 56:641–648.
- EdelsteinM. PimkinM. PalaginI. EdelsteinI. StratchounskiL, 2003. Prevalence and molecular epidemiology of CTX-M extended-spectrum β-lactamase-producing Escherichia coli and Klebsiella pneumoniae in Russian hospitals. Antimicrob. Agents Ch. 47:3724–3732.
- EFSA, 2011. The European Union Summary Report on antimicrobial resistance in zoonotic and indicator bacteria from humans, animals and food in the European Union in 2009. EFSA J. 9:2154.
- European Commission, 2007. Commission Regulation of 5 December 2007 amending Regulation (EC) No 2073/2005 on microbiological criteria for foodstuffs, 1441/2007/EC. In: Official Journal, L 322/12, 07-12-2007.
- FranzC.M.A.P. HolzapfelW.H. StilesM.E., 1999. Enterococci at the crossroads of food safety? Int. J. Food Microbiol. 47:1–24.
- GiraffaG., 2002. Enterococci from foods. FEMS Microbiol. Rev. 26:163–171.
- HolkoI. BisovaT HolkovaZ. KmetV., 2006. Virulence markers of Escherichia coli strains isolated from traditional cheese made from unpasteurised sheep milk in Slovakia. Food Control 17:393–396.
- HusičkováV. ChromáM. KolářM. HricováK. StosováT KantorL DubravaL, 2011. Analysis of ESBL- and AmpC-positive Enterobacteriaceae at the Department of Neonatology, University Hospital Olomouc. Curr. Microbiol. 62:1664–1670.
- ISO, 2003. Microbiology of food and animal feeding stuffs. Preparation of test samples, initial suspension and decimal dilutions for microbiological examination. Part 3: Specific rules for the preparation of fish and fishery products. ISO norm EN 6887-3/03. International Organization for the Standardization, Geneva, Switzerland.
- JurkovičD. KrižkováL DušinskýR. BelicováA. SojkaM. KrajcovicJ. EbringerL, 2006. Identification and characterization of enterococci from bryndza cheese. Lett. Appl. Microbiol. 42:553–559.
- LaukováA. MarcinčákováM. StromfováV., 2004. Použitie metodiky PCR na detegovanie kmeñov Enterococcus faecium, izolátov z plnotucnej zimnej bryndze a zo sudového ovcieho syra. pp 251–252 in Proc 25th Conf. on Food Safety, Štrbské Pleso, Slovak Republic.
- LittleC.L. RhoadesJ.R. SagooS.K. HarrisJ. GreenwoodM. MithaniV. GrantK. McLauchlinJ., 2008. Microbiological quality of retail cheese made from raw, thermized or pasteurized milk in the UK. Food Microbiol. 25:304–312.
- LukášováJ. ŠustačkováA., 2003. Review article enterococci and antibiotic resistance. Acta Vet. Brno 72:315–323.
- MabilatC CourvalinP., 1990. Development of oligotyping for characterization and molecular epidemiology of TEM β-Lactamases in members of the family Enterobacteriaceae. Antimicrob. Agents Ch. 34:2210–2216.
- MannuL PabaA. DagaE. ComunianR. ZanettiS. DuprèI. SechiLA, 2003. Comparison of the incidence of virulence determinants and antibiotic resistance between Enterococcus faecium strains of dairy animal and clinical origin. Int. J. Food Microbiol. 88:291–304.
- M’ZaliF.H. HeritageJ. Gascoyne-BinziD.M. DentonM. ToddN.J. HawkeyP.M., 1997. Transcontinental importation into the UK of Escherichia coli expressing a plasmid-mediated AmpC - type β-lactamase exposed during an outbreak of SHV-5 extended - spectrum β-lactamase in a Leeds hospital. J. Antimicrob. Chemoth. 40:823–831.
- OgierJ.C. SerrorP., 2008. Safety assessment of dairy microorganisms: the Enterococcus genus. Int. J. Food Microbiol. 126:291–301.
- PesaventoG. CalonicoC. DucciB. MagnaniniA. Lo NostroA., 2014. Prevalence and antibiotic resistance of Enterococcus spp. isolated from retail cheese, ready-to-eat salads, ham, and raw meat. Food Microbiol. 41:1–7.
- RamosM.M.B. Gaetti-JardimE.C. Gaetti-JardimE.Junior, 2009. Resistance to tetracycline and β-lactams and distribution of resistance markers in enteric microorganisms and pseudomonads isolated from the oral cavity. J. Appl. Oral Sci. 17:13–18.
- RibeiroT. OliveiraM. FraquezaM.J. LaukováA. EliasM. TenreiroR. BarretoA.S. Semedo-LemsaddekT., 2011. Antibiotic resistance and virulence factors among enterococci isolated from Chouriço, a traditional Portuguese dry fermented sausage. J. Food Protect 74:465–469.
- RiceL.B. BellaisS. CariasL.L. Hutton-ThomasR. BonomoR.A. CaspersP. PageM.G. GutmannL., 2004. Impact of specific pbp5 mutations on expression of β-lactam resistance in Enterococcus faecium. Antimicrob. Agents Ch. 48:3028–3032.
- RomeroL. LópezL. Rodríguez-BañoJ. Ramón HernándezJ. Martínez-MartínezL. PascualA., 2005. Long-term study of the frequency of Escherichia coli and Klebsiella pneumoniae isolates producing extended-spectrum β-lactamases. Clin. Microbiol. Infec. 11:625–631.
- ScheetzM.H. KnechtelS.A. MalczynskiM. PostelnickM.J. QiC., 2008. Increasing incidence of linezolid-intermediate or - resistant, vancomycin-resistant Enterococcus faecium strains parallels increasing linezolid consumption. Antimicrob. Agents Ch. 6:2256–2259.
- TeuberM. MeileL. SchwarzF., 1999. Acquired antibiotic resistance in lactic acid bacteria from food. Anton. Leeuw. Int. J. G. 76:115–137.
