Abstract
Aim: To investigate antimicrobial resistance mechanisms in a cluster of colistin-resistant Klebsiella pneumoniae. Methods: Antimicrobial susceptibility was tested by disk diffusion and broth microdilution. Whole-genome sequencing and genome analysis were performed. Results: The eight colistin-resistant K. pneumoniae isolates belonged to three different clones (ST11, 14 and 231). The eptA and arnT genes from lipid modification pathway had novel (R157S in arnT and Q319R in eptA) and rare mutations (V39L, R152H, S260L and A279G in eptA). Several substitutions were also identified in mgrB, pmrB, phoP and phoQ genes. The mcr genes were absent in all isolates. Isolates had variants from existing classes of fosA gene. Conclusion: Complex combination of mutations might have led to colistin resistance, which suggests that continuous surveillance of molecular mechanisms is required.
Lay abstract
This study focused on identifying antimicrobial resistant mechanisms behind colistin resistance in clinical Klebsiella pneumoniae isolates. Novel and rare mutations were identified in the genes involved in the mechanism of colistin resistance. In addition, novel variants of fosfomycin resistance genes were identified in the study isolates. These findings provide an argument for continuous surveillance of colistin resistance.
Klebsiella pneumoniae is an important nosocomial pathogen with increasing multi drug resistance capability [Citation1]. At present, there is a limited selection of treatment options for carbapenem-resistant Enterobacteriaceae (CRE) infections. There is now a renewed interest in old antimicrobial agents such as polymyxins and fosfomycin.
Data on the activity of fosfomycin against K. pneumoniae carbapenemase (KPC)-producing K. pneumoniae and New Delhi metallo-β-lactamase (NDM)-1-producing Enterobacteriaceae are limited. Use of intravenous fosfomycin monotherapy was proven to effectively control K. pneumoniae bacteraemia but can be limited due to its potential resistance development on treatment [Citation2]. Colistin and polymyxin B are known as the most active antimicrobials against CRE [Citation3]. However, in the past few years, there have been sporadic reports of colistin-resistant, CRE cases from various parts of the world including Greece, Israel, South Korea, Singapore and the USA. The exact mechanism(s) of colistin resistance in Enterobacteriaceae remain to be unveiled.
Resistance to colistin is mediated mainly via alteration in the lipopolysaccharides of bacterial outer membrane. The alterations include mutations in lipid A modifying genes. The most commonly reported mutations were in the mgrB gene and therefore were not transferable through horizontal gene transfer [Citation4]. However, in 2015, the first plasmid-mediated colistin resistance gene (mcr-1) was reported [Citation5], which belongs to the phosphoethanolamine transferase enzyme family (Ept A). The mcr-1 was identified in Escherichia coli from human patients and animals in China. In 2016, another study reported the mobilizable colistin resistance gene, mcr-2 from porcine and bovine E. coli isolates in Belgium [Citation6].
Various fosfomycin-modifying enzymes have been identified that act by inactivating the drug. FosA, FosB and FosX are the commonly reported metalloenzymes, while FomA and FomB are kinases. FosA was initially found from a plasmid in Serratia marcescens associated with TN2921 transposon [Citation7], while other related FosA type enzymes being reported are FosA3, FosA4, FosA5 and FosC2 [Citation8].
In this study, we performed whole-genome shotgun sequencing of a cluster of colistin-resistant K. pneumoniae isolates from North India to identify the molecular mechanism.
Materials & methods
Isolates studied
A cluster of eight K. pneumoniae isolates from clinical samples (blood, bronchoalveolar lavage and urine) resistant to colistin were chosen for complete molecular characterization, using PCR and next-generation sequencing.
Antimicrobial susceptibility testing
Disc diffusion
All eight isolates were screened for antimicrobial susceptibility by Kirby–Bauer method using amikacin (30 μg), chloramphenicol (30 μg), tetracycline (30 μg), gentamicin (10 μg), ciprofloxacin (5 μg), cefotaxime (30 μg), cefoxitin (30 μg), ceftazidime (30 μg), cefpodoxime (10 μg), piperacilllin-tazobactam (100/10 μg), cefoperazone-sulbactam (75/30), netilmicin (30 μg), imipenem (10 μg), meropenem (10 μg) and tigecycline (15 μg), according to guidelines suggested by CLSI M100-S25, 2015. Quality control strains used were E. coli ATCC 25922 for all antibiotics concurrently in all the batches. Tigecycline results were interpreted according to the US FDA criteria.
MIC testing
MIC values were determined for meropenem and colistin by broth microdilution method. E-test was performed for fosfomycin MIC using strips with glucose 6-phosphate (bioMérieux, Marcy-l'Etoile, France). E. coli ATCC 25922 and Pseudomonas aeruginosa ATCC 27853 were used as quality control strains for MIC determination of meropenem, fosfomycin and colistin with the expected ranges of 0.008–0.06 μg/ml, 0.5–2 μg/ml and 0.25–2 μg/ml for E. coli and 0.12–1 μg/ml, 2–8 μg/ml and 0.5–4 μg/ml for P. aeruginosa, respectively. The interpretive criterion provided by CLSI 2015 for susceptible, intermediate and resistant strains were ≤4, 8 and ≥16 μg/ml for meropenem, and ≤64, 128 and ≥256 for fosfomycin, respectively. As per EUCAST 2015, isolates with ≤2 and >2 μg/ml MIC were recorded as susceptible and resistant for colistin, respectively.
PCR for screening of plasmid-mediated colistin resistance genes
Isolation of total DNA was performed using QIAamp DNA mini kit as per manufacturer's instructions (Qiagen, Hilden, Germany). The amplification of colistin resistance genes mcr-1 and mcr-2 [Citation5] & [Citation6] was performed using Veriti Thermal cycler (Applied Biosystems, CA, USA).
Next-generation sequencing
Isolates were further analyzed by whole genome sequencing. Genomic DNA was extracted with QIAamp DNA mini kit (Qiagen, Hilden, Germany). Whole genome sequencing was performed using Ion Torrent (PGM) sequencer with 400-bp read chemistry (Life Technologies, CA, USA) according to manufacturer's instructions. The data were assembled de novo using AssemblerSPAdes version 5.0.0.0 embedded in Torrent suite server version 5.0.3. The sequence annotation was performed in PATRIC, the bacterial bioinformatics database and analysis resource [Citation9], Rapid Annotation using Subsystem Technology (RAST) pipeline [Citation10] and NCBI Prokaryotic Genome Automatic Annotation Pipeline. Downstream analysis was done in the Center for Genomic Epidemiology server (www.cbs.dtu.dk/services), RAST and PATRIC. The sequence data were used to perform relativeness analysis by eBURST V3, and UPGMA dendogram was generated using MEGA 7. This Whole Genome Shotgun project has been deposited at DDBJ/ENA/GenBank.
Statistical analysis
Genome coverage and other parameters were calculated using SPSS 16.0 and Microsoft Excel 2007 (IL, USA).
Results
Antimicrobial susceptibility
The resistance pattern for the colistin-resistant K. pneumoniae isolates (n = 8) were as given in . All eight isolates were resistant to cefpodoxime, cefotaxime, ceftazidime, cefoxitin, ciprofloxacin, gentamicin, amikacin, netilmicin and cefoperazone/sulbactam. Isolates except PM5186 were resistant to meropenem and all eight were resistant to colistin by broth microdilution. MICs for colistin ranged from 4–16 μg/ml. Isolate PM716 was resistant to fosfomycin while the remaining were either intermediate or susceptible ().
Table 1. Phenotypic susceptibility testing and polymerase chain reaction data of colistin-resistant Klebsiella pneumoniae.
Genome analysis
Raw read assembly of the genome data presented 105–160 contigs (≥ 500 bp). The genome coverage of these isolates were about 32x–51x. The coding sequences (CDSs) of the genomes range from 5859 to 6744, rRNAs from 11 to 14, tRNAs from 64 to 73. The annotation revealed multiple antimicrobial resistance genes ranging from 26 to 38 from ARDB database, and 75–97 from CARD database (www.patricbrc.org). Similarly, for virulence genes, the virulence factor database (VFDB) and Victors database revealed the presence of 81–106 and 177–188 genes, respectively (www.patricbrc.org) (). Whole genome sequencing of all eight isolates were deposited in Genbank/DDBJ under the accession numbers as follows: PM565 - MNPB00000000; PM1842 - MNPC00000000; PM1995 - MNPD00000000; PM138 - MNPG00000000; PM716 - MNPH00000000; PM1134 - MNPF00000000; PM5186 - MNPE00000000 and PM1168 - MNPA00000000.
Table 2. Whole genome characteristics of colistin-resistant Klebsiella pneumoniae.
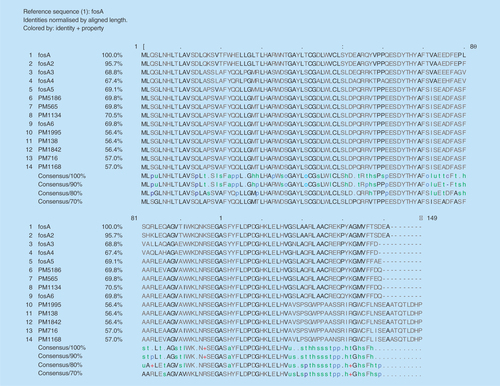
The sequence type of the isolates were found to be ST11 for PM565, PM1842, PM1995, PM138, ST14 for PM716, PM1134, and ST231 for PM5186 and PM1168 as analysed by MLST 1.8 tool (https://cge.cbs.dtu.dk//services/MLST/). ResFinder 2.1 (www.cbs.dtu.dk/services) returned multiple antimicrobial resistance genes for most of the antibiotic classes (). Interestingly fosfomycin, fluoroquinolone, aminoglycoside and β-lactam resistant determinants were found in all eight isolates. The fosA genes observed in these isolates were different from the existing six variants and reported for the first time in this study (). However, plasmid mediated colistin resistance determinants mcr-1 and mcr-2 were not found in any of the isolates.
Table 3. Antimicrobial resistance genes and plasmid profiles of colistin-resistant Klebsiella pneumoniae.
Plasmid analysis
Plasmids screening was performed using PlasmidFinder 1.3 (https://cge.cbs.dtu.dk//services/PlasmidFinder/). On analysis of plasmids using PlasmidFinder 1.3 (www.cbs.dtu.dk/services), IncFII(K) and IncFIB(pQil) were found in all isolates in addition to few other plasmids ().
Mutational analysis
Multiple mutations were observed in the genes responsible for lipid A modification and Ara-4 N pathway in K. pneumoniae isolates (). Interestingly, novel (R157S in arnT & Q319R in eptA) and rare mutations (V39L, R152H, S260L, A279G in eptA) were observed in the isolates studied. R157S in arnT was observed in all the isolates, whereas Q319R in eptA was observed in PM565, PM1995, PM138 and PM1842. It is important to note that there were deletions of three amino acids LLG at 521, 522 and 523 (). PM1168 and PM5186 had mutations in mgrB gene, V1A and L24H, respectively.
Table 4. Cumulative results of various mutations (amino acid) found upon whole genome sequencing analysis of colistin-resistant Klebsiella pneumoniae.
Discussion
Currently, there are increasing reports of CRE which results in less choice of antimicrobials for therapy. Fosfomycin is gaining interest for the treatment of carbapenem-resistant K. pneumoniae [Citation2]. In this scenario, resistance to fosfomycin is an alarming threat to those treating infections by Enterobacteriaceae, especially nosocomial pathogens.
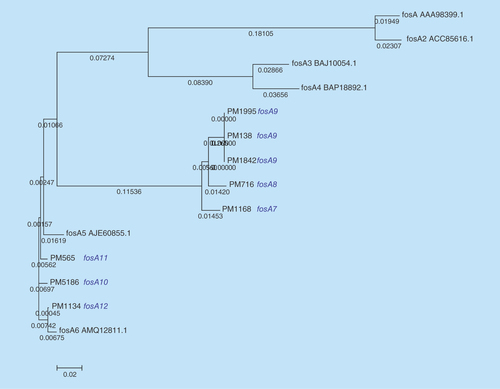
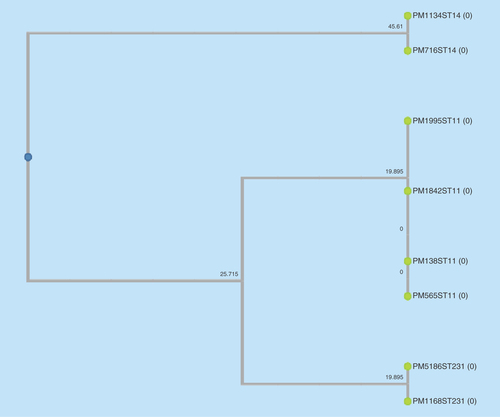
The eight selected colistin-resistant K. pneumoniae isolates were observed to be of three different clonal types (ST11, ST14 and ST231) as observed by eBURST analysis and UPGMA dendogram ( & ). These were the commonly reported sequence types previously reported from India [Citation11]. Also, among the seven meropenem-resistant isolates, bla OXA producers; bla OXA-232 (n = 5) and bla OXA181 (n = 1) were commonly seen followed by bla NDM-1 (n = 2).
Figure 3. Dendrogram of colistin-resistant Klebsiella pneumoniae isolates to show the clonal similarity using dendroUPGMA.
Polymyxins are known to serve as drug of choice for carbapenem-resistant K. pneumoniae either alone or in combination. Polymyxin acts like cationic detergents and disrupts the cytoplasmic membrane by attacking phosphate groups of membrane phospholipid; this ultimately leads to leakage of cytoplasmic contents and death of cell [Citation12]. In this regard, lipid A modification genes were largely known to be involved in chromosomal-mediated colistin resistance. Among the seven genes which are known to be involved in the lipid A modifications (paqP, pmrA, pmrB, phoP, phoQ, eptA and eptB), only four genes (pmrA, pmrB, phoP and phoQ) were extensively discussed in the literature. In K. pneumoniae, mutations including G53C, E35A in pmrA [Citation13]; S85R, T140P, T157P, S205P [Citation13], T157P [Citation14], T157P and S208N with deletion of three nucleotide at 14 and 209 in pmrB [Citation15]; L26Q in phoP [Citation13]; S174N and L384Q in phoQ [Citation15] were previously reported.
In this study, mutations were observed in ten genes (paqP, pmrB, phoP, phoQ and eptA of lipid A modifications and arnA_DH/FT, arnB, arnC, arnT and pmrJ of lipid A-Ara4N pathway) which includes novel (R157S in arnT & Q319R in eptA) and rare mutations (V39L, R152H, S260L and A279G in eptA) which might be conferring for colistin resistance. Recently, in one of our studies, we have observed novel mutations in eptA gene of lipid A modification pathway and arnT gene of lipid A-Ara4N pathway among cluster of isolates from South India [Citation4]. Also, two study isolates (PM1168 and PM5186) exhibited mutations in mgrB gene, the most commonly reported genetic determinant for colistin resistance. L24H observed in PM5186 was previously reported by Cannatelli et al. [Citation16], whereas V1A (GTG - GCC) was not previously reported. However, no major change was observed in colistin MIC levels for the isolates with and without mutation in mgrB. The role of observed mutations in colistin resistance development should be further analyzed with confirmatory tests.
It is also worth noting that there were deletions of LLG aminoacids in eptA gene in one isolate (PM1842). The arnT gene belonging to L-Ara4N moiety and eptA were known to be responsible for attachment of modified arabinose to lipid A 4′-phosphate group. This reduces bacterial susceptibility towards polymyxin and cationic antimicrobial peptides [Citation17].
Interestingly, plasmid-mediated colistin resistance genes mcr-1 and mcr-2 were not seen in these isolates. In spite of their absence, the isolates were resistant to colistin (with MICs 8 and 16 μg/ml) indicating the clinical importance of chromosomal mutations in the lipid A modification pathway.
In addition, intravenous fosfomycin had been proposed as a treatment option for systemic infections by resistant K. pneumoniae [Citation18]. However, resistance may develop to fosfomycin during treatment. Resistance to fosfomycin involves various mechanisms, majorly chromosomal-mediated and plasmid-mediated. Transferable plasmids with fosfomycin-resistant determinants result in accelerated dissemination of fosfomycin resistance. Also, fosA and fosB were reported to be responsible for plasmid-mediated resistance, whereas fosX was cited to be responsible for chromosomal-mediated resistance [Citation18]. The fosA gene encodes a glutathione S-transferase and fosB encodes an L-cysteine thiol transferase, while fosX encodes an epoxide hydrolase [Citation19]. Among these fosA seems to be widely reported in K. pneumoniae isolates [Citation20–22]. To date, six variants of fosA have been reported worldwide which includes fosA (NC_011617.1), fosA2 (ACC85616.1), fosA3 (NC_019073.1), fosA4 (WP_034169466.1), fosA5 (NC_022374.1) and fosA6 (AMQ12811.1). However, to the best of our knowledge, reports are lacking at variant level identification of fosA from India. In this study we observed clusters of K. pneumoniae isolates with novel fosA variants. The variant numbers (PM1168 – fosA7; PM716 – fosA8; PM138, PM1995, PM1842 – fosA9; PM5186 – fosA10; PM565 – fosA11 and PM1134 – fosA12) were assigned based on the phylogenetic variation of fosA genes (). The isolate with a fosA8 gene had a high MIC of 1024 μg/ml for fosfomycin. However, all other variants of fosA genes reported in this study were noted to be either susceptible or moderately susceptible to fosfomycin. In addition, eight more K. pneumoniae isolates were screened for fosA genes, where all eight were positive for the gene but phenotypically susceptible to fosfomycin. Further studies are required to understand the mechanisms behind fosfomycin resistance and the non-functional variants.
Conclusion
Overall the study reports novel and rare mutations in the arnT gene of the Ara-4 N pathway and the eptA gene of lipid A modifications. The complex combination of such mutations leads to high MIC levels for colistin. The result of the study provide an argument for continuous surveillance of the molecular mechanism behind the colistin resistance.
Future perspective
Most recently, colistin resistance is rapidly increasing among K. pneumoniae. The major mechanism reported for colistin resistance is mutations in lipid A modification genes, in which several novel mutations are being reported. Functional validation of such mutations might reveal the level of resistance with each mutation. Plasmid-mediated colistin resistance is seen predominantly in animals, while chromosomal-mediated resistance is higher in humans. It is important to better understand resistance mechanisms – either chromosomal- or plasmid-mediated – and the trend of plasmid-mediated resistance will help us to delineate transmission dynamics of animal to human spread. This information will facilitate the appropriate containment of colistin-resistant pathogen infections.
To the best of our knowledge, variants of fos A from India have not yet been characterized. The study reports novel variants of fos A genes at amino acid level from colistin-resistant Klebsiella pneumoniae. The variant numbers (PM1168 – fosA7; PM716 – fosA8; PM138, PM1995, PM1842 – fosA9; PM5186 – fosA10 and PM565 – fosA11) were assigned based on the phylogenetic variation of fosA genes.
The study also reports novel mutations in arnT gene of Ara-4 N pathway and rare mutations in eptA gene of lipid A modification pathway involved in contributing to colistin resistance.
The plasmid-mediated colistin resistance genes mcr-1 and mcr-2 were absent in all eight K. pneumoniae isolates.
The bla OXA-232 was seen in most of the isolates, conferring resistance to carbapenem.
IncFII(K) and IncFIB(pQil) plasmids were seen predominantly in all isolates.
The common sequence types observed from this study were ST-11 followed by ST-14 and ST-231.
Authors’ contributions
P Mathur and B Veeraraghavan were involved in conceptualization of the study, study design and implementation. NK Devanga Ragupathi, FY Inbanathan, S Khurana and N Bhardwaj involved in laboratory analysis and data collection. B Veeraraghavan, NK Devanga Ragupathi, FY Inbanathan, S Khurana, S Sagar and A Gupta were involved in interpretation and conclusion of the study results. B Veeraraghavan, P Mathur, NK Devanga Ragupathi and FY Inbanathan, prepared the manuscript along with inputs from S Khurana, N Bhardwaj, S Kumar, S Sagar and A Gupta. Manuscript was critically reviewed by P Mathur, B Veeraraghavan, NK Devanga Ragupathi and FY Inbanathan.
Ethical conduct of research
The authors state that they have obtained appropriate institutional review board approval or have followed the principles outlined in the Declaration of Helsinki for all human or animal experimental investigations. In addition, for investigations involving human subjects, informed consent has been obtained from the participants involved.
Financial & competing interests disclosure
The authors have no relevant affiliations or financial involvement with any organization or entity with a financial interest in or financial conflict with the subject matter or materials discussed in the manuscript. This includes employment, consultancies, honoraria, stock ownership or options, expert testimony, grants or patents received or pending, or royalties.
No writing assistance was utilized in the production of this manuscript.
References
- Halaby T, Kucukkose E, Janssen AB et al. Genomic characterization of colistin heteroresistance in Klebsiella pneumoniae during a nosocomial outbreak. Antimicrob. Agents Chemother. 60(11), 6837–6843 (2016).
- Michalopoulos AS, Livaditis JG, Gougoutas V. The revival of fosfomycin. Int. J. Infect. Dis. 15, e732–e739 (2011).
- Gales AC, Jones RN, Sader HS. Contemporary activity of colistin and polymyxin B against a worldwide collection of Gram-negative pathogens: results from the SE N TRY Antimicrobial Surveillance Program (2006–2009). J. Antimicrob. Chemother. 66, 2070–2074 (2011).
- Pragasam AK, Shankar C, Veeraraghavan B et al. Molecular mechanisms of colistin resistance in Klebsiella pneumoniae causing bacteremia from India - a first report. Front. Microbiol. 7, 2135 (2016).
- Liu YY, Wang Y, Walsh TR et al. Emergence of plasmid-mediated colistin resistance mechanism MCR-1 in animals and human beings in China: a microbiological and molecular biological study. Lancet Infect. Dis. 16(2), 161–168 (2015).
- Xavier BB, Lammens C, Ruhal R et al. Identification of a novel plasmid-mediated colistin-resistance gene, mcr-2, in Escherichia coli, Belgium, June 2016. Euro Surveill. 21(27), doi: 10.2807/1560-7917.ES.2016.21.27.30280 (2016).
- Suárez JE, Mendoza MC. Plasmid-encoded fosfomycin resistance. Antimicrob. Agents Chemother. 35, 791–795 (1991).
- Silver LL. Fosfomycin: mechanism and resistance. Cold Spring Harb. Perspect. Med. 7(2), a025262 (2017).
- Wattam AR, Abraham D, Dalay O et al. PATRIC, the bacterial bioinformatics database and analysis resource. Nucl. Acids Res. 42(D1), D581–D591 (2014).
- Overbeek R, Olson R, Pusch GD et al. The SEED and the rapid annotation of microbial genomes using subsystems technology (RAST). Nucl. Acids Res. 42(Database issue), D206–D214 (2014).
- Giske CG, Fröding I, Hasan CM et al. Diverse sequence types of Klebsiella pneumoniae contribute to the dissemination of blaNDM-1 in India, Sweden, and the United Kingdom. Antimicrob. Agents Chemother. 56(5), 2735–2738 (2012).
- Finch RG, Greenwood D, Whitley RJ, Norrby SR. Antibiotic and Chemotherapy (9th Ed). Elsevier Health Sciences, Amsterdam, The Netherlands (2010).
- Olaitan AO, Morand S, Rolain JM. Mechanisms of polymyxin resistance: acquired and intrinsic resistance in bacteria. Front. Microbiol. 5, 643 (2014).
- Jayol A, Poirel L, Brink A, Villegas MV, Yilmaz M, Nordmann P. Resistance to colistin associated with a single amino acid change in protein PmrB among Klebsiella pneumonia isolates of worldwide origin. Antimicrob. Agents Chemother. 58, 4762–4766 (2014).
- Choi MJ, Park YK, Peck KR, Ko KS. Mutant prevention concentrations of colistin used in combination with other antimicrobial agents against Acinetobacter baumannii, Klebsiella pneumoniae and Pseudomonas aeruginosa clinical isolates. Int. J. Antimicrob. Agents. 44, 475–476 (2014).
- Cannatelli A, Giani T, D'Andrea MM et al. MgrB inactivation is a common mechanism of colistin resistance in KPC-producing Klebsiella pneumoniae of clinical origin. Antimicrob. Agents Chemother. 58, 5696–5703 (2014).
- Raetz CRH, Reynolds CM, Trent MS, Bishop RE. Lipid A modification systems in gram-negative bacteria. Annual rev. biochem. 76, 295–329 (2007).
- Karageorgopoulos DE, Wang R, Yu XH, Falagas ME. Fosfomycin: evaluation of the published evidence on the emergence of antimicrobial resistance in gram negative pathogens. J. Antimicrob. Chemother. 67(2), 255–268 (2012).
- Rigsby RE, Fillgrove KL, Beihoffer LA, Armstrong RN. Fosfomycin resistance proteins: a nexus of glutathione transferases and epoxide hydrolases in a metalloenzyme superfamily. Methods Enzymol. 401, 367–379 (2005).
- Hou J, Huang X, Deng Y et al. Dissemination of the fosfomycin resistance gene fosA3 with CTX-M-Lactamase genes and rmtB carried on IncFII plasmids among Escherichia coli isolates from pets in china. Antimicrob. Agents Chemother. 56, 2135–2138 (2011).
- Xu X, Chen C, Lin D et al. The fosfomycin resistance gene fosB3 is located on a transferable, extrachromosomal circular intermediate in clinical enterococcus faecium isolates. PLoS ONE 8, e78106 (2013).
- Tseng SP, Wang SF, Kuo CY et al. Characterization of fosfomycin resistant extended-spectrum β-lactamase-producing Escherichia coli isolates from human and pig in Taiwan. PLoS ONE 10, e0135864 (2015).