Abstract
SCLIP, a microtubule-destabilizing phosphoprotein, is known to be involved in the development of the central nervous system (CNS). It has been well established that there are notable parallels between normal development and tumorigenesis, especially in glioma. However, no studies have examined the significance of SCLIP in gliomagenesis. To address this, we investigated the expression of SCLIP and its roles in the development of gliomas. Notably, we found that SCLIP was highly expressed in various grades of glioma samples, as compared with normal brain tissues. Overexpression of SCLIP dramatically stimulated tumor cell migration and invasion as well as proliferation and downregulation of SCLIP showed opposite effects, establishing an important oncogenic role for this gene. Furthermore, we revealed that STAT3 was required to maintain SCLIP stability, suggesting that overexpression of STAT3 may be a critical step to facilitate microtubule dynamics and subsequently promotes migration and invasion of glioma cells. Taken together, our findings demonstrate that SCLIP plays an important role in glioma pathology, and may represent a novel therapeutic strategy against human glioma.
Abbreviations
| SCLIP | = | SCG10 (superior cervical ganglia protein 10)-like protein Op18, Oncoprotein 18 |
| HRP | = | horseradish peroxidase |
| GBM | = | glioblastoma |
| MTT | = | 3-[4,5-dimethylthiazol-2-yl] -2,5-diphenyl-tetrazolium bromide |
| RT-PCR | = | reversed transcription polymerase chain reaction |
| IHC | = | immunohistochemical |
| BrdU | = | Bromodeoxyuridine |
| ELISA | = | enzyme-linked immunosorbent assay |
| WHO | = | World Health Organization |
| CHX | = | cycloheximide |
Introduction
Gliomas are the most common primary brain tumors in adults. They were classified as low-grade I and II and high-grade III and IV according to WHO classification scheme.Citation1 Unfortunately, low-grade glioma has an inherent tendency to progress to high-grade glioma. Despite considerable advances in our understanding of these tumors, the median survival for patients diagnosed with GBM has remained about 1–2 years.Citation2 Because of disappointing results with conventional therapies, there is a critical need for new molecular targets to significantly improve patients outcome in this incurable disease.
It is well-established that control of the subcellular cytoskeleton including microtubule dynamics may account for tumor proliferation, apoptosis, invasion and metastasis.Citation3,4 A critical property of microtubules, known as dynamic instability, involves rapid switch between periods of polymerization (growth) and depolymerization(shrinkage) at microtubule ends.Citation5 It has been demonstrated that several microtubue/tubulin-interactive proteins increase microtubule turnover in tumor cells, strongly indicating that specific targeting the expression of genes that regulates microtubule dynamics represents a promising option for tumor therapy.
Stathmin family proteins are important signal transduction molecules and regulators which play an important role in regulating the dynamics of the microtubules.Citation6 This family includes the cytosolic Op18/stathmin (Op18) and neural growth-associated protein termed SCLIP, SCG10 and RB3, which have structural and functional homologies. Although all members of this family share conserved “stathmin-like domain,” the location and different number of phosphorylation sites as well as sequence variations may be responsible for their diverse function in different cell types.Citation7 Like other members of stathmin's family phosphoprotein, SCLIP is a microtubule-destabilizing protein, acting either by sequestering free tubulin dimers or directly inducing microtubule-catastrophe.Citation8 It has been demonstated that SCLIP has critically important roles in several cellular processes, including neurite outgrowth, dendritic differentiation of Purkinje cell as well as cell cycle progression.Citation9-11
Mounting evidences support the roles for stathmin in the regulation of cell migration, division, and growth cone guidance by influencing the association of microtubule with the actin cytoskeleton.Citation12 The above studies strongly indicate that the stathmin protein family is likely to participate in many cellular processes indispensable for tumor progression such as motility and survival. Indeed, stathmin is highly expressed in numerous solid tumors such as gastric cancer, breast cancer and hepatocellular carcinoma.Citation13-15 Elevated expression of stathmin is strongly associated with tumor cells survival, migration, invasion and chemoresistance. While stathmin cancer connection has been extensively documented, very limited data exist regarding the expression levels of SCLIP in tumor tissues. Our previous study has shown that SCLIP interacted with signal transducer and activator of transcription 3(STAT3), which was constitutively activated in glioma samples and glioma cell lines as demonstrated by our recently published paper.Citation16,17 However, the biological roles of SCLIP in tumorigenesis remain largely unknown.
Here, we focused on SCLIP expression and function in gliomagenesis. Using RT-PCR,western blot and IHC analyses, we demonstrated for the first time that SCLIP was frequently overexpressed in human glioma tissues as compared to normal human brain. Functionally, we show that the genetic knockdown of SCLIP, using shRNA constructs, could inhibit glioma cell growth, migration and invasion. Consistently, SCLIP overexpression showed opposite effects. Moreover, we also demonstrated that SCLIP up-regulation could be mediated by overexpression of STAT3. Our findings provide further insight for the oncogenic role of SCLIP in mediating gliomagenesis.
Results
SCLIP was overexpressed in glioma tissues
To determine whether the SCLIP gene was up-regulated in gliomas, we performed qRT-PCR to explore SCLIP mRNA levels in a test set of 55 samples of primary glioma. SCLIP mRNA levels were significantly elevated in glioma samples in comparison with normal brain tissues (P < 0.05, ). Consistently, western blot data also showed a widespread increase in SCLIP protein level in glioma samples when compared to normal brain tissues (p < 0.05, ). To further evaluate the protein level of SCLIP in glioma samples, IHC staining was performed in 10 normal brain tissues and various grades of gliomas which include 13 grade II, 16 grade III and 26 grade IV tissue samples. In the control tissue, most normal brain cells were weakly or even negatively expressed SCLIP protein (). However, increased expression of SCLIP was frequently detected in glioma samples particular in high-grade tumors as compared with normal brain tissues (). The immunostained tissue sections were evaluated using a semiquantitative scoring method, which was based on the intensity of cytoplasmic SCLIP staining. The data are summarized in . Statistically, the difference in SCLIP protein levels between the malignant specimens and nontumor specimens was highly significant. In addition, the expression of SCLIP was also examined in 5 primary glioblastoma cells, C6 and primary normal rat astrocyte. Moderate to high levels of SCLIP were detected in primary glioblastoma cells and C6 glioma cells compared with normal rat astrocytes with no detectable SCLIP expression (Fig.S1).
Table 1. Expression of SCLIP in gliomas
Figure 1. SCLIP levels were increased in glioma samples compared with normal brain tissues. (A)and B). RT-PCR was used to determine the mRNA levels of SCLIP in normal brain tissues (NB) and glioblastomas(Grade IV), anaplastic astrocytoma(Grade III), anaplastic ependymoma(Grade III), diffuse astrocytoma(Grade II) and oligodendroglioma(Grade II). β-actin was used as a loading control. #P < 0.05. low-grade: Grade II; high-grade: Grade III and IV. (C)and D) Western blots were performed to assess the protein levels of SCLIP in normal brain tissues, high-grade and low-grade gliomas. #P < 0.05.
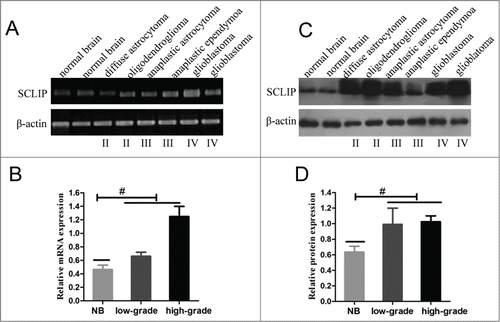
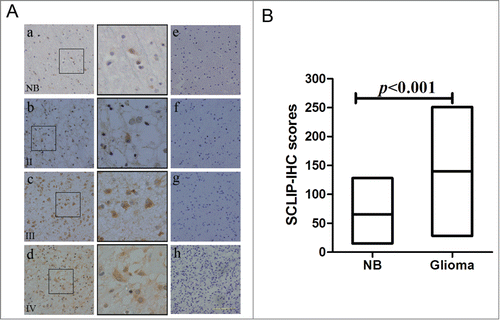
Figure 2. Immunostaining of normal brain and glioma sections with SCLIP antibody. (A) Immunohischemistry showed that SCLIP was weakly localized in most human normal brain cells (a, NB). SCLIP expression was evidently detected in primary glioma cells grades II(b) and strongly observed in glioma samples grade III (c) and IV (d). The SCLIP-positive cells were magnified to show their histological appearance (black box). Negative control of normal brain and gliomas samples (e, f, g and h) has the same incubation conditions as the tested group except that the primary antibody was omitted. Sections were counterstained with hematoxylin (blue). Scale bar = 25 μm. (B) Average number of SCLIP-positive cells was significantly higher in glioma samples (gliomas) than control normal brain tissue (NB). The expression scores of SCLIP implement expression intensity and percentages of positive cells (score=staining intensity×% positive cells).
SCLIP knockdown inhibited glioblastoma cell growth and proliferation
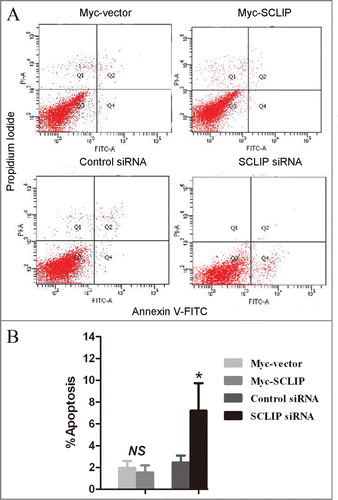
Given that the upregulated SCLIP was detected in the vast majority of glioma samples, it may possess oncogenic functions in the development of gliomas. To test the possibility, we transfected SCLIP siRNA, Myc-SCLIP as well as control plasmids into U87 and U251 glioblastoma cells (). As shown in , MTT assay showed that SCLIP knockdown significantly reduced the viability of U87 and U251 cells as compared with the control group, and overexpression of SCLIP showed opposite effect. In accordance with this, SCLIP overexpression was sufficient to induce glioma cell proliferation whereas SCLIP attenuation significantly suppressed cell proliferation (). To investigate whether SCLIP reduction affected apoptosis of U251 cells, apoptotic cells were studied by Annexin V/propidium iodide staining. Flow cytometry results showed that apoptotic cells significantly increased in U251 cells transfected with SCLIP siRNA compared with the cells transfected with control siRNA (). In contrast, there was no significant difference in the percentage of apoptotic cells between cells transfected with Myc-SCLIP and control cells (). The similar results were also obtained in U87 cells (data not shown).
Figure 3. Growth effects of SCLIP on U251 and U87 cells. (A) Expression of SCLIP in U251 and U87 cells transfected with indicated plasmids was determined by protein gel blotting analysis. (B) Cell viability in U251 and U87 cells after SCLIP knockdown and SCLIP overexpression was assessed by MTT assays. Myc-SCLIP vs. Myc-vector, #P<0.05; SCLIPsiRNA vs. Control siRNA, *P<0.05. (C) Cell proliferation was measured using bromodeoxyuridine ELISA assay. At least 3 experiments were run, each in triplicate. Myc-SCLIP vs. Myc-vector, #P<0.05; SCLIPsiRNA vs. Control siRNA, *P < 0.05.
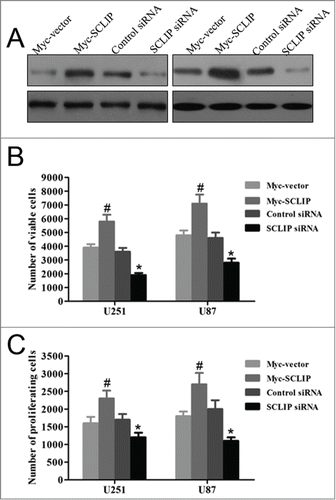
Figure 4. SCLIP knockdown enhanced apoptosis in U251 cells. (A) Apoptotic rates of U251 cells transfected with the indicated plasmids were assessed by Annexin V-FITC/propodium iodide staining. (B) Statistical analysis indicated that percentage of apoptotic cells in SCLIP siRNA group were greater than that in the Control siRNA group. *p < 0.05 vs. Control siRNA; NS: no significant difference.
SCLIP knockdown in glioblastoma cells impaired cell migration and invasion
SCLIP proteins are important players in the regulation of cell motility. Therefore, we examined whether SCLIP affected glioma cell migration and invasion. In the wound migration assay, silencing of SCLIP had a significant inhibition on the speed of wound closure in both U87 and U251 cells compared with the control siRNA, overexpression of SCLIP had an opposite role (). Similarly, SCLIP knockdown significantly reduced cell invasion compared with the control group. Conversely, SCLIP overexpressoion in glioma cells significantly promoted invasion (
Figure 5. SCLIP promotes migration of U251 and U87 cells. (A) Migration of indicated cell lines was conducted with cell wound healing assay as described in Materials and Methods. (B) Quantification of cell migration from the indicated cell lines was performed. Values are the average of triple determinations with the SD indicated by error bars. Myc-SCLIP vs. Myc-vector, #P<0.05; SCLIP siRNA vs. Control siRNA, *P <0.01.
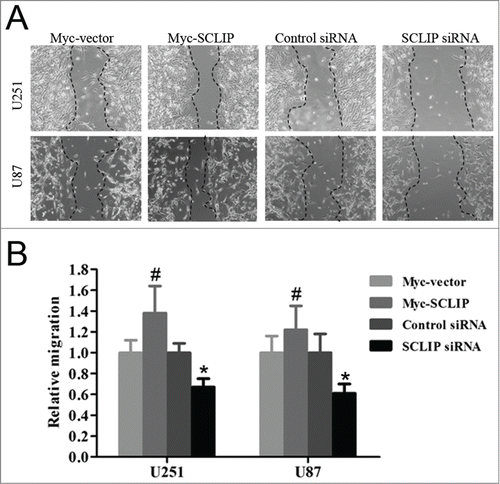
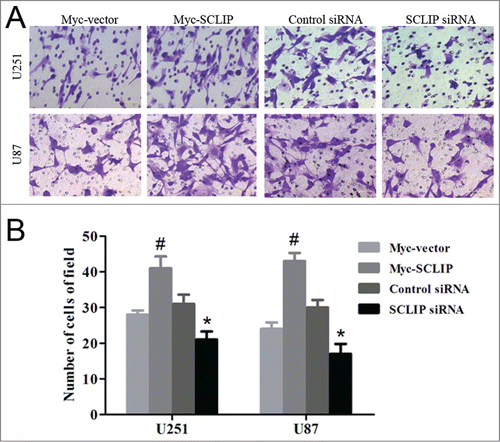
Figure 6. SCLIP induces invasion of U251 and U87 cells. (A) Images of indicated cell lines were taken from the Matrigel invasion assay. (B) Quantification of indicated invasive cells from 3 independent experiments. Myc-SCLIP vs. Myc-vector, #P<0.01; SCLIPsiRNA vs. Control siRNA, *P<0.05.
STAT3 was essential for stabilization of SCLIP protein levels
Constitutive activation of STAT3 has been found in the majority of human tumor specimens and tumor cell lines.Citation18 Consistently, we demonstrated that STAT3 protein levels were markedly upregulated in various glioma samples as compared with normal brain tissues (
Figure 7. STAT3 was involved in stabilization of SCLIP protein levels (A) Western blot showed that STAT3 expression in samples from low-grade (diffuse astrocytoma and oligodendroglioma) and high-grade (anaplastic ependymoa and glioblastoma) as well as normal brain tissue (NB). STAT3 protein level was significantly increased in various grades of gliomas compared to NB. (B) Expression of endogenous SCLIP and STAT3 was detected by immunofluorescence and co-localization was determined by merging the images. Scale bar indicates 20 μm. (C) U251 and U87 cells stably transfected with STAT3siRNA were blotted for STAT3 and SCLIP protein. β-actin was used as an internal control. (D) U251 cells were stably transfected with STAT3siRNA or control siRNA. These cells were treated with CHX(5 μg/ml), or an ethanol control for the times indicated and further blotted for SCLIP protein.
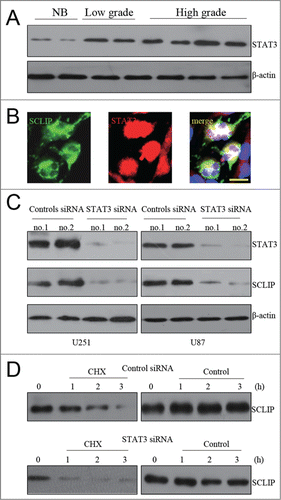
The above result strongly hinted that STAT3 may be essential for maintaining SCLIP levels in glioma cells. To test this hypothesis, U251 cells expressing control or STAT3 siRNA were pretreated with CHX to inhibit protein synthesis for measuring SCLIP protein stability. Immunoblotting analysis showed that turnover of SCLIP protein was accelerated in U251 cells expressing STAT3 siRNA as compared with control siRNA-expressing cells (
Discussion
SCLIP, one key important member of Stathmin family proteins, acts as potent regulators of microtubules stability.Citation8 Although it is well established that SCLIP plays an important role in the development of central nervous system, little information has been reported about SCLIP expression and its roles in gliomas. In normal astrocyte but not neuron, sclip was rarely detected.Citation19 Interestingly, SCLIP was expressed at extraordinarily high levels in human malignant gliomas. More importantly, SCLIP silencing significantly inhibited glioblastoma cells migration, invasion and proliferation. In addition, we provided evidence that STAT3 was involved in stabilizing SCLIP protein to maintain its levels in glioblastoma cells.
Compared with normal brain tissues, we noticed a robust increase in both SCLIP mRNA and protein level in glioma samples. No significant correlation between SCLIP expression and glioma grades was found because the sample size (n = 55) was too small to attempt reasonable statistical correlation. However, this is the first published data to demonstrate that SCLIP was up-regulated in glioma tissues,especially in high-grade samples. Consistently, strong expression of SCLIP was also observed in primary glioblastoma cells and C6 cell lines whereas its expression was not detected in normal rat astrocytes. These results indicated that SCLIP may play an important role in driving aggressive phenotypes in gliomas. Consistently, both U251 and U87 glioblastoma cell lines displayed strong SCLIP expression. Nevertheless, glioblastoma-associated neurons (with high-level SCLIP) might also make a substantial contribution to the total SCLIP levels observed in glioma patients, because a recent report has demonstrated that mature neurons also can give rise to gliomagenesis by genetic mutations.Citation20 Similar to the results described in the present study, increased expression of SCLIP was reported previously in non-small-cell lung cancer and their several cell lines.Citation21
As a functional molecule in CNS, high expression of SCLIP has been found in neurons and is involved in neuron differentiation.Citation9,22 In contrast, little is known about the function of the SCLIP gene in tumor cell biology. To unveil the functional roles of SCLIP in the carcinogenesis of gliomas, tests were carried out with up- and down-regulation of SCLIP in the U251 and U87 glioblastoma cell lines. Our study demonstrated that depletion of SCLIP by shRNA caused a modest inhibtion in cellular proliferation and cell viability and its overexpression showed opposite roles. Furthermore, we demonstrated that depletion of SCLIP in glioma cell lines significantly induced apoptosis. Conversely, overexpression of SCLIP had no such effect. Therefore, the apoptosis induced by SCLIP knockdown may be partly responsible for inhibition of cell viability and proliferation mediated by reduced SCLIP. Collectively, balance between survival and death may be mainly disrupted due to abnormal expression of SCLIP. This dysregulated balance is likely to confer a considerable survival benefit to glioma cells. This is in agreement with the role of SCLIP in other cell lines.Citation21
GBM is the most common and most malignant primary brain tumor in humans. Invasion of GBM cells is the major reason for the lack of lasting success with surgical therapy and for tumor recurrence.Citation23 Hence, defining the mechanism controlling invasion is essential to improve cancer survival. It is well documented that tumor cell migration and invasion rely on the appropriate control of the subcellular cytoskeleton including microtubule dynamics.Citation24 Given the fact that SCLIP is a microtubule destabilizing protein, we reasoned that SCLIP may function in the migration and invasion of glioblastoma cells. As expected, suppression of SCLIP significantly reduced migration and invasion of both U251 and U87 glioma cells in vitro. It has been demonstrated that Stathmin reduction showed a significant inhibitory effect in glioma cells.Citation25 All these data plus ours suggest that Stathmin family proteins may have common roles in development and progression of gliomas. Consistent with our observations, overexpression of other MT destabilizing protein, such as Stathmin, has been investigated to be associated with lymphatic invasion, lymph node metastasis and poor prognosis of human cancers.Citation26
It has been demonstrated that STAT3 was strongly expressed in gliomas, where blocking it often leads to growth arrest and/or cell death.Citation27 Consistently, our results also revealed that STAT3 was highly increased in glioma samples as compared with normal brain tissues as demonstrated by Western blot analysis. Interestingly, our previous publication has demonstrated an interaction between SCLIP and STAT3 in various cells.Citation17 Based on the above data plus overexpression of both SCLIP and STAT3 detected in gliomas, we thus speculated that the aberrant STAT3 might regulate the expression of SCLIP in glioblastoma cells. As expected, efficient inhibition of STAT3 in both cell lines reduced SCLIP expression. In addition, STAT3 silencing accelerated SCLIP protein turnover. Collectively, our data suggest that STAT3 is associated with SCLIP stabilization to maintain its levels in glioblastoma cells. Co-localization of STAT3 and SCLIP in glioblastoma cells as demonstrated by immunoinfluorescence supports the above results. While Stathmin, the other important Stahmin family member, has been regulated by different modes such as by the tumor suppressor gene P53 and small noncoding miRNA-223, the up-stream regulators of SCLIP remains poorly understood.Citation28,29 In the present study, induction of SCLIP by STAT3 may represent a novel regulatory level based on the protein stability.
In summary, our study showed that SCLIP is overexpressed in primary glioma specimens as well as glioblastoma cell lines. In addition, our findings indicate that decreased SCLIP expression inhibited glioblastoma cell growth, migration and invasion in vitro. Furthermore, we demonstrated that STAT3 is required for maintaining SCLIP stability. Altogether, these data strongly point to SCLIP as a promising target for biological therapies in tumors, at least in glioblastoma.
Material and methods
Cell lines and regents
Human glioblastoma cell lines U87, U251 and rat glioma cell line C6 were obtained from Cell Bank of Type Culture Collection of Chinese Academy of Sciences (Shanghai, China). Cells were cultured in Dulbecco's modified Eagle's medium (Life Technologies, Carlsbad,CA), 10% fetal bovine serum (HyClone, Logan, UT, USA).
Tumor samples
Glioma specimens from patients with primary gliomas and normal brain tissues from trauma patients were collected during surgical procedures at the Department of Neurosurgery, Qilu Hospital, Shandong University (Jinan, China). Collection was carried out in accordance with the National Regulation of Clinical Sampling in China.
Primary culture of GBM and rat cortical astrocyte
Primary cells were obtained from tumor samples from patients with newly diagnosed GBM operated at the Department of Neurosurgery, Qilu hospital, Shandong University, China. The resected tissues were washed twice with D-Hank's solution under aseptic conditions. After clearing out of visible blood vessel and necrotic tissue, the tissue was minced with a sterile scalpel and chemically dissociated in Papain (Worthington Biochemical Corporation). The dissociated tissue was filtered through a 70 μm cell strainer and the remaining cells were cultured routinely in Dulbecco's modified Eagle's medium, 20% fetal bovine serum.
Astrocyte cultures were prepared cerebral hemispheres of 1- or 2-day-old Sprague-Dawley rat pups. Briefly, cerebral cortices were dissected and the meninges removed. The tissue was chopped finely and incubated in calcium/magnesium-free MEM containing 0.05% trypsin and 0.1% EDTA for 30 min at 37°C. The cells were trituated using serological flame-polished pipettes. After centrifugation, the obtained pellet was resuspended in the final medium composed of DMEM, 10% FBS, 100 U/ml penicillin, and 100 μg/ml streptomycin. After reaching confluency, the flasks were shaken to remove contaminating cells. First, the flasks were preshaken for 1.5 h at 225 rpm on an orbital shaker at 37°C. After preshaking, the medium was refreshed, and shaking was then continued for 2 d with a serum-free medium. After 2 d of shaking, only the astrocytes remained in the flasks.
Flow cytometry assay
Cells were transfected with various plasmids. After 48 h after transfection, the cells were treated with annexin V and propidium iodide (Abcam, Cambridge, MA, USA). The cells were incubated for 15 min and then subjected to flow cytometry.
Semiquantitative RT-PCR analysis
Total RNA was extracted with the Qiagen RNA Easy kit in combination with QIAshredder columns according to the manufacturer's protocol (Qiagen, Valencia, CA). RNA was quantitated spectrophotometrically and 2 μg of RNA were used to make cDNA using Thermoscript RT-PCR Reaction System (Invitrogen, Carlsbad, CA) Levels of expression were measured at least 3 times independently for each gene and normalized to the expression level of β-actin for cell lines and to the averaged expression levels of β-actin and SCLIP for primary tumors and normal tissues.
The primer sequences are as follows: Human SCLIP: 5′-AGC TGC TAT TAT TGA ACG TC-3′(forward); 5′-AAC CTG GAG TTC CTT GTT CC-3′(reverese). Human β-actin: 5′-GGCATCGTGATGGACTCCG-3′ (forward). 5′-GCTGGAA-GGTGGACAGCGA-3′ (reverse)
Western blot
Proteins were separated by 10% sodium dodecyl sulfated-polyacrylamide, and transferred to a polyvinylidene difluoride membrane and probed with primary antibodies. The membranes were subsequently probed with HRP conjugated secondary antibodies followed by development using an ECL detection system (Pierce, Rockford, IL, USA). The following primary antibodies were used: anti-β-actin (Sigma-Aldrich, St Louis, Missouri, USA); anti-SCLIP (kindly provided by Professor Xinmin Cao, Signal Transduction Laboratory, Institute of Molecular and Cell Biology, Singapore); anti-STAT3 (Cell Signaling and Technologies, Boston, MA, USA); Secondary antibodies HRP-conjugated polyclonal goat anti-mouse (Jackson ImmunoResearch) and polyclonal rabbit anti-goat immunoglobulins (Dako) were also used.
Immunohistochemical analysis
Formalin–fixed paraffin-embedded tissue sections were deparaffinized, rehydrated and subjected to antigen retrieval. The sections were stained with a polyclonal antibody against human SCLIP at a dilution of 1:100 using standard procedures. Similar tissue sections immunostained with nonspecific IgG were used as negative controls. Subsequently, sections were counterstained with hematoxylin. The average number of SCLIP-positive cells was quantified in 20 fields randomly from each sample section at ×40 objective. Staining was classified using a 3-tiered system according to the percentage of positive cells and staining intensity as described previously.Citation30 The tumor samples were scored for SCLIP as follows: +, <30% of tumor cells are immunopositive; ++, 30-60% of tumor cells are immunopositive; and +++, >60% of tumor cells are immunopositive. Scoring of staining intensity was conducted in a blinded manner to prevent bias from knowledge of clinical data.
Plasmids, siRNA and generation of stably transfected cells
Transfection of plasmids and siRNAs into glioblastoma cells was done by using Lipofectamine 2000 (Invitrogen, NY, USA). The Myc–SCLIP plasmids, Psuper. neo.GFP.KD-SCLIP (SCLIP siRNA), Psuper.neo.GFP (control scrambled siRNA) were kindly provided by Professor Xinmin Cao (Signal Transduction Laboratory, Institute of Molecular and Cell Biology, Singapore). The plasmids were then transfected into U251 and U87 cells using Lipofectamin 2000 as recommended by manufacturer's protocols. Stable transfectants were selected by growing cells in 400 μg/ml G418 (Sigma, St. Louis, MO) for one week and maintained with 100 μg/ml of G418. Surviving cells were pooled and analyzed by western blot.
Cell viability assay
To determine cell viabilities and proliferative abilities, cells were initially seeded in 96-well plates. Cell viability was measured by MTT assay as previously described.Citation21 Twelve replicate samples were taken for each time point, and each experiment was repeated at least 3 times. The cell proliferation assay was utilized using BrdU (Bromodeoxyuridine) ELISA (Roche Diagnostics, Indianapolis, IN) according to the manufacturers’ instructions.
Immunofluorescence
Transfected cells were washed, fixed with methanol, and permeabilized with 0.1% TritonX-100, and blocked in 1% BSA. Cells were incubated with primary antibody overnight at 4°C, followed by secondary antibody for 1 hour at room temperature. The samples were washed again, then mounted with VectaShield plus 4′,6-diamidino-2-phenylindole (DAPI) mounting medium (Vector Laboratories). Images were acquired with a Zeiss Axioplan 2 imaging microscope and Axio Vision Release 4.6 software (Carl Zeiss Micro-Imaging Inc.).
Wound healing assay and Transwell invasion assay
Glioblastoma cells were seeded a 6-well plate and grown to confluence. Cell monolayers were then wounded by dragging a 200-μL pipette tip and rinsed with PBS. The surface area of the scratched surface was quantified after wounding and again for 24 hours on a Nikon BioStation IM (NikonInstruments, Inc.). Cells were then photographed again at 3 randomly selected sites per well.
Glioblastoma cells were seeded at a density of 1.0 × 10Citation4 per well in the upper chamber of the insert and cells were allowed to invade through the Matrigel (R&D system, USA). Medium containing 10% fetal bovine serum was added to the lower chamber as a chemoattractant. The top surface of the insert was scraped using a cotton swab. The cells on the lower surface of the membrane were stained with 0.1% crystal violet in methanol and counted under a microscope in 5 predetermined fields. All assays were independently repeated at least in triplicate.
Statistical analysis
Descriptive statistics were generated for all quantitative data with presentation of mean ± standard deviation (SD). A one-way analysis of variance (ANOVA) was used to compare mRNA and protein levels between groups. The average number of SCLIP -positive cells was compared using the unpaired Student's t-test. Two-dimensional area determination for migration assay was performed using the digital image analysis software Axiovision LE (Zeiss). The significance levels were defined as P < 0.05.
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
987037_Supplementary_Materials.zip
Download Zip (2.2 MB)Funding
This work was supported by Natural Science Foundation of Shandong province (No.ZR2012HQ010, 201203049); National Natural Science Foundation of China (No. 2012M521351, 8117244; 81372720; 81402059).
Supplemental Material
Supplemental data for this article can be accessed on the publisher's website.
References
- Figarella-Branger D, Colin C, Coulibaly B, Quilichini B, Maues De Paula A, Fernandez C, Bouvier C. [Histological and molecular classification of gliomas]. Rev Neurol (Paris) 2008; 164:505-15; PMID:18565348; http://dx.doi.org/10.1016/j.neurol.2008.03.011
- Stupp R, Hegi ME, Mason WP, van den Bent MJ, Taphoorn MJ, Janzer RC, Ludwin SK, Allgeier A, Fisher B, Belanger K, et al. Effects of radiotherapy with concomitant and adjuvant temozolomide versus radiotherapy alone on survival in glioblastoma in a randomised phase III study: 5-year analysis of the EORTC-NCIC trial. Lancet Oncol 2009; 10:459-66; PMID:19269895; http://dx.doi.org/10.1016/S1470-2045(09)70025-7
- Singh P, Rathinasamy K, Mohan R, Panda D. Microtubule assembly dynamics: an attractive target for anticancer drugs. IUBMB Life 2008; 60:368-75; PMID:18384115; http://dx.doi.org/10.1002/iub.42
- Kaverina I, Straube A. Regulation of cell migration by dynamic microtubules. Semin Cell Dev Biol 2011; 22:968-74; PMID:22001384; http://dx.doi.org/10.1016/j.semcdb.2011.09.017
- Gardner MK, Zanic M, Howard J. Microtubule catastrophe and rescue. Curr Opin Cell Biol 2013; 25:14-22; PMID:23092753; http://dx.doi.org/10.1016/j.ceb.2012.09.006
- Cassimeris L. The oncoprotein 18/stathmin family of microtubule destabilizers. Curr Opin Cell Biol 2002; 14:18-24; PMID:11792540; http://dx.doi.org/10.1016/S0955-0674(01)00289-7
- Curmi PA, Gavet O, Charbaut E, Ozon S, Lachkar-Colmerauer S, Manceau V, Siavoshian S, Maucuer A, Sobel A Stathmin and its phosphoprotein family: general properties, biochemical and functional interaction with tubulin. Cell Struct Funct 1999; 24:345-57; PMID:15216892; http://dx.doi.org/10.1247/csf.24.345
- Charbaut E, Curmi PA, Ozon S, Lachkar S, Redeker V, Sobel A. Stathmin family proteins display specific molecular and tubulin binding properties. J Biol Chem 2001; 276:16146-54; PMID:11278715; http://dx.doi.org/10.1074/jbc.M010637200
- Poulain FE, Chauvin S, Wehrle R, Desclaux M, Mallet J, Vodjdani G, Dusart I, Sobel A. SCLIP is crucial for the formation and development of the Purkinje cell dendritic arbor. J Neurosci 2008; 28:7387-98; PMID:18632943; http://dx.doi.org/10.1523/JNEUROSCI.1942-08.2008
- Mahapatra NR, Taupenot L, Courel M, Mahata SK, O'Connor DT. The trans-Golgi proteins SCLIP and SCG10 interact with chromogranin A to regulate neuroendocrine secretion. Biochemistry 2008; 47:7167-78; PMID:18549247; http://dx.doi.org/10.1021/bi7019996
- Poulain FE, Sobel A. The "SCG10-like protein" SCLIP is a novel regulator of axonal branching in hippocampal neurons, unlike SCG10. Mol Cell Neurosci 2007; 34:137-46; PMID:17145186; http://dx.doi.org/10.1016/j.mcn.2006.10.012
- Belletti B, Baldassarre G. Stathmin: a protein with many tasks. New biomarker and potential target in cancer. Expert Opin Ther Targets 2011; 15:1249-66; PMID:21978024; http://dx.doi.org/10.1517/14728222.2011.620951
- Ke B, Wu LL, Liu N, Zhang RP, Wang CL, Liang H. Overexpression of stathmin 1 is associated with poor prognosis of patients with gastric cancer. Tumour Biol 2013; 34:3137-45; PMID:23760979; http://dx.doi.org/10.1007/s13277-013-0882-0
- Baquero MT, Hanna JA, Neumeister V, Cheng H, Molinaro AM, Harris LN, Rimm DL Stathmin expression and its relationship to microtubule-associated protein tau and outcome in breast cancer. Cancer 2012; 118:4660-9; PMID:22359235; http://dx.doi.org/10.1002/cncr.27453
- Hsieh SY, Huang SF, Yu MC, Yeh TS, Chen TC, Lin YJ, et al. Stathmin1 overexpression associated with polyploidy, tumor-cell invasion, early recurrence, and poor prognosis in human hepatoma. Mol Carcinog 2010; 49:476-87; PMID:20232364
- Zhang Y, Hao H, Zhao S, Liu Q, Yuan Q, Ni S, Wang F, Liu S, Wang L, Hao A, et al. Downregulation of GRIM-19 promotes growth and migration of human glioma cells. Cancer Sci 2011; 102:1991-9; PMID:21827581; http://dx.doi.org/10.1111/j.1349-7006.2011.02059.x
- Ng DC, Lim CP, Lin BH, Zhang T, Cao X. SCG10-like protein (SCLIP) is a STAT3-interacting protein involved in maintaining epithelial morphology in MCF-7 breast cancer cells. Biochem J 2010; 425:95-105; PMID:19824884; http://dx.doi.org/10.1042/BJ20091213
- Johnston PA, Grandis JR. STAT3 signaling: anticancer strategies and challenges. Mol Interv 2011; 11:18-26; PMID:21441118; http://dx.doi.org/10.1124/mi.11.1.4
- Ozon S, El Mestikawy S, Sobel A. Differential, regional, and cellular expression of the stathmin family transcripts in the adult rat brain. J Neurosci Res 1999; 56:553-64; PMID:10369222
- Friedmann-Morvinski D, Bushong EA, Ke E, Soda Y, Marumoto T, Singer O, Ellisman MH, Verma IM Dedifferentiation of neurons and astrocytes by oncogenes can induce gliomas in mice. Science 2012; 338:1080-4; PMID:23087000; http://dx.doi.org/10.1126/science.1226929
- Singer S, Malz M, Herpel E, Warth A, Bissinger M, Keith M, Muley T, Meister M, Hoffmann H, Penzel R, et al. Coordinated expression of stathmin family members by far upstream sequence element-binding protein-1 increases motility in non-small cell lung cancer. Cancer Res 2009; 69:2234-43; PMID:19258502; http://dx.doi.org/10.1158/0008-5472.CAN-08-3338
- Kang SW, Shin YJ, Shim YJ, Jeong SY, Park IS, Min BH. Clusterin interacts with SCLIP (SCG10-like protein) and promotes neurite outgrowth of PC12 cells. Exp Cell Res 2005; 309:305-15; PMID:16038898; http://dx.doi.org/10.1016/j.yexcr.2005.06.012
- Vehlow A, Cordes N. Invasion as target for therapy of glioblastoma multiforme. Biochim Biophys Acta 2013; 1836:236-44; PMID:23891970
- Gundersen GG, Wen Y, Eng CH, Schmoranzer J, Cabrera-Poch N, Morris EJ, Chen M, Gomes ER. Regulation of microtubules by Rho GTPases in migrating cells. Novartis Found Symp 2005; 269:106-16; discussion 16-26, 223-30; PMID:16358406; http://dx.doi.org/10.1002/047001766X.ch10
- Ngo TT, Peng T, Liang XJ, Akeju O, Pastorino S, Zhang W, Kotliarov Y, Zenklusen JC, Fine HA, Maric D, et al. The 1p-encoded protein stathmin and resistance of malignant gliomas to nitrosoureas. J Natl Cancer Inst 2007; 99:639-52; PMID:17440165; http://dx.doi.org/10.1093/jnci/djk135
- Byrne FL, Yang L, Phillips PA, Hansford LM, Fletcher JI, Ormandy CJ, McCarroll JA, Kavallaris M. RNAi-mediated stathmin suppression reduces lung metastasis in an orthotopic neuroblastoma mouse model. Oncogene 2014; 33:882-90; PMID:23396365; http://dx.doi.org/10.1038/onc.2013.11
- Luwor RB, Stylli SS, Kaye AH. The role of Stat3 in glioblastoma multiforme. J Clin Neurosci 2013; 20:907-11; PMID:23688441; http://dx.doi.org/10.1016/j.jocn.2013.03.006
- Masciarelli S, Fontemaggi G, Di Agostino S, Donzelli S, Carcarino E, Strano S, et al. Gain-of-function mutant p53 downregulates miR-223 contributing to chemoresistance of cultured tumor cells. Oncogene 2014; 33:1601-8; PMID:23584479; http://dx.doi.org/10.1038/onc.2013.106
- Kang W, Tong JH, Chan AW, Lung RW, Chau SL, Wong QW, et al. Stathmin1 plays oncogenic role and is a target of microRNA-223 in gastric cancer. PLoS One 2012; 7:e33919; PMID:22470493; http://dx.doi.org/10.1371/journal.pone.0033919
- Du Z, Jia D, Liu S, Wang F, Li G, Zhang Y, et al. Oct4 is expressed in human gliomas and promotes colony formation in glioma cells. Glia 2009; 57:724-33; PMID:18985733; http://dx.doi.org/10.1002/glia.20800
