Abstract
eIF2B is a multisubunit protein that is critical for protein synthesis initiation and its control. It is a guanine nucleotide exchange factor (GEF) for its GTP-binding protein partner eIF2. eIF2 binds initiator tRNA to ribosomes and promotes mRNA AUG codon recognition. eIF2B is critical for regulation of protein synthesis via a conserved mechanism of phosphorylation of eIF2, which converts eIF2 from a substrate to an inhibitor of eIF2B GEF. In addition, inherited mutations affecting eIF2B subunits cause the fatal disorder leukoencephalopathy with Vanishing White Matter (VWM), also called Childhood Ataxia with Central nervous system Hypomyelination (CACH). Here we review findings which reveal that eIF2B is a decameric protein and also define a new function for the eIF2B. Our results demonstrate that the eIF2Bγ subunit is required for eIF2B to gain access to eIF2•GDP. Specifically it displaces a third translation factor eIF5 (a dual function GAP and GDI) from eIF2•GDP/eIF5 complexes. Thus eIF2B is a GDI displacement factor (or GDF) in addition to its role as a GEF, prompting the redrawing of the eIF2 cycling pathway to incorporate the new steps. In structural studies using mass spectrometry and cross-linking it is shown that eIF2B is a dimer of pentamers and so is twice as large as previously thought. A binding site for GTP on eIF2B was also found, raising further questions concerning the mechanism of nucleotide exchange. The implications of these findings for eIF2B function and for VWM/CACH disease are discussed.
Introduction
Roles of eIF2, eIF5 and eIF2B in protein synthesis
The GTP-binding protein (G-protein) eIF2 (eukaryotic initiation factor 2) functions during protein translation initiation by delivering initiator methionyl tRNA to the ribosome. This is a fundamental process that occurs in all eukaryotic cells and ensures that protein synthesis originates at the correct AUG start codon on each mRNA. Translation initiation is a complex multistep process. In addition to eIF2, there are at least 11 other translation factors that interact with mRNAs and/or ribosomal subunits to ensure appropriate mRNA selection and translation initiation. An overview of the entire pathway is beyond the scope of this review and interested readers are directed to other recent reviews.Citation1,2 Here we consider 2 factors that directly control eIF2 activities: eIF5 and eIF2B.
Similar to other G-proteins, eIF2 is cycled between inactive (GDP-bound) and active (GTP-bound) states and this G-protein cycle drives successive rounds of translation initiation. eIF5 and eIF2B are key players in these processes. In its active (GTP-bound) conformation, eIF2 interacts with methionyl initiator tRNA (Met-tRNAi) to form a ternary complex (TC)Citation3 and delivers it to the small (40S) ribosomal subunit. Other translation factors dictate mRNA selection and promote scanning of the 5’ leader sequence to locate the start codon. During AUG selection by the ribosome, eIF2 bound GTP is hydrolysed by the GTPase accelerating protein (GAP) activity of eIF5 and Pi is released.Citation4 Pi release reduces eIF2 affinity for Met-tRNAiCitation3,4 and triggers release of both factors from the ribosome, allowing formation of the full 80S ribosome and translation elongation to begin. eIF2 is therefore released from ribosomes in its inactive (GDP-bound) state in complex with eIF5.Citation5 In this eIF2•GDP/eIF5 complex eIF5 acts as a GDP dissociation inhibitor (GDI) maintaining eIF2 in its inactive GDP-bound state.Citation6 To participate in a subsequent round of Met-tRNAi recruitment to the ribosome, eIF2 must be reactivated to the GTP form. This process is carried out by the guanine nucleotide exchange factor (GEF) eIF2B.Citation7 What was not known is how eIF2B gained access to eIF2 to promote its reactivation, when eIF2 is bound to eIF5. In a recent paperCitation8 we have now demonstrated that eIF2B has a second function and can itself promote release of eIF5 from eIF2. In G protein 3-letter acronyms, this function is described as GDF (for GDI displacement factor). Such factors have previously been described for other G-proteins that have a GDI component.Citation9-11 There has been only one other published study identifying a dual function GEF-GDF protein. This is SidM/DrrA, a protein encoded by the pathogen Legionella pneumophila.
The study makes significant progress toward explaining why eIF2B is such a complicated multisubunit protein and defines eIF2B as a multifunctional protein required for reactivation of eIF2, being both a GDF and a GEF. Here we review the major findings that led us to propose a new model for translation initiation and its control that accounts for the activities of eIF5 and eIF2B and which speculates on the role of GDF mutations in human disease. In addition we review a second study that reveals greater complexity to this translation factor as it is shown that eIF2B is a decamer rather than a pentamer factor.Citation12
eIF2B displaces eIF5 from eIF2•GDP
Prior to ∼2005, models of the role of eIF5 in translation initiation were confined to its roles in AUG codon selection and GTPase activation. Roles that required interactions with active eIF2 bound to GTP and Met-tRNAi and not with with inactive eIF2. Similarly eIF2B was known to interact with eIF2•GDP and to function as a GEF (Pavitt 2005; Pavitt and Proud 2009). Thus a simple model with GAP and GEF activities described the control of the eIF2 G protein cycle. This model is depicted in .
Figure 1. Models for eIF2B functions in protein synthesis initiation. (A) Original model depicting eIF2-GTP-Met-tRNAi recruitment to the ribosome and its release following GTP hydrolysis to eIF2-GDP upon mRNA AUG start codon recognition. Here eIF2B perfoms a single GEF step to reactivate eIF2. (B) Revised model for eIF2 recycling accounting for eIF5 GDI and eIF2B GDF functions. (C) New model showing the impact of eIF2α phosphorylation on eIF2B and eIF5. For further explanations, including step numbering, refer to the main text.
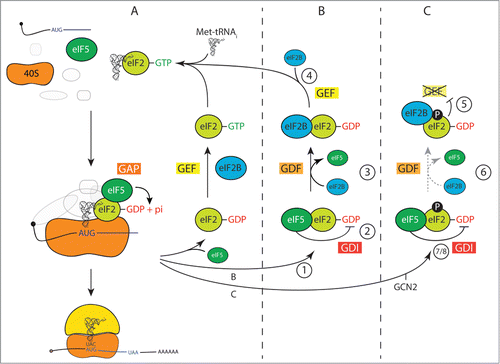
However observations from several labs suggested that a more complex eIF2 cycle may operate in cells. eIF5 was found to bind to eIF2•GDP with equal affinity as to eIF2•GTP•Met-tRNAi (∼23 nM).Citation4,5,13 In addition by comparing the relative abundance of factors and their interactions in complexes purified from yeast cells, Asano and colleagues uncovered that eIF5 and eIF2 form a complex that is in greater abundance than the fraction of eIF2 bound to Met-tRNAi.Citation5,13 These observations led to the idea that eIF5 bound eIF2•GDP in vivo may therefore have additional functions. We showed by a combination of biochemistry and yeast genetics that eIF5 does have a second function with eIF2•GDP where it functions to stabilize GDP-binding to eIF2. This GDI activity antagonises GDP release from eIF2 and was therefore expected antagonise eIF2B GEF.Citation8
How does eIF2B overcome antagonism by eIF5? For continued translation eIF2 must become reactivated by eIF2B GEF. Both eIF2 and eIF5 are equally abundant, whereas eIF2B is roughly ten-fold less abundant.Citation13 The eIF5 carboxy terminal domain (CTD) is critical for its interaction with eIF2 and for its GDI functionCitation6,14 and is a close structural mimic of the eIF2B GEF domain, that resides at the CTD of the largest eIF2B subunit (eIF2Bϵ).Citation15-17 Both eIF5 and eIF2Bϵ CTDs are proposed to interact with eIF2 in a mutually exclusive manner and so to compete with each other for interaction with eIF2. Taken together this posed a conundrum of how eIF2B gained access to eIF2 when eIF2 is bound to eIF5.
We tested the idea that eIF2B had a separate function to displace eIF5 from eIF2•GDP/eIF5 complexes prior to its known role as a GEF, as this seemed the most logical solution to the problem. eIF2B is a particularly complicated factor, assembled of subunits α-ϵ, encoded by 5 distinct genes, of which only one is critically required for its GEF function. So it seemed plausible that one of the others was important for eIF5 displacement. In agreement with this prediction, we used a steady state protein-protein interaction assay to demonstrate that eIF2B can efficiently dissociate the eIF2/eIF5 complex, but that the isolated eIF2Bϵ could not. Further studies showed that eIF2Bγ and ϵ together were necessary for GDF activity. A second assay that we used was a coupled kinetic assay that measured rates of GDP release from eIF2•GDP/eIF5. This agreed that the eIF2Bγϵ sub-complex was required for efficient GDP release when eIF5 was included in the assay. One key element to our study was the identification of single amino acids that are important for GDF activity, but not the GEF function. We screened eIF2Bγ mutations originally isolated in yeast in the 1970s and 1980s, before the gene (yeast GCD1) was cloned and sequenced. Phenotypically the eIF2Bγ mutants were not distinguishable from eIF2Bϵ mutations that impair GEF activity: they impair general translation and cause slow-growth and derepress the translation of GCN4, a translationally controlled transcription factor critical for responses to amino acid starvation (see below for discussion of eIF2B regulation). Both eIF2Bγ mutants analyzed biochemically (G11V and L480Q substitutions) did not interfere significantly with eIF2 binding or GEF activity, however both impaired eIF5 displacement/GDF activity. Taken together the study shows that GDF function is important for normal cell growth and cell division at optimal rates and that it is biochemically separate to the previously described GEF activity: a new function for eIF2B and a new step in protein synthesis pathway.Citation8
Roles of eIF5-GDI and eIF2B-GDF in eIF2 responses to stress
A wide variety of stimuli and cellular stresses cause eIF2 to be targeted by various protein kinases (for example Gcn2p in Saccharomyces cerevisiae, and GCN2, PERK, PKR and HRI in mammalian cells). All phosphorylate eIF2 at the same position, serine 51 within α subunit of eIF2.Citation1,18 The resulting phosphorylated eIF2 (eIF2αP) acts a competitive inhibitor of eIF2B, restricting GEF activity and reactivation of eIF2.Citation8 This applies a brake, lowering levels of active eIF2 leading to a decrease in general protein synthesis initiation. At the same time certain mRNAs are up-regulated, including specific mRNAs required for the cellular stress response. One well studied class of mRNAs that increase expression following phosphorylation of eIF2 are GCN4 in yeast and ATF4 in mammalian cells. Both possess short ORFs upstream of the main coding region that normally limit the flow of ribosomes to the main coding AUG. eIF2αP promotes ribosomes to bypass the inhibitory upstream ORF(s).Citation1,19
Our studies identifying eIF5 GDI and eIF2B GDF functions revealed that mutations that impair each function have opposing impacts on translational control. Our earlier work showed that the eIF5 GDI mutant W391F is resistant to the inhibitory eIF2αP.Citation6 Thus eIF5-GDI is required for eIF2αP to fully inhibit eIF2B and permit translation of GCN4. In contrast the eIF2B-GDF mutants impair the ability of eIF2B to access eIF2 and this defect leads to constitutive expression of GCN4. By in vitro kinetic studies we were also able to demonstrate that phosphorylation of eIF2 does not prevent eIF2B-GDF, but does prevent eIF2B-GEF. Taking all the findings together we can refine the model for eIF2 recycling that includes GDI, GDF and GEF activities and shows their individual importance for the regulation of protein synthesis by eIF2αP.
Refining the model of eIF2 recycling and its control by eIF2 phosphorylation
The identification of eIF5 GDI activity and eIF2B GDI displacement activity have altered our perception of how eIF2 is recycled and regulated in yeast cells. This has allowed us to refine the original model () and propose a new, more complex, model that is depicted in . The elements that we have identified as important for each activity are conserved in mammals including man, so we suspect that the findings in yeast may also be important in mammalian systems, but this has not yet been demonstrated.
Our revised model for eIF2 recactivation () is:
During translation initiation and following AUG codon recognition by Met-tRNAi-bound eIF2, eIF2•GDP is released with eIF5 from the ribosome/ mRNA (48S) complex so that that large subunit can join and translation elongation can commence.
eIF5 stabilises the eIF2 bound GDP to maintain eIF2 inactivity.Citation6 As eIF2B is considerably less abundant than eIF2 or eIF5 (∼10 fold), this eIF2•GDP/eIF5 complex forms an abundant cellular pool.Citation5
eIF2B GDF activity means it can readily access eIF2 from the inactivated eIF2•GDP/eIF5 pool, displacing eIF5.Citation8
eIF2B can then reactivate eIF2 by guanine nucleotide exchange,Citation8 permitting Met-tRNAi binding and a new round of protein synthesis.
If eIF2B can readily displace eIF5, why the need for the additional GDI and GDF steps? Our data shows that eIF5 GDI is primarily important under conditions when eIF2 is phosphorylated.
Model for eIF2αP regulation ():
When eIF2 is phosphorylated eIF2B binds with high affinity to eIF2α via contacts made to the α, β or δ subunits of eIF2B.Citation20 This prevents eIF2B GEF activity.Citation8
As increasing amounts of eIF2B become trapped in complex with eIF2αP, there is little or no free eIF2B to interact with eIF2•GDP/eIF5. So with limiting free eIF2B, eIF2B GDF activity diminishes.Citation8
Continued protein synthesis initiation causes a backlog of released eIF2•GDP/eIF5 to form, increasing the cellular pool eIF2•GDP/eIF5Citation8
eIF5 GDI acts to prevent spontaneous eIF2B-independent nucleotide exchange, which would otherwise bypass the effectiveness of the eIF2αP regulatory loop that has evolved to act as a brake on protein synthesis initiation.
Implications of eIF2B GDF in disease
Mutations in all eIF2B subunits cause a fatal inherited leukodystrophy. Called Leukoencephalopathy with Vanishing White Matter (VWM) or Childhood Ataxia with Central Nervous System Hypomyelination (CACH).Citation21 The disease is characterized by a progressive loss of brain white matter. The affected cells are glial cells (astrocytes and oligodendrocytes), which comprise the blood-brain barrier and form myelin sheaths to insulate neuronal axons. Well over a hundred different eIF2B missense mutations have been associated with the disorder and various causes of disease suggested by biochemical analyses. Many mutations impact on the stability of the eIF2B complex, others appear to alter eIF2 interactions.Citation22,23 However some mutations cause severe disease, yet apparently do not affect eIF2 interactions or GEF activity.Citation24 Drawing parallels with our findings in the yeast system it seems plausible that some VWM/CACH mutations will impact on eIF2B GDF. Indeed one of the eIF2Bγ mutants identified (G12V) is analogous to a human mutation (EIF2B3-G11V). The yeast eIF2Bγ GDF mutations studied impair eIF5 displacement from eIF2, cause severe growth impairment and hinder translational control. However they do not significantly impact on eIF2 interaction in vitro or eIF2B GEF activity in the standard assay used.Citation8 In the standard GEF assay eIF2 and labeled GDP and is mixed with either purified eIF2B or a cell extract.Citation22,25 Cell extract assays using immortalized lymphocytes from patient serum have been most commonly used to assess clinical samples.Citation26-28 Our study would suggest that supplementing these assays with a concentration of eIF5 equimolar to eIF2 would be a useful modification. Such an assay should report on both GDF and GEF defects in patient cells in a single assay.
eIF2B is a dimer of pentamers
The finding that eIF2B has an additional function as a GDF, goes some way to explaining why it is such a complicated protein. However, recent observations highlight that there much remains to be understood. Two studies have shown that eIF2B purified from humanCitation29 or yeastCitation12 cells is a dimer and so has 10 rather than 5 subunits. In the yeast study evidence was presented primarily from nano-electrospray mass spectrometry (n-EMS) of intact proteins and further refined using lysine cross-linking approach and surface accessibility measurements. n-EMS is a technique that can preserve non-covalent interactions between proteins in the gas phase of the mass spectrometer and informs on protein size and subunit stability. Experiments showed that eIF2B has a mass approaching 600 KDa and is a dimer of α-ϵ pentamers.Citation12 The α subunit could be readily lost from the complex and complexes produced lacking α subunits formed stable β-ϵ dimers. Because the α subunit is necessary for regulation of eIF2B by phosphorylation of eIF2α (see above), this result suggests that dimerization and phospho-regulation by this conserved pathway are not linked.Citation12 In contrast the study of human eIF2B suggested dimer formation was weakened by mutation or loss of the eIF2Balpha subunit and that it may contribute to dimer formation, as suggested by the prior eIF2Bα crystal structureCitation30 where an α-α dimer interface was evident. While it is clear isolated eIF2Bα can form a dimer,Citation29,31 it remains less clear if this happens within the intact eIF2B complex, and the data obtained with yeast proteins suggests that other elements are required for dimer formation.
The yeast structural study further focused on the γ and ϵ subunits, which are the key subunits for both GEF and the new GDF function described in the sections above. These subunits share homology with a family of sugar-pyrophosphorylase enzymes. Pyrophophosphorylases including potato ADP-glucose pyrophosphorylase form a homo-tetrameric structure.Citation32,33 Extrapolating to eIF2B suggests that a γ2ϵ2 subunit arrangement may be possible. However the γϵ purified complex was mainly a dimer by n-EMS, not a tetramer, although a tetramer could be stabilized by acetronitrile, suggesting that a γ2ϵ2 could contribute to the dimer interface. Alternatively the dimer interface could be mediated by the β and δ subunits. Our functional analyses of the isolated γϵ complexes show they retain full GDF and full GEF activities,Citation8 yet without forming a stable dimer.Citation12 So it appears unlikely that tetramer formation is required for these known activities of eIF2B. This leaves open the question of why eIF2B forms a dimer and what contribution it makes to eIF2B activities. Further studies will hopefully shed light on these aspects of eIF2B function and regulation.
eIF2B is a GTP-binding protein
A final twist to the eIF2B story is that eIF2B itself can bind GTP. GTP-binding is not usual for GEFs and the standard mechanism proposed for small GTPase GEFs is that they bind and cause GDP release, stabilizing a nucleotide free form, prior to GTP binding from the free GTP pool in the solvent.Citation34 However applying enzyme kinetic methods to eIF2B suggested a role for a second nucleotide binding prior to the release of the outgoing GDP.Citation35,36 What was less clear is where the second nucleotide was bound. Nika et alCitation36 found that GTP could bind directly to eIF2B, but did not indicate where. As pyrophosphorylase enzymes can bind specific nucleotides, it seems plausible that the homologous γ and/or ϵ subunits bind GTP. As eIF2Bϵ is the primary GEF subunit, we previously performed mutagenesis of key residues of eIF2Bϵ, but this failed to provide strong evidence for its involvement in GTP-binding.Citation33 Now using n-EMS and MS with 6-Thio-GTP and UV cross-linking, Gordiyenko et al show that GTP or Thio-GTP binds to eIF2Bγ.Citation12
What is the role of GTP-binding to eIF2Bγ? Three possible options are: firstly, GTP-binding to eIF2Bγ may play a direct role in the GEF reaction. Secondly, GTP binding may have an allosteric regulatory role. Or finally GTP-binding may contribute to an unrelated, currently unknown, function of eIF2B. Evidence in favor of the first of these options comes from lysine-lysine cross-linking studies. Specifically a crosslink between eIF2Bγ K249 (within the pyrophosphorylase-like domain) and eIF2γ K113 (within the G domain) was found when a bis(sulfosuccinimidyl)suberate cross-linker was mixed with eIF2 and eIF2B.Citation12 The crosslink places the GTP-binding domains of each protein in close proximity. As eIF2Bϵ alone can mediate GDP-release from eIF2,Citation25,37 GTP transfer from eIF2B to eIF2γ is not critical for this step. However it is possible that it is important for efficient nucleotide exchange and GTP binding to eIF2. Such a function may help explain the stimulatory effect eIF2Bγ has on eIF2B-catalyzed GEF activity.Citation8,38
In conclusion, eIF2B is a multifunctional protein required for protein synthesis and its control. Recent advances have shown that eIF2B is more than just a GEF. In addition our improved understanding of eIF2B complexity lays foundations for future studies of this fascinating protein.
Additional information
Funding
References
- Jackson RJ, Hellen CU, Pestova TV. The mechanism of eukaryotic translation initiation and principles of its regulation. Nat Rev Mol Cell Biol 2010; 11:113-27; PMID:20094052; http://dx.doi.org/10.1038/nrm2838
- Hinnebusch AG, Lorsch JR. The mechanism of eukaryotic translation initiation: new insights and challenges. Cold Spring Harb Perspect Biol 2012; 4:29-55; http://dx.doi.org/10.1101/cshperspect.a011544
- Erickson FL, Hannig EM. Ligand interactions with eukaryotic translation initiation factor 2: role of the γ-subunit. EMBO J 1996; 15:6311-20: PMID:8947054
- Algire MA, Maag D, Lorsch JR. Pi release from eIF2, not GTP hydrolysis, is the step controlled by start-site selection during eukaryotic translation initiation. Mol Cell 2005; 20:251-62; PMID:16246727; http://dx.doi.org/10.1016/j.molcel.2005.09.008
- Singh CR, Lee B, Udagawa T, Mohammad-Qureshi SS, Yamamoto Y, Pavitt GD, Asano K. An eIF5/eIF2 complex antagonizes guanine nucleotide exchange by eIF2B during translation initiation. EMBO J 2006; 25:4537-46; PMID:16990799; http://dx.doi.org/10.1038/sj.emboj.7601339
- Jennings MD, Pavitt GD. eIF5 has GDI activity necessary for translational control by eIF2 phosphorylation. Nature 2010; 465:378-81; PMID:20485439; http://dx.doi.org/10.1038/nature09003
- Pavitt GD. eIF2B, a mediator of general and gene-specific translational control. Biochem Soc Trans 2005; 33:1487-92; PMID:16246152; http://dx.doi.org/10.1042/BST20051487
- Jennings MD, Zhou Y, Mohammad-Qureshi SS, Bennett D, Pavitt GD. eIF2B promotes eIF5 dissociation from eIF2•GDP to facilitate guanine nucleotide exchange for translation initiation. Genes Dev 2013; 27:2696-707; http://dx.doi.org/10.1101/gad.231514.113
- Pfeffer SR. Rab GTPases: master regulators of membrane trafficking. Curr Opin Cell Biol 1994; 6:522-6; PMID:7986528; http://dx.doi.org/10.1016/0955-0674(94)90071-X
- Dirac-Svejstrup AB, Sumizawa T, Pfeffer SR. Identification of a GDI displacement factor that releases endosomal Rab GTPases from Rab-GDI. EMBO J 1997; 16:465-72; PMID:9034329; http://dx.doi.org/10.1093/emboj/16.3.465
- Sivars U, Aivazian D, Pfeffer S. Purification and properties of Yip3/PRA1 as a Rab GDI displacement factor. Methods Enzymol 2005; 403:348-56; PMID:16473601; http://dx.doi.org/10.1016/S0076-6879(05)03030-2
- Gordiyenko Y, Schmidt C, Jennings MD, Matak-Vinkovic D, Pavitt GD, Robinson CV. eIF2B is a decameric guanine nucleotide exchange factor with a gamma2epsilon2 tetrameric core. Nat Commun 2014; 5:3902; PMID:24852487; http://dx.doi.org/10.1038/ncomms4902
- Singh CR, Udagawa T, Lee B, Wassink S, He H, Yamamoto Y, Anderson JT, Pavitt GD, Asano K. Change in nutritional status modulates the abundance of critical pre-initiation intermediate complexes during translation initiation in vivo. J Mol Biol 2007; 370:315-30; PMID:17512538; http://dx.doi.org/10.1016/j.jmb.2007.04.034
- Asano K, Krishnamoorthy T, Phan L, Pavitt GD, Hinnebusch AG. Conserved bipartite motifs in yeast eIF5 and eIF2Bepsilon, GTPase- activating and GDP-GTP exchange factors in translation initiation, mediate binding to their common substrate eIF2. EMBO J 1999; 18:1673-88; PMID:10075937; http://dx.doi.org/10.1093/emboj/18.6.1673
- Boesen T, Mohammad SS, Pavitt GD, Andersen GR. Structure of the catalytic fragment of translation initiation factor 2B and identification of a critically important catalytic residue. J Biol Chem 2004; 279:10584-92; PMID:14681227; http://dx.doi.org/10.1074/jbc.M311055200
- Bieniossek C, Schutz P, Bumann M, Limacher A, Uson I, Baumann U. The crystal structure of the carboxy-terminal domain of human translation initiation factor eIF5. J Mol Biol 2006; 360:457-65; PMID:16781736; http://dx.doi.org/10.1016/j.jmb.2006.05.021
- Alone PV, Dever TE. Direct binding of translation initiation factor eIF2gamma-G domain to its GTPase-activating and GDP-GTP exchange factors eIF5 and eIF2B epsilon. J Biol Chem 2006; 281:12636-44; PMID:16522633; http://dx.doi.org/10.1074/jbc.M511700200
- Baird TD, Wek RC. Eukaryotic initiation factor 2 phosphorylation and translational control in metabolism. Adv Nutr 2012; 3:307-21; PMID:22585904; http://dx.doi.org/10.3945/an.112.002113
- Pavitt GD, Ron D. New Insights into Translational Regulation in the Endoplasmic Reticulum Unfolded Protein Response. Cold Spring Harb Perspect Biol 2012; 4:163-75.
- Krishnamoorthy T, Pavitt GD, Zhang F, Dever TE, Hinnebusch AG. Tight binding of the phosphorylated alpha subunit of initiation factor 2 (eIF2alpha) to the regulatory subunits of guanine nucleotide exchange factor eIF2B is required for inhibition of translation initiation. Mol Cell Biol 2001; 21:5018-30; PMID:11438658; http://dx.doi.org/10.1128/MCB.21.15.5018-5030.2001
- Pavitt GD, Proud CG. Protein synthesis and its control in neuronal cells with a focus on vanishing white matter disease. Biochem Soc Trans 2009; 37:1298-310; PMID:19909266; http://dx.doi.org/10.1042/BST0371298
- Li W, Wang X, Van Der Knaap MS, Proud CG. Mutations linked to leukoencephalopathy with vanishing white matter impair the function of the eukaryotic initiation factor 2B complex in diverse ways. Mol Cell Biol 2004; 24:3295-306; PMID:15060152; http://dx.doi.org/10.1128/MCB.24.8.3295-3306.2004
- Richardson JP, Mohammad SS, Pavitt GD. Mutations causing childhood ataxia with central nervous system hypomyelination reduce eukaryotic initiation factor 2B complex formation and activity. Mol Cell Biol 2004; 24:2352-63; PMID:14993275; http://dx.doi.org/10.1128/MCB.24.6.2352-2363.2004
- Liu R, van der Lei HD, Wang X, Wortham NC, Tang H, van Berkel CG, Mufunde TA, Huang W, van der Knaap MS, Scheper GC, et al. Severity of vanishing white matter disease does not correlate with deficits in eIF2B activity or the integrity of eIF2B complexes. Human mutation 2011; 32:1036-45; PMID:21560189; http://dx.doi.org/10.1002/humu.21535
- Gomez E, Pavitt GD. Identification of domains and residues within the epsilon subunit of eukaryotic translation initiation factor 2B (eIF2Bepsilon) required for guanine nucleotide exchange reveals a novel activation function promoted by eIF2B complex formation. Mol Cell Biol 2000; 20:3965-76; PMID:10805739; http://dx.doi.org/10.1128/MCB.20.11.3965-3976.2000
- Fogli A, Schiffmann R, Hugendubler L, Combes P, Bertini E, Rodriguez D, Kimball SR, Boespflug-Tanguy O. Decreased guanine nucleotide exchange factor activity in eIF2B-mutated patients. Eur J Hum Genet 2004; 12:561-6; PMID:15054402; http://dx.doi.org/10.1038/sj.ejhg.5201189
- Horzinski L, Huyghe A, Cardoso MC, Gonthier C, Ouchchane L, Schiffmann R, Blanc P, Boespflug-Tanguy O, Fogli A. Eukaryotic initiation factor 2B (eIF2B) GEF activity as a diagnostic tool for EIF2B-related disorders. PloS One 2009; 4:e8318; PMID:20016818; http://dx.doi.org/10.1371/journal.pone.0008318
- de Almeida RA, Fogli A, Gaillard M, Scheper GC, Boesflug-Tanguy O, Pavitt GD. A yeast purification system for human translation initiation factors eIF2 and eIF2Bepsilon and their use in the diagnosis of CACH/VWM disease. PloS One 2013; 8:e53958; PMID:23335982; http://dx.doi.org/10.1371/journal.pone.0053958
- Wortham NC, Martinez M, Gordiyenko Y, Robinson CV, Proud CG. Analysis of the subunit organization of the eIF2B complex reveals new insights into its structure and regulation. FASEB J 2014; 28:2225-37; PMID:24532666; http://dx.doi.org/10.1096/fj.13-243329
- Hiyama TB, Ito T, Imataka H, Yokoyama S. Crystal structure of the alpha subunit of human translation initiation factor 2B. J Mol Biol 2009; 392:937-51; PMID:19631657; http://dx.doi.org/10.1016/j.jmb.2009.07.054
- Bogorad AM, Xia B, Sandor DG, Mamonov AB, Cafarella TR, Jehle S, Vajda S, Kozakov D, Marintchev A. Insights into the Architecture of the eIF2Balpha/beta/delta Regulatory Subcomplex. Biochemistry 2014; 53:3432-45; PMID:24811713; http://dx.doi.org/10.1021/bi500346u
- Jin X, Ballicora MA, Preiss J, Geiger JH. Crystal structure of potato tuber ADP-glucose pyrophosphorylase. EMBO J 2005; 24:694-704; PMID:15692569; http://dx.doi.org/10.1038/sj.emboj.7600551
- Reid PJ, Mohammad-Qureshi SS, Pavitt GD. Identification of intersubunit domain interactions within eukaryotic initiation factor (eIF) 2B, the nucleotide exchange factor for translation initiation. J Biol Chem 2012; 287:8275-85; PMID:22238343; http://dx.doi.org/10.1074/jbc.M111.331645
- Cherfils J, Zeghouf M. Regulation of small GTPases by GEFs, GAPs, and GDIs. Physiol Rev 2013; 93:269-309; PMID:23303910; http://dx.doi.org/10.1152/physrev.00003.2012
- Manchester KL. Catalysis of guanine nucleotide exchange on eIF2 by eIF2B: can it be both a substituted enzyme and a sequential mechanism? Biochem Biophys Res Commun 2001; 289:643-6; PMID:11726195; http://dx.doi.org/10.1006/bbrc.2001.6010
- Nika J, Yang W, Pavitt GD, Hinnebusch AG, Hannig EM. Purification and kinetic analysis of eIF2B from Saccharomyces cerevisiae. J Biol Chem 2000; 275:26011-7; PMID:10852917; http://dx.doi.org/10.1074/jbc.M003718200
- Gomez E, Mohammad SS, Pavitt GD. Characterization of the minimal catalytic domain within eIF2B: the guanine-nucleotide exchange factor for translation initiation. EMBO J 2002; 21:5292-301; PMID:12356745; http://dx.doi.org/10.1093/emboj/cdf515
- Pavitt GD, Ramaiah KV, Kimball SR, Hinnebusch AG. eIF2 independently binds two distinct eIF2B subcomplexes that catalyze and regulate guanine-nucleotide exchange. Genes Dev 1998; 12:514-26; PMID:9472020; http://dx.doi.org/10.1101/gad.12.4.514
