Abstract
The predominant function of the tumor suppressor p53 is transcriptional regulation. It is generally accepted that p53-dependent transcriptional activation occurs by binding to a specific recognition site in promoters of target genes. Additionally, several models for p53-dependent transcriptional repression have been postulated. Here, we evaluate these models based on a computational meta-analysis of genome-wide data. Surprisingly, several major models of p53-dependent gene regulation are implausible. Meta-analysis of large-scale data is unable to confirm reports on directly repressed p53 target genes and falsifies models of direct repression. This notion is supported by experimental re-analysis of representative genes reported as directly repressed by p53. Therefore, p53 is not a direct repressor of transcription, but solely activates its target genes. Moreover, models based on interference of p53 with activating transcription factors as well as models based on the function of ncRNAs are also not supported by the meta-analysis. As an alternative to models of direct repression, the meta-analysis leads to the conclusion that p53 represses transcription indirectly by activation of the p53-p21-DREAM/RB pathway.
Abbreviations
| CDE | = | cell cycle-dependent element |
| CHR | = | cell cycle genes homology region |
| DREAM | = | DP, RB-like, E2F4, and MuvB complex |
| ChIP | = | chromatin immunoprecipitation |
| NF-Y | = | Nuclear factor Y |
| cdk | = | cyclin-dependent kinase |
| HPV | = | human papilloma virus |
Introduction
Initially, p53 was falsely described as an oncogene. About a decade after its discovery, p53 was found to be a tumor suppressor.Citation1,2 Despite 35 years of research and an ever growing number of publications, currently over 70,000 listed in PubMed, the central function of p53 as a transcriptional regulator still holds a major contradiction. It remains unresolved how p53 binding results in activation of one target gene and repression of another.
Following the discovery of p53's first transcriptional targets, many more genes were claimed to harbor p53 binding sites and thus to be potential targets resulting in an “expanding universe of p53 targets”.Citation3,4 In recent years, genome-wide analyses led to the discovery of novel p53 target genes by combining p53 chromatin occupancy data with gene expression analyses.Citation5-9 Hundreds of genes were identified as novel direct p53 targets. For a long time the search for direct p53 target genes often was undertaken without distinguishing significant regulation from experimental noise, similar to the assignment of function to large parts of the genome despite the substantial lack of conservation in these genomic regions by the ENCODE Consortium.Citation10
While reproducibility is a hallmark of scientific discovery, results from a substantial fraction of published work remain irreproducible.Citation11 A general problem appears to be that today's science is strongly biased for significant positive findings encouraging researchers to overinterpret small effects and inflate associations.Citation12 One method to clarify contradictions is meta-analysis of data from independent experiments.Citation11
In this study, we employ a meta-analysis on p53's transcriptional network employing data on 19,736 known protein-coding genes from several independent genome-wide studies to evaluate models of transcriptional regulation by p53. Six major mechanisms of p53-dependent transcriptional regulation are currently accepted in the literature:Citation13-17
direct activation of target genes following p53 binding to a p53 response element (RE)
direct repression of target genes after p53 binding to p53 REs, including variations such as head-to-tail elements or p53 REs with inverted dinucleotide cores
direct repression of target genes through p53 binding via adaptor proteins, in particular NF-Y
indirect repression via direct activation of p21 by p53 and subsequent formation of pocket protein/E2F complexes such as RB/E2F and DREAM
indirect repression through interference with transcriptional activators, in particular NF-Y, Sp1 and TBP
indirect repression of target genes via non-coding RNAs (ncRNAs), with mir34a, lincRNA-p21 and PANDA as prominent examples
We provide a comprehensive overview on original research findings and compare them to results from the meta-analysis. With this comparison we test the previously proposed models on p53-dependent transcriptional regulation. Important findings from the meta-analysis are supported by experimental validation. In general, our analysis resolves major contradictions and leads to a paradigm shift.
Results and Discussion
Computational meta-analysis on binding and regulation by p53
To evaluate the function of p53 as a transcription factor we have performed a computational meta-analysis from several independent experiments to minimize the influence of laboratory-specific effects and bias in study design.Citation11,18 Data from 6 genome-wide analyses of p53-dependent gene expression were extracted.Citation7,Citation19-23 In each study a gene can be identified as activated (positive score; +1) or repressed (negative score; −1) by p53. By calculating the sum over all analyses, Expression Scores ranging from −6 to +6 were assigned to genes, forming 13 gene groups (Table S1). Thus, the Expression Score represents direction of regulation as well as confidence of classification. By matching these data with transcription factor binding analyses, it is possible to evaluate whether activated or repressed genes are enriched for binding of a transcription factor such as p53. In case that the transcription factor is a repressor, its binding is expected to be substantially enriched at genes in negative Expression Score groups compared to genes in Expression Score group 0. We used 6 genome-wide p53 binding studiesCitation6-9,Citation24,25 and observed that 13.4% of all known protein-coding genes were identified as bound by p53.
Next, we compared the distribution of p53-bound genes across Expression Score groups to a theoretical uniform distribution of 13.4% (). A uniform distribution would be expected if there is no correlation between p53 binding and p53-dependent regulation.
Figure 1. Solely genes activated by p53 are found enriched for p53 binding. A regulation score, named Expression Score, ranging from −6 to +6 was assigned to 19,736 known protein-coding genes from 6 genome-wide p53-dependent gene expression analyses.Citation7,Citation19-23 (A) All ChIP-peaks from 6 genome-wide p53 binding studies, that were identified in at least 2 studies, were allocated to the nearest gene.Citation6-9,Citation24,25 Out of the 19,736 genes, 13.4% were assigned at least one such p53 ChIP-peak. The percentage of genes with a p53 ChIP-peak in a specific Expression Score group is displayed by the black line. The blue line indicates a theoretical uniform distribution of ChIP-peak-containing genes across the 13 Expression Score groups. (B) The percentage of default p53 targets (Table S2) in each Expression Score group is given by the black line. The theoretical uniform distribution of default p53 targets (n = 171 or 0.8% of 19,736 genes) across the 13 Expression Score groups is indicated by the blue line.
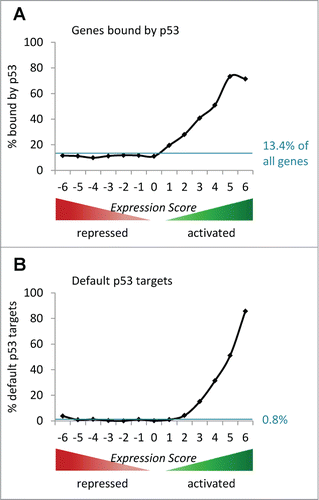
In contrast to most current modelsCitation13-17 but in agreement with observations made in recent genome-wide studies,Citation9,Citation26-28 solely genes activated by p53 are found enriched for p53 binding (; ). Thus, these data strongly suggest that p53 does not act as a direct transcriptional repressor.
Default p53 target genes
The authors of 2 recent genome-wide studies argue that a “default program” of p53 targets can be found that is shared regardless of cell type or treatment.Citation7,9 Based on the criteria that a target gene is bound and regulated by p53, we collated information describing individual p53 targets from about 300 reports (Table S2).Citation19,20,23,Citation29-324 This compilation was then complemented with data from 5 genome-wide studies on target genes bound and also regulated by p53.Citation5-9 Furthermore, we have correlated 2 genome-wide p53 binding studiesCitation24,25 with the 6 genome-wide gene expression studiesCitation7,Citation19-23 identifying additional target genes. This meta-analysis yielded potential direct p53 targets of which 892 are assigned as activated, 384 repressed, and 10 ambiguously regulated genes (Table S3). However, most genes in this compilation were observed in one study but were not confirmed in any other report. Many p53 target genes that were described in the literature earlier could not be confirmed in genome-wide approaches.
With this data collection, we included essentially all targets that might have been missed by single studies (false negatives). Yet, combining data sets in order to limit false negatives, inflates detection of false positives. One has to consider that each study can contain false positives and false negatives because of imperfect experimental conditions.Citation18 Therefore, after extending the data set on direct p53 targets, we defined limits to identify “default” targets. Genes detected in only one study have a high potential of being false positive hits and are most likely not part of the default program. Thus, from the published studies we derived weighted data sets to assign Default Target Scores to each direct p53 target gene. We considered a gene as a default target that was reported in at least 3 data sets, which corresponds to a Default Target Score > 2. We found 157 (17.6%) of all activated direct p53 target genes to meet these criteria (Table S3). Highest Default Target Scores were reached by many well established p53 target genes, all of which are activated by p53 (Table S3), such as CDKN1A (p21),Citation72 BTG2,Citation54 GADD45A,Citation112 BAX,Citation45 and MDM2.Citation122-124 In contrast, only 15 (3.9%) of the direct p53 target genes which have been described as repressed by p53 were assigned a Default Target Score > 2 (Table S3). Thus, the average Default Target Score of potentially repressed p53 targets is much lower compared to the score of activated target genes. Additionally, we evaluated the distribution of all default p53 target genes across the Expression Score groups (). Only genes activated by p53 were found enriched for default p53 targets. Taken together, in addition to looking solely at p53 binding as described above (), also data on default p53 targets substantiates the view that p53 does not directly repress its targets ().
Concordantly, recent genome-wide studies on p53 targets acknowledged a low abundance of p53-bound targets among repressed genes and entertained the possibility that repression by p53 may be largely indirect.Citation9,Citation26-28 Nevertheless, 90 reports describe 91 genes in detail as transcriptionally downregulated by direct binding of p53 (Table S2). The observations reported in these articles require further consideration.
Experimental validation of meta-analysis data
The meta-analysis data stand in contrast to the mechanisms of direct transcriptional repression by p53 and the regulation reported for many potential p53 targets (Table S2). Thus, we retested 18 genes for binding and regulation by p53 that were described to be directly repressed by p53, namely ABCB1 (MDR1),Citation325 BCL2,Citation245,326 BNIP3,Citation251 CCNB1,Citation254,327 CD44,Citation262 CDC20,Citation328 CDK1 (CDC2),Citation258 CRYZ,Citation260 HSPA8,Citation260 ID2,Citation275 LASP1,Citation281 MAD1L1 (MAD1),Citation260,329 ME1, ME2, ME3,Citation285 NEK2,Citation289 PTK2 (FAK),Citation306 and TPT1 (TCTP).Citation320
We tested p53 binding in chromatin immunoprecipitation assays (ChIP) followed by real-time PCR. Gene regulation by p53 was assayed by reverse transcriptase reaction followed by real-time PCR. If available, we used the published primers for PCR (; Fig. S2). No p53 binding was observed at the GAPDHS gene which served as a negative control. Binding of p53 was observed at the positive controls of CDKN1A (p21) and MDM2 (). Most importantly, at all other regions tested no significant p53 binding was observed (). Thus, the p53 response elements (RE) reported for the genes listed above can neither be confirmed by genome-wide studies nor by direct experimental re-analysis.
Figure 2. Experimental validation of data from the meta-analysis. (A) p53 protein binding to reported p53 response regions in untreated anddoxorubicin-treated HCT116 cells was tested by ChIP. A fragment of the GAPDHS promoter served as a negative control while CDKN1A and MDM2 served as positive controls. (B) mRNA expression in HCT116 cells treated with doxorubicin or nutlin3a for 24 h. Cells treated with DMSO served as a control. The log2 fold-expression from doxorubicin- or nutlin3a-treated cells compared to DMSO control cells is displayed as. GAPDH, L7, and U6 served as negative controls, while CDKN1A, MDM2, and PPM1D were employed as positive controls. Significance of expression was tested against U6 expression levels using paired Student's t-test. Experiments were performed with 2 biological replicates and 2 technical replicates each (n = 4). *P ≤ 0.05; **P ≤ 0.01;***P ≤ 0.001.
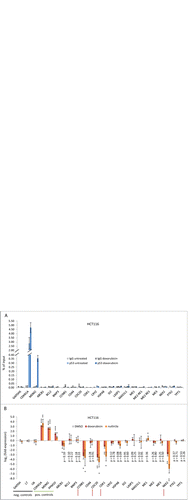
Although ABCB1, CD44, CDK1, MAD1L1, ME2, and PTK2 were found in genome-wide studies to bind p53 within 25 kb of their transcriptional start sites (TSS), the regions detected in genome-wide studies do not overlap with reported p53 REs (Table S1).Citation258,260,262,285,306,325,329 Therefore, all our results confirm data from the genome-wide studies and the meta-analysis. We asked how the discrepancies could arise between genome-wide data with the confirmatory results presented here and the observations from the reports mentioned above. Most discrepancies are explained by the use of real-time PCR instead of traditional PCR to evaluate binding of p53 in ChIP assays. Relative quantification is necessary to evaluate binding of a protein to one locus compared to non-bound regions. However, traditional PCR hardly allows relative quantifications often leading to erroneous results.
Expression of mRNA from these 18 genes depending on p53 was examined in doxorubicin- or nutlin3a-treated HCT116 cells compared to DMSO treatment. GAPDH mRNA, L7 mRNA, and U6 RNA served as negative controls not regulated by p53. The positive controls CDKN1A (p21), MDM2, and PPM1D were significantly upregulated upon treatment with doxorubicin or nutlin3a (). In contrast, only CCNB1, CDC20, CDK1, and NEK2 were significantly repressed after treatment with doxorubicin and nutlin3a, while ABCB1, BCL2, BNIP3, CRYZ, HSPA8, ID2, LASP1, MAD1L1, ME1, ME2, ME3, PTK2, and TPT1 were not significantly regulated by both treatments (). Again, these results confirm data from genome-wide studies and the meta-analysis, but do not support observations from the reports on direct transcriptional repression (; Tables S1 and S2). These discrepancies might largely stem from insufficient controls and overinterpretation of small effects.Citation11 In most reports criteria for p53 target genes were not met that were formulated 2 decades ago.Citation3 In addition to the p53 targets that were not confirmed by our re-analysis (), reports of directly repressed p53 targets in mouse such as NANOG,Citation288 PPARGC1A (PGC1a), and PPARGC1B (PGC1b)Citation300 are also not supported by human genome-wide data (Table S2).
Since CCNB1, CDC20, CDK1, and NEK2 are repressed but not bound by p53 (), we asked whether mechanisms other than direct repression have been postulated for the p53-dependent regulation of these genes. All 4 genes were shown to be repressed by the p53 target and CDK-inhibitor p21.Citation330-334 Furthermore, p53-dependent repression of CCNB1, CDC20, and CDK1 was shown to depend on the pocket proteins p107 and p130,Citation335 which also contrasts direct transcriptional repression by p53. In agreement with the reported p53-dependent repression via p21, we found that doxorubicin-induced repression of CCNB1, CDC20, CDK1, and NEK2, but not activation of MDM2 and PPM1D, is essentially lost in HCT116 p21−/- cells (Fig. S3).
Taken together, in most cases binding of p53 as well as p53-dependent regulation were not confirmed. Therefore, the reported mechanisms of direct transcriptional repression by p53 are unlikely of importance.
Challenging models of direct repression
Early in the history of p53 research, numerous genes were found to be repressed upon p53 induction.Citation336 For a long time the question remained open how binding of a transcription factor such as p53 can result in activation of one target gene and repression of another.
One of the proposed models is based on a head-to-tail p53 RE that had been described as a repressive element in the ABCB1 (MDR1) promoter.Citation325 Later, related elements were found to bind p53 and mediate downregulation of genes such as NANOG, CD44 and TPT1 (TCTP).Citation262,288,320 However, these results were never confirmed in any genome-wide study.Citation5-9 Moreover, NANOG, ABCB1, CD44 and TPT1 were actually found not to be repressed by p53 (Expression Scores ≥ 0) (; Table S1 and S2). Therefore, investigating their regulation could not yield a mechanism for p53-dependent transcriptional repression in the first place. Additionally, retesting the proposed p53 REs of ABCB1, CD44, and TPT1 provided evidence that these loci are not detected as bound by p53 when using ChIP followed by real-time PCR.
The authors of one report claimed to have found a dinucleotide core code underlying the p53 RE that determines whether a target gene is activated or repressed by p53 binding.Citation337 Based on their finding, the authors re-analyzed 162 published p53 REs and described 20 of them to be falsely assigned as either activators or repressors. However, the discrepancies included re-assignment of well established p53 targets such as BTG2 and PLK2.Citation54,163,337 One explanation of this discrepancy could be that in the experiments p53 REs were tested in an artificial promoter context. Importantly, a recent genome-wide search for a preference of the dinucleotide core in repressed versus activated genes did not yield data to support this model.Citation8 Thus, the dinucleotide core model was disproved, and we refrained from including these results in our analysis.
The third model of direct repression proposes p53 binding to its target promoter via proteins that are general activators of the gene. The transcription factor NF-Y is the most prominent example serving as an adaptor for p53 binding to repressed target promoters.Citation243,255,338 Fourteen genes were described as being controlled by this mechanism (Table S2). Searching in the genome-wide p53 target studies,Citation5-9 only one gene was confirmed in a single study, although the locus of p53 binding does not overlap with the CCAAT-box.Citation6 Furthermore, NF-Y-binding CCAAT-boxes were not found to be enriched at loci bound by p53.Citation8 One might argue that ChIP studies are less efficient if the target protein does not directly bind to the DNA, although the method has been used successfully with other indirectly bound transcription factors such as FoxM1, p130, RB, and LIN9.Citation339-342 However, arguing against indirect ChIPs similarly questions the initial findings that are all based on the same method. Thus, adaptor function of NF-Y recruiting p53 to repress target genes cannot be considered a general mechanism.
Similarly, examining genome-wide data from all 91 p53 targets published as directly repressed, only 5 (5.5%) could be confirmed by at least one genome-wide p53 target study, which resembles the typical false discovery rate of genome-wide studies (Table S2). Yet, 21 (23.1%) were actually observed to be activated instead of being repressed (Expression Score > 0) (Table S2).
Taken together, results from the meta-analysis falsify the models involving direct transcriptional repression through p53. Target genes that were reported to be directly repressed by p53 are either not repressed by p53 after all, not bound by p53 at the proposed p53 RE, or both. This inevitably leads to the conclusion that p53 is not a direct repressor of transcription.
Indirect repression through p53-p21-DREAM or -RB/E2F pathways
Many genes downregulated by p53 are cell cycle genes (Table S1). Researchers argued for a long time whether p53-dependent transcriptional regulation of cell cycle genes requires direct binding of p53 or occurs indirectly. One well known example is the p53-dependent regulation of the CDC25C phosphatase gene. Initially, CDC25C was published to be activated as a direct target of p53.Citation343 Later, the gene was shown to be actually repressed by p53 signaling Citation344 and that p21 is required for indirect downregulation.Citation333 Then, CDC25C was claimed to be both, downregulated by the p53-p21 pathway and by direct interaction of p53 with the promoter.Citation257 Another study supported the model of direct repression by p53,Citation258 while two other reports described indirect downregulation of CDC25C via p107/p130/E2F4.Citation335,345 Thus, over a period of 15 y the proposed mechanism for p53-dependent regulation of CDC25C changed from direct activation of transcription over direct repression to indirect downregulation.
The history of CDC25C regulation shows that in addition to direct also indirect repression of p53 target genes has been suggested. Even prior to these reports, p53-dependent downregulation of many cell cycle genes, including CCNB1, CDC20, CDK1, and NEK2 (; Fig. S3), was shown to depend on p21 (WAF1, CIP1, CDKN1A).Citation330-333,Citation346-350 Similar to p21, RB was suggested to be involved in p53-dependent transcriptional repression of genes such as CCNA2, CCNB1, CDK1, CHEK1, FOXM1, MAD2L1, PCNA, PLK1, and TERT.Citation332,335,348,Citation350-353
Recently, attention has shifted to the p53-p21-DREAM pathway.Citation354-357 The mammalian DREAM complex consists of the pocket proteins p107 or p130, the transcription factors E2F4 or E2F5 and the dimerization partner DP1, as well as the MuvB core composed of RBBP4 and the LIN proteins LIN9, LIN37, LIN52 and LIN54.Citation341,358,359 The DREAM components E2F4 and p107/p130 have repeatedly been reported to participate in p53-dependent downregulation of cell cycle genes.Citation335,345,346,350,352,360,361
In order to evaluate the proposed indirect repression mechanism involving p21, DREAM, or RB/E2F, we searched the literature and found 88 genes that were described to be indirectly regulated by p53 through this mechanism (Table S4).Citation9,98,248,257,Citation330-335,Citation345-357,Citation360-371 Impressively, 83 (94.3%) genes were confirmed as repressed (Expression Score ≦-1) (Table S4). Therefore, in contrast to the direct repression models, the mechanism of indirect repression employing p21, DREAM, or RB/E2F is supported by the genome-wide expression studies.
Next, we evaluated whether genome-wide protein binding to these 88 genes is in agreement with this mechanism. To this end, ChIP-Chip data on DREAM binding Citation341 and ChIP-Seq data on p130 and RB binding Citation340 were used. We found that 79 (89.8%) of the 88 genes were indeed shown to bind DREAM, p130, or RB (Table S4). Furthermore, we evaluated the distribution of DREAM-, p130-, or RB-bound genes across the Expression Score groups (; Table S1). In fact, we find DREAM, p130, or RB binding to be highly enriched at p53-repressed target genes. As an example, 306 (76.7%) of 399 genes that are found to be repressed by p53 in at least 4 expression studies (Expression Score ≦-4) are found to bind DREAM, p130, or RB in proximity of their transcription start site (; Table S1). Interestingly, binding of DREAM or p130 appears to correlate stronger with repression by p53 than binding of RB (). With CCNB2 as an example for cell cycle genes, the complete pathway from induction of DNA damage over activation of p21 through p53 and finally to downregulation of the target was presented as a mechanism that involves binding of DREAM including its component p130 to specific elements in the promoter.Citation355,372 In summary, these data strongly support the notion that DREAM, p130, or RB mediates p53-dependent repression.
Figure 3. Indirect repression through p53-p21-DREAM or -RB/E2F pathways. (A) The percentage of genes bound by DREAM in proximity to their transcriptional start site (TSS) in each Expression Score group is displayed.Citation341,408 The theoretical uniform distribution across the 13 Expression Score groups of genes bound by DREAM is indicated by the blue line (3.5% of 19,736 genes). (B) Displayed for each Expression Score group is the percentage of genes bound by RB in proximity to their TSS.Citation340 The blue line indicates a theoretical uniform distribution of genes bound by RB (4.1% of 19,736 genes) across the 13 Expression Score groups. (C) The percentage of genes bound by p130 in proximity to their TSS is shown for each Expression Score group.Citation340 A theoretical uniform distribution of genes bound by p130 (15.2% of 19,736 genes) across the 13 Expression Score groups is indicated. (D) Compilation of targets displayed in (A-C). The blue line indicates a theoretical uniform distribution of genes bound by DREAM, p130, or RB (16.1% of 19,736 genes) across the 13 Expression Score groups. The red area marks the fraction of genes bound by DREAM, p130, or RB in Expression Score groups −6, −5 and −4 (76.7 % of 399 genes).
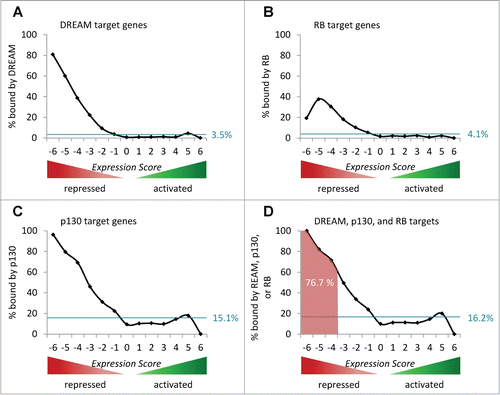
Lately, E2F7 attracted much attention as another possible factor in mediating p53-dependent transcriptional repression of cell cycle genes.Citation98 This report described that G1/S genes such as E2F1, DHFR, RRM2, and E2F8 require E2F7 for p53-dependent downregulation. While the initial study suggested that downregulation of all targets also requires p21,Citation98 it was observed in another study that repression of GBJ2 and E2F8 depends on E2F7 but not on p21.Citation9 However, a more recent study concluded that a contribution of E2F7 to p53-dependent downregulation of target genes such as E2F1 is unlikely.Citation345 Unfortunately, the authors did not discuss these contradictory results although there is an overlap in authorship with the initial study.Citation98,345 Thus, it is difficult to conclude whether E2F7 contributes to p53-dependent gene regulation. Nevertheless, we included E2F7 ChIP-Seq data Citation373 to investigate whether E2F7 target genes are repressed by p53. In general, our data support the possibility that E2F7 participates in p53-dependent transcriptional repression (Fig. S4). However, essentially all E2F7 target genes are also bound by DREAM, p130, or RB (Table S1). This suggests that a p53-dependent repression via E2F7 occurs, if at all, only in conjunction with DREAM, p130, or RB. In conclusion, the results uncover a dominant role of the p53-p21-DREAM/RB pathway in p53-dependent transcriptional repression.
Lessons learned from network perturbations by viral oncoproteins
Oncogenic viruses often interfere with the p53 pathway.Citation21 In addition to targeting p53, many viruses interfere with pocket protein/E2F complexes such as RB/E2F and DREAM. Human papilloma virus (HPV) employs E6 and E7 oncoproteins to selectively target p53 and pocket protein complexes, respectively.Citation374 Importantly, RB/E2F and DREAM are disrupted by HPV E7.Citation357,375,376 Thus, one would expect that the expression of genes directly targeted by p53 is impaired by HPV E6 but not by E7 expression. In contrast, genes targeted by RB/E2F or DREAM downstream of the p53 pathway are expected to be deregulated similarly by HPV E6 and E7. Therefore, we investigated genome-wide expression data after induction of HPV16/18 E6 and HPV16/18 E7 Citation21 (Table S1). Indeed, we find prominent p53 targets such as CDKN1A, MDM2, BAX, FAS, BTG2, and PLK2 to be downregulated upon induction of HPV E6, while they show no regulation or a slight upregulation after induction of HPV E7 (Table S1). Thus, their p53-dependent regulation is not impaired by HPV E7. In contrast, established targets of the p53-p21-DREAM-CDE/CHR pathway such as CCNB2, KIF23, and PLK4 are upregulated upon induction of HPV E6 and are also upregulated by HPV E7 (Table S1). Next, we investigated whether this is a general phenomenon of genes directly activated by p53 in contrast to genes indirectly repressed via the p53-p21-DREAM/RB pathway. We find 469 genes that are upregulated by HPV E6, which display an Expression Score ≦-2, and bind DREAM, p130, or RB (Table S1). Interestingly, solely 14 (3.0%) of these genes display a significantly divergent expression (>2.5-fold or negative ratio) after HPV E6 compared to E7 expression. In contrast, 119 genes are downregulated by HPV E6, which show an Expression Score ≧2, and bind p53. Most interestingly, 50 (42.0%) of these genes display a significantly divergent expression (>2.5-fold or negative ratio) by HPV E6 compared to E7 (Table S1). This 14-fold increase of gene numbers regulated by HPV E7 in addition to E6 among pocket protein target genes is highly significant (P < 10−27) and thus substantiates the model that p53 can directly activate its target genes while p53-dependent repression largely occurs via the p53-p21-DREAM/RB pathway.
Evaluating alternative models of indirect repression
Among the first models trying to explain p53-dependent transcriptional repression, interference of p53 with the TATA-box binding protein (TBP) and its associated factors was proposed.Citation377,378 Another model involves displacement of NF-Y (CBF) binding to CCAAT-boxes by p53, which was observed at the HSPA4 (hsp70) promoter.Citation379 The model was supported by the finding that the NF-Y subunit C interacts with p53 in vitro and in vivo.Citation255 Furthermore, this model was extended toward a possible direct p53-NF-Y-CCAAT repression model with the observation that p53 binds to several CCAAT-box-containing cell cycle genes.Citation255,338 However, as outlined in the chapters above, direct p53 binding to target promoters most likely does not lead to repression but solely to activation. Consistent with this notion, a genome-wide motif search at p53 binding regions did not find TATA-, CCAAT- or GC-boxes to be enriched.Citation8
Yet, several reports describe that transcriptional repression of target genes by p53 is lost upon mutation of CCAAT-boxes. Thus, we searched the literature for reports of indirect repression involving interference of p53 with activating transcription factors such as NF-Y (Table S5).Citation191,309,333,377,Citation379-402 We asked whether target genes are possibly repressed through NF-Y-bound CCAAT-boxes after p53 activation. It was observed that downregulation by p53 is lost after CCAAT elements were destroyed in the promoters of genes such as CCNB2,Citation255,384 CDK1 (CDC2),Citation403 CDC20,Citation334 and TOP2A.Citation397 We and others observed a loss of p53-dependent repression and falsely interpreted that CCAAT-boxes bound by NF-Y are involved. In these reports it was not considered that mutation of CCAAT-boxes essentially inactivates promoters. Thus, the inactive promoters could not be repressed any further. In support of this interpretation, it is well established that NF-Y-bound CCAAT-boxes are essential for activity of the respective genes.Citation404,405 This is further supported by the observation that recruitment of RNA polymerase II depends on intact CCAAT-boxes.Citation406
Many of the cell cycle genes activated through CCAAT-boxes also carry phylogenetically conserved cell cycle-dependent elements (CDE) and cell cycle genes homology regions (CHR) in their promoters which are responsible for cell cycle-dependent transcriptional regulation.Citation405,407 It has been shown that DREAM binds to CDE and CHR elements.Citation408 Importantly, p53-dependent repression of these genes is controlled by DREAM binding to CDE and CHR sites.Citation355-357 Consistently, instead of losing activity by altering the CCAAT-boxes, destruction of CDE and CHR elements leads to derepression of genes such as CCNB2,Citation355 CDK1 (CDC2),Citation349,360 CDC20,Citation363 and TOP2A.Citation349
In addition to NF-Y, Sp1 has also repeatedly been implicated in mediating p53-dependent repression (Table S5). Similar to the observations on NF-Y-mediated regulation, it was described that repression by p53 can depend on Sp1 binding sites, namely GC-boxes. The Survivin (BIRC5) gene served as an example where promoter activity is lost upon GC-box mutation.Citation381,382 As shown for promoters regulated by CCAAT-boxes, also Survivin possesses a phylogenetically conserved CHR downstream of its Sp1 sites.Citation408,409 Considering DREAM-mediated repression via CHRs,Citation355-357 it is likely that also in the case of Survivin the CHR mediates p53-depedent repression. Concordantly, binding of DREAM components was shown to mediate repression of Survivin upon induction of p53.Citation354
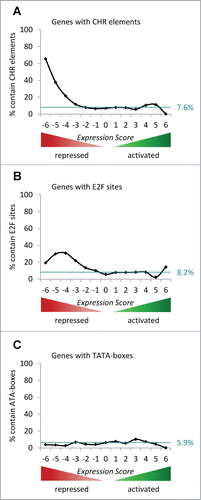
In order to evaluate a possible general function of CCAAT-, GC-, TATA-boxes, CHRs, and E2F sites in p53-dependent transcriptional control, we investigated the distribution of genes harboring such phylogenetically conserved elements across the Expression Score groups. CHR elements which bind DREAM Citation408 and E2F sites that recruit RB/E2F complexes Citation410-412 are also enriched at genes repressed by p53 ( and B). Consistent with this notion, DREAM, p130, and RB binding are strongly enriched at genes downregulated by p53 (). In contrast, TATA-box-containing genes are not accumulated in groups of genes activated or repressed by p53 ().
Figure 4. Genes repressed by p53 are enriched for CHRs which bind DREAM and E2F sites which recruit RB/E2F complexes. (A) The percentage of genes possessing a phylogenetically conserved CHR element in proximity to their TSS in each Expression Score group is displayed. The theoretical uniform distribution across the 13 Expression Score groups of genes with a phylogenetically conserved CHR element is indicated by the blue line (12.1% of 19,736 genes). (B) The percentage of genes harboring a phylogenetically conserved E2F site in proximity to their TSS in each Expression Score group is displayed. The theoretical uniform distribution across the 13 Expression Score groups of genes possessing a phylogenetically conserved E2F sites is indicated by the blue line (8.2% of 19,736 genes). (C) The percentage of genes with a phylogenetically conserved TATA-box in proximity to their TSS in each Expression Score group is displayed. The theoretical uniform distribution across the 13 Expression Score groups of genes holding a phylogenetically conserved TATA-box is indicated by the blue line (5.9% of 19,736 genes).
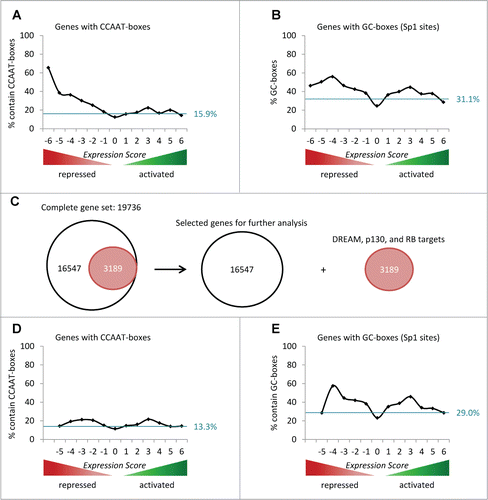
It is established that NF-Y and Sp1 often activate E2F and DREAM/CHR target genes.Citation334,357,405,Citation413-415 Thus, it is not surprising that CCAAT- and GC-boxes are overrepresented at target genes repressed by p53 ( and B). However, when removing all DREAM-, p130-, and RB-bound genes from the analysis, we observe that CCAAT- and GC-box enrichment is essentially lost in the group of genes downregulated compared to genes activated by p53 (–E). These results lead to the conclusion that CCAAT- and GC-boxes do not mediate repression by p53 independently of DREAM, p130, or RB. Still, it is unknown why the transcription factors NF-Y and Sp1 particularly often activate genes that are regulated by pocket protein complexes such as DREAM.
Figure 5. CCAAT- and GC-boxes do not mediate repression by p53 independent of DREAM, p130, or RB. (A) The percentage of genes harboring a phylogenetically conserved CCAAT-box in proximity to their TSS in each Expression Score group is displayed. The theoretical uniform distribution across the 13 Expression Score groups of genes with a phylogenetically conserved CCAAT-box is indicated by the blue line (15.9% of 19,736 genes). (B) The percentage of genes holding a phylogenetically conserved GC-box (Sp1 site) in proximity to their TSS in each Expression Score group is displayed. The theoretical uniform distribution across the 13 Expression Score groups of genes possessing a phylogenetically conserved GC-box (Sp1 site) is indicated by the blue line (31.1% of 19,736 genes). (C) All genes bound by DREAM, p130, or RB (n = 3,189) are removed from the total set of 19,736 genes for further analyses. (D) The percentage of genes harboring a phylogenetically conserved CCAAT-box in proximity to their TSS in each Expression Score group is displayed. The theoretical uniform distribution across the 13 Expression Score groups of genes with a phylogenetically conserved CCAAT-box is indicated by the blue line (13.3% of 16,547 genes). (E) The percentage of genes possessing a phylogenetically conserved GC-box (Sp1 site) in proximity to their TSS in each Expression Score group is displayed. The theoretical uniform distribution across the 13 Expression Score groups of genes holding a phylogenetically conserved GC-box (Sp1 site) is indicated by the blue line (29.0% of 16,547 genes).
Taken together, gene regulation by interference of p53 with activating transcription factors is, if at all, an exception.
ncRNAs in p53's transcriptional network: major players or minor influence?
The most prominent examples of ncRNAs in p53's transcriptional network are mir34a,Citation416 lincRNA-p21,Citation417 and PANDA.Citation418 The original studies on mir34a and lincRNA-p21 were performed in mouse cells.Citation416,417 Here, we are limited to draw conclusions for p53's transcriptional network in human by comparing results of the meta-analysis with major findings from the initial ncRNA studies.
The original study on mir34a explicitly described mir34a-dependent downregulation of Cdk4, Ccne2, and Met via their 3′UTR.Citation416 Indeed, CDK4 and CCNE2 are found to be repressed by p53. However, both genes are also targeted by pocket proteins making it difficult to distinguish the influence of mir34a from that of the pocket proteins (Table S1). In contrast, Met does not bind pocket proteins and even showed the strongest repression by mir34a in the initial study.Citation416 This observation was confirmed by another report.Citation419 Interestingly, human MET appears not to be repressed by p53 (Expression Score = 2) (Table S1). Thus, the influence of mir34a on p53's transcriptional program is not the same between mouse and human. Importantly, experiments on mir34a-c triple knockout mice showed that the mir34 family is not necessary for p53 function.Citation420 Considering these observations, the mir34 family appears to have only a minor influence on p53-dependent transcription.
The initial study on lincRNA-p21 explicitly reported Vcan, Cxcr6, Hus1, Kdm6b (Jmjd3), Zbtb20, Atf2, Rb1, Lpp, Pdlim2, and Usp25 to be repressed by lincRNA-p21 and hnRNP-K in response to p53.Citation417 However, solely ATF2 and USP25 show a slightly negative Expression Score of -1, while VCAN, CXCR6, HUS1, KDM6B, ZBTB20, RB1, LPP, and PDLIM2 are not found to be repressed by p53 in human (Expression Scores ≧ 0) (Table S1). Considering these large discrepancies between mouse and human, it appears unlikely that p53-dependent repression via lincRNA-p21 and hnRNP-K plays a major role in human. Concordantly, the authors of a very recent study investigating lincRNA-p21 knockout mice concluded that lincRNA-p21 unlikely has genome-wide regulatory functions.Citation421
In addition to mir34a and lincRNA-p21, the PANDA ncRNA was observed to be p53-dependently induced.Citation418 PANDA was described to interfere with NF-YA upon induction of p53. However, as outlined above, genes regulated by NF-Y/CCAAT-boxes are not generally repressed by p53 (). Consistently, the authors observed only 224 genes to be induced upon PANDA knockdown,Citation418 although 1412 genes are downregulated after NF-YA was targeted directly by shRNA.Citation422 Moreover, FAS, PIDD (LRDD), APAF1, and BIK were explicitly reported to be downregulated by the p53-PANDA-NF-YA pathway.Citation418 In contrast, expression of FAS, PIDD (LRDD), and APAF1 was not found to be deregulated upon depletion of NF-YA by shRNA, while BIK even was observed to be activated.Citation422 Thus, one can conclude that the p53-PANDA-NF-YA pathway does not generally influence gene transcription, but regulates, if at all, only a few promoters in certain cell types. In the initial study, PANDA was shown to fine-tune the p53-dependent transcription of pro-apoptotic target genes in human fetal fibroblasts.Citation418
Taken together, a major contribution of well known ncRNAs to p53's transcriptional program is not evident. The transcriptional influence of the ncRNAs discussed above appears to be, if at all, limited to fine-tuning expression of a few genes in certain cell types.
Conclusions and Perspective
Our results resolve the longstanding question on how p53 binding can activate one target gene and repress another. Most surprisingly, results from the computational meta-analysis do not support models involving direct transcriptional repression through p53. Experimental validation supports the conclusions from the meta-analysis. Thus, the previously reported regulation of several target genes appear questionable (Table S2). Generally, binding and regulation are not necessarily cause and consequence, considering that not every binding event leads to regulation and that regulation can be indirect.
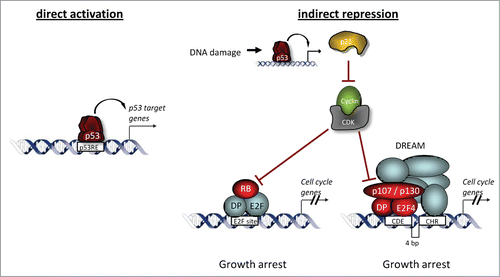
As an alternative to direct repression, the results show that p53-dependent repression occurs indirectly and is largely mediated by activation of the p53-p21-DREAM/RB pathway (). Other reported indirect pathways such as ncRNAs appear to be, if at all, either an exception or to merely mediate fine-tuning of p53's transcriptional program.
Figure 6. The tumor suppressor p53 is not a direct repressor of transcription, it solely activates its target genes upon binding to DNA. In order to activate transcription, the p53 tetramer binds to the p53 RE of its target gene. The transcription factor p53 acts as repressor by activation of the p53-p21-DREAM/RB pathway ultimately leading to indirect p53-dependent transcriptional repression.
In summary, with direct activation and indirect downregulation via the p53-p21-DREAM/RB pathway only 2 out of the previously reported 6 major mechanisms of p53-dependent regulation are supported by the meta-analysis (). Future research will have to show whether there are still other mechanisms that are of general importance mediating p53-dependent transcription.
Materials and Methods
Computational meta-analysis on binding and regulation by p53
Expression data on known protein-coding genes were extracted from 6 studies on p53-dependent regulation.Citation7,Citation19-23 The expression values of the analyzed genes were compiled and classified into repressed (−1), induced (+1), and not-regulated (0) by p53. For every gene the Expression Score was calculated as the sum of the classifications of the individual studies. Expression Scores range from −6 to 6, where “6” means found as induced by p53 in all studies and “−6” means classified as repressed by p53 in all 6 studies. Thus, the Expression Score describes the direction of regulation as well as the confidence of the classification (Table S1).
Due to the fact that the data originate from different sources, all studies must be evaluated and filtered with individual thresholds for log-fold change and/or p-values. We aimed not to alter criteria that were used in the original studies. However, if a study yielded many more regulated genes compared to a related study, we slightly adjusted thresholds in p-values and expression fold-changes to yield data sets of similar size. The following thresholds were used for the 6 studies: For the data from the study by Böhlig et al. (kindly provided by Levin Böhlig) the p-value must not exceed 0.05, the log-fold change has to exceed 1 to be classified as "induced" and undercut −1 for classification "repressed".Citation19 For the data from the Nikulenkov et al. study (kindly provided by Galina Selivanova) 0.5 and -0.5 are used as thresholds of the log-fold change and 0.05 of the p-value.Citation7 The data on differentially expressed genes after expression of HPV-16 E6 or HPV-18 E6 from the study by Rozenblatt-Rosen et al. (GSE38467) were filtered with log-fold change of 0.25 and an adjusted p-value of 0.05.Citation21 The data from the report by Kracikova et al. (GSE30753) were filtered solely with an adjusted p-value of 0.05, the same criteria were used in the original study.Citation22 The thresholds for the Goldstein et al. data (GSE30137) were set to an absolute log-fold change of 0.5 and an adjusted p-value of 0.05.Citation23 The expression data from Rashi-Elkeles et al. represent a meta-study on different data sets. For filtering, the sum of the Z-values from the individual studies is used; larger than 10 is counted as "increased", less than -10 is counted as "repressed".Citation20
For every gene, the genomic location is shown, i.e. chromosome, strand, transcription start and stop, and start and stop of the coding sequence. Primarily, the annotations of the canonical transcripts for the human genome version hg19 were taken from the UCSC Genome Browser database.Citation423 Only in cases where no annotation was available at the UCSC Genome Browser database, the annotation from Ensembl human genome version GRCh37 was used.Citation424 Additionally, mappings to the different database identifiers are provided if available including UCSC canonical transcript ID, Ensembl gene ID, HUGO gene symbol, and Affymetrix microarray IDs (Table S1).
ChIP peaks from 6 genome-wide p53 binding studies were annotated 25 kb around the TSS.Citation6-9,Citation24,25 In 4 of the 6 studies, ChIP peaks originate from several experiments. In case of 2 data sets in one report, all ChIP peaks were included in our analysis.Citation6,8 To reduce the number of false positive annotated p53 ChIP peaks, we filtered for peaks which occurred in at least 2 data sets in cases where 3 or 4 experiments were performed.Citation7,9 Furthermore, ChIP peaks occurring in more than one experiment from the same study were merged into one peak using BEDTools.Citation425 All ChIP peaks from the 6 studies that overlap by at least one base pair were merged. From this set of p53 ChIP peaks, only those peaks were selected for further analysis that were found in at least 2 studies. For each gene, the location of the p53 peaks is annotated for each study as well as the p53 ChIP Score showing the number studies for which peaks in the promoter of this gene were found (Table S1).
Search for phylogenetically conserved binding motifs
Several binding sites were annotated in the promoter regions of the genes. CHR (TTTGAA, TTTAAA, CTTGAA, TAGGAA), E2F (TTSSSSS), TATA (TATATA, TATAA), CCAAT (CCAAT), and SP1 (GGGCGG, GGCGGG) sites were searched in the region of 200 bp around the TSS on both strands that were not extended into the coding sequence or genes located upstream of the TSS. PhastCons conservation scores Citation426 obtained from the multiz46 alignment of placental mammalia Citation427 were used to calculate average phylogenetic sequence conservation. Only those hits were annotated that have an average PhastCons conservation score of at least 0.8 (Table S1).
Meta-analysis of “default targets”
An extensive literature search for potential direct p53 target genes was performed that started with 2 reviews Citation13,428 and includes about 300 reports in total (Table S2). We included all target genes that were reported as differentially expressed upon p53 induction and bound by p53 in proximity of their locus (Table S2). All reported p53 target genes were compiled and classified as repressed (−1) or activated (+1) by p53 (Table S3).
Additionally, we included potential p53 target genes from genome-wide studies that combined p53 binding data (ChIP-PET, ChIP-chip, ChIP-seq) with p53-dependent expression data from microarray analyses.Citation5-9 Two studies contained 2 data sets each from ChIP-seq combined with expression data following 2 different treatments to activate p53.Citation6,Citation8 We included the 2 data sets of each study separately in our analysis. As both data sets originate from experiments with similar conditions, we assigned a lower score (0.75) when a gene was found as p53 target gene in these data sets in order to not overweigh the study's influence on our meta-analysis (Table S3). Next, we combined 2 genome-wide p53 binding data sets,Citation24,25 that previously had not been compared to expression data, with 6 genome-wide p53 dependent expression studies.Citation7,Citation19-23 From this combination, we included genes as potential p53 targets that were identified as bound by p53 in at least one of the binding studies and as regulated in at least one expression study, assigning a score of 0.25 for each study in which the gene was identified as bound or regulated by p53 (Tables S1 and S3).
For every gene the Default Target Score was calculated as the sum of the scores from the individual data sets. Thus, it represents the direction of regulation as well as the confidence of the classification. We considered a gene as a “default target” that was reported in at least 3 data sets, which corresponds to a Default Target Score greater than 2 (Table S3).
DREAM, p130, RB, and E2F7 binding data
The promoter regions 200 bp upstream and downstream from the TSS, but not extending into the coding sequence or genes located upstream of the TSS, were overlayed with peaks from 4 ChIP-chip experiments measuring binding of E2F4, p130, LIN9, and LIN54 proteins as indicators for DREAM complex binding Citation341 as described previously.Citation408 ChIP-seq peaks for DNA bound by p130, RB,Citation340 and E2F7 Citation373 were overlayed with an extended promoter region of 1000 bp around the TSS. Again, the promoter regions were truncated to not overlap with the coding sequence or genes located upstream of the TSS. ChIP peaks for p130 and RB were restricted to those with a false discovery rate ≤ 0.1 (Table S1).
Cell culture, FACS, chromatin immunoprecipitation, RNA extraction, and semi-quantitative real-time PCR
Experiments were performed as described previously.Citation357
Primer
Real-time PCR primer for ChIP analyses
GAPDHS: for 5′-AGACCAGCCTGAGCAAAAGA-3′, rev 5′-CTAGGCTGGAGTGCAGTGGT-3′;Citation356, Citation357 CDKN1A: for 5′-CTGAGCCTCCCTCCATCC-3′, rev 5′-GAGGTCTCCTGTCTCCTACCATC-3′;Citation356, Citation357 MDM2: for 5′-TCGGGTCACTAGTGTGAACG-3′, rev 5′-TGAACACAGCTGGGAAAATG-3′; ABCB1: for 5′-TTATCCCAGTACCAGAGGAGGA-3′, rev 5′-TGCTTTGGAGCCATAGTCAT-3′; BCL2: for 5′-ATCCTTCCCAGAGGAAAAGC-3′, rev 5′-ATCAAGTGTTCCGCGTGATT-3′; BNIP3: for 5′-AGCGTTTCTGGGGCGCACCTTG-3′, rev 5′-GGGACTGGGAGGCACTTTTCAGAGGA-3′;Citation251 CCNB1: for 5′-CCTGATTTTCCCATGAGAGG-3′, rev 5′-GGATCACACATTAGCAACGGG-3′;Citation254 CD44: for 5′-TTTACGGTTCGGTCATCCTC-3′, rev 5′-TGCTCTGCTGAGGCTGTAAA-3′;Citation262 CDC20: for 5′-TAAAGCCCCAAGGGGATAAG-3′, rev 5′-CGTGTGTTTGTCTCGTTTGC-3′; CDK1: for 5′-AACTGTGCCAATGCTGGGAG-3′, rev 5′-AGCCAGCTTTGAAGCCAAGT-3′;Citation258 CRYZ: for 5′-TCCACCATGATTGTGAGACC-3′, rev 5′-CAAACATTTACCTGACACCCA-3′;Citation260 HSPA8: for 5′-TGGGTAGATGGGTCCTTCAT-3′, rev 5′-AATAGTGCCCATCACCTCCT-3′;Citation260 ID2: for 5′-GAACGCGGAAGAACCAAG-3′, rev 5′-GGCTCGGCTCAGAATGAA-3′; LASP1: for 5′-AGCGTTCAGGAGGATCCAA-3′, rev 5′-AGCGCTCTCAGGCTGACT-3′; MAD1L1: for 5′-ACTGGGAAGGTAGCCTAGTAGCATA-3′, rev 5′-AGCCTCCTCGGACAAACTTGC-3′;Citation260 ME1: for 5′-GGAAACTGCACCAACTGTGA-3′, rev 5′-TAAACATGCGGGTTGGCTAT-3′; ME2-RE1: for 5′-GTTGCCCAGGCTGGAGTG-3′, rev 5′-CTGTAATCCCAGCACTTT-3′;Citation285 ME2-RE3: for 5′-AAGTTGGAGACCACCCTGTG-3′, rev 5′-GCTAGAGTGCAGTGGCATGA-3′; ME3: for 5′-GTTGCGATCCCGTGGCTG-3′, rev 5′-ACCGCAGGTCAGACTGAC-3′;Citation285 NEK2: for 5′-TGCAACCCCATGCTCTGTTAC-3′, rev 5′-TCACGCCTATAATCCTAGCAC-3′;Citation289 PTK2: for 5′-CTCCAACCTCGCCTTTTGC-3′, rev 5′-GGGACTTAGAAGTCCACTGG-3′;Citation306 TPT1: for 5′-TAGGGAGCGCCCCGAGAGTT-3′, rev 5′-GTGACGTGGCACGAAGAG-3′.Citation320
Real-time PCR primer for expression analyses
GAPDH: for 5′-GACCCCTTCATTGACCTCAAC-3′, rev 5′-CACGACGTACTCAGCGCC-3′; U6: for 5′-AACGCTTCACGAATTTGCGT-3′, rev 5′-CTCGCTTCGGCAGCACA-3′;Citation357 L7: for 5′-GCACTATCACAAGGAATATAGGCAG-3′, rev 5′-CCCATGCAATATATGGCTCTAC-3′;Citation356 CDKN1A: for 5′-GGAAGACCATGTGGACCTGT-3′, rev 5′-GGATTAGGGCTTCCTCTTGG-3′; MDM2: for 5′-GTGAATCTACAGGGACGCCA-3′, rev 5′-CTGATCCAACCAATCACCTGAA-3′;Citation351 PPM1D: for 5′-CAACTGCCAGTGTGGTCATC-3′, rev 5′-CGATTCACCCCAGACTTGTT-3′; ABCB1: for 5′-CATGATGCTGGTGTTTGGAG-3′, rev 5′-AGGCACCAAAATGAAACCTG-3′; BCL2: for 5′-ACTTGTGGCCCAGATAGGCACCCAG-3′, rev 5′-CGACTTCGCCGAGATGTCCAGCCAG-3′;Citation245 BNIP3: for 5′-TCCTCTTTAAACACCCGAAGCGCA-3′, rev 5′-ATCCGATGGCCAGCAAATGAGAGA-3′;Citation251 CCNB1: for 5′-AAGAGCTTTAAACTTTGGTCTGGG-3′, rev 5′-CTTTGTAATGCCTTGATTTACCATG-3′;Citation254 CD44: for 5′-CCACGTGGAGAAAAATGGTC-3′, rev 5′-CATTGGGCAGGTCTGTGAC-3′;Citation262 CDC20: for 5′-CGCCAACCGATCCCACAG-3′, rev 5′-CAGGTTCAAAGCCCAGGC-3′;Citation328 CDK1: for 5′-TGGGGTCAGCTCGTTACTCA-3′, rev 5′-CACTTCTGGCCACACTTCATTTA-3′;Citation258 CRYZ: for 5′-GAGTGATAGTTGTTGGCAGCAGAG-3′, rev 5′-TGCTGAAATTCCTCCTTGGTTG-3′;Citation260 HSPA8: for 5′-GCCGTTTGAGCAAGGAAGACA-3′, rev 5′-CAGCAGTCTGATTCTTATCAAGCC-3′;Citation260 ID2: for 5′-TCAGCCTGCATCACCAGAGA-3′, rev 5′-CTGCAAGGACAGGATGCTGAT-3′;Citation275 LASP1: for 5′-GTATCCCACGGAGAAGGTGA-3′, rev 5′-TGTCTGCCACTACGCTGAAA-3′;Citation281 MAD1L1: for 5′-CAGGGTGACTATGACCAGAGCAG-3′, rev 5′-TCAGCTCTGCCACCTCCTTG-3′;Citation260 ME1: for 5′-GGATTGCACACCTGATTGTG-3′, rev 5′-TCTTCATGTTCATGGGCAAA-3′; ME2: for 5′-ATGGGCTTGTACCAGAAACG-3′, rev 5′-TGCTGCAAGAAGACCTGCTA-3′; ME3: for 5′-CAGCAGAGTGACCTGGACAA-3′, rev 5′-CTTCTGGCCAAGAATTCAGC-3′; NEK2: for 5′-AGTGCAAGGACCTGAAGAAAAG-3′, rev 5′-TCAATATCTGACAGGGCTTGAG-3′;Citation289 PTK2: for 5′-GTGCTCTTGGTTCAAGCTGGAT-3′, rev 5′-ACTTGAGTGAAGTCAGCAAGATGTGT-3′;Citation306 TPT1: for 5′-GATCGCGGACGGGTTGT-3′, rev 5′-TTCAGCGGAGGCATTTCC-3′.Citation320
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Author Contributions
MF conceived the study. LS and MF performed the computational analyses. MF performed the experiments. KE supervised the study. MF and KE, including segments provided by LS, wrote the manuscript. All authors read and approved the final article.
949083_Supplemental_Materials.zip
Download Zip (11.3 MB)Acknowledgments
We are indebted to Carola Koschke and Andrea Rothe for expert technical assistance and Andreas Lösche and Kathrin Jäger at the IZKF Leipzig core unit for performing FACS analyses. We thank Levin Böhlig and Galina Selivanova for providing pre-analyzed genome-wide gene expression data, Thorsten Stiewe for pre-analyzed hg19 ChIP data, Bert Vogelstein for the kind gift of HCT116 cell lines, Gerd A. Müller and Marianne Quaas for helpful discussions, and Christine E. Engeland for critical reading of the manuscript.
Supplemental Material
Supplemental data for this article can be accessed on the publisher's website.
Additional information
Funding
References
- Lane DP. Benchimol S. p53: oncogene or anti-oncogene? Genes Dev 1990; 4:1-8; PMID:2137806
- Levine AJ, Oren M. The first 30 years of p53: growing ever more complex. Nat Rev Cancer 2009; 9:749-58; http://dx.doi.org/10.1038/nrc2723; PMID:19776744
- Ko LJ, Prives C. p53: puzzle and paradigm. Genes Dev 1996; 10:1054-72; PMID:8654922
- Menendez D, Inga A, Resnick MA. The expanding universe of p53 targets. Nat Rev Cancer 2009; 9:724-37; PMID:19776742; http://dx.doi.org/10.1038/nrc2730
- Wei CL, Wu Q, Vega VB, Chiu KP, Ng P, Zhang T, Shahab A, Yong HC, Fu Y, Weng Z, et al. A global map of p53 transcription-factor binding sites in the human genome. Cell 2006; 124:207-19; PMID:16413492
- Smeenk L, van Heeringen SJ, Koeppel M, Gilbert B, Janssen-Megens E, Stunnenberg HG, Lohrum M. Role of p53 serine 46 in p53 target gene regulation. PLoS One 2011; 6(3):e17574; PMID:21394211; http://dx.doi.org/10.1371/journal.pone.0017574
- Nikulenkov F, Spinnler C, Li H, Tonelli C, Shi Y, Turunen M, Kivioja T, Ignatiev I, Kel A, Taipale J, et al. Insights into p53 transcriptional function via genome-wide chromatin occupancy and gene expression analysis. Cell Death Differ 2012; 19:1992-2002; PMID:22790872; http://dx.doi.org/10.1038/cdd.2012.89
- Menendez D, Nguyen TA, Freudenberg JM, Mathew VJ, Anderson CW, Jothi R, Resnick MA. Diverse stresses dramatically alter genome-wide p53 binding and transactivation landscape in human cancer cells. Nucleic Acids Res 2013; 41:7286-301; http://dx.doi.org/10.1093/nar/gkt504
- Schlereth K, Heyl C, Krampitz AM, Mernberger M, Finkernagel F, Scharfe M, Jarek M, Leich E, Rosenwald A, Stiewe T. Characterization of the p53 cistrome - DNA binding cooperativity dissects p53‘s tumor suppressor functions. Plos Genetics 2013; 9(8):e1003726; PMID:23966881; http://dx.doi.org/10.1371/journal.pgen.1003726
- Graur D, Zheng Y, Price N, Azevedo RB, Zufall RA, Elhaik E. On the immortality of television sets: ``function'' in the human genome according to the evolution-free gospel of ENCODE. Genome Biol Evol 2013; 5:578-90; PMID:23431001; http://dx.doi.org/10.1093/gbe/evt028
- Ioannidis JP. Why most published research findings are false. PLoS Med 2005; 2:e124; PMID:16060722
- Fanelli D. Negative results are disappearing from most disciplines and countries. Scientometrics 2012; 90:891-904; http://dx.doi.org/10.1007/s11192-011-0494-7
- Riley T, Sontag E, Chen P, Levine A. Transcriptional control of human p53-regulated genes. Nat Rev Mol Cell Biol 2008; 9:402-12; PMID:18431400; http://dx.doi.org/10.1038/nrm2395
- Vousden KH, Prives C. Blinded by the Light: The Growing Complexity of p53. Cell 2009; 137:413-31; PMID:19410540; http://dx.doi.org/10.1016/j.cell.2009.04.037
- Beckerman R, Prives C. Transcriptional regulation by p53. Cold Spring Harb Perspect Biol 2010; 2:a000935; PMID:20679336; http://dx.doi.org/10.1101/cshperspect.a000935
- Böhlig L, Rother K. One function–multiple mechanisms: the manifold activities of p53 as a transcriptional repressor. J Biomed Biotechnol 2011; 2011:464916; PMID:21436991; http://dx.doi.org/10.1155/2011/464916
- Rinn JL, Huarte M. To repress or not to repress: this is the guardian's question. Trends Cell Biol 2011; 21:344-53; PMID:21601459; http://dx.doi.org/10.1016/j.tcb.2011.04.002
- Leek JT, Scharpf RB, Bravo HC, Simcha D, Langmead B, Johnson WE, Geman D, Baggerly K, Irizarry RA. Tackling the widespread and critical impact of batch effects in high-throughput data. Nat Rev Genet 2010; 11 733-9; PMID:20838408; http://dx.doi.org/10.1038/nrg2825
- Böhlig L, Friedrich M, Engeland K. p53 activates the PANK1miRNA-107 gene leading to downregulation of CDK6 and p130 cell cycle proteins. Nucleic Acids Res 2011; 39:440-53; PMID:20833636; http://dx.doi.org/10.1093/nar/gkq796
- Rashi-Elkeles S, Elkon R, Shavit S, Lerenthal Y, Linhart C, Kupershtein A, Amariglio N, Rechavi G, Shamir R, Shiloh Y. Transcriptional modulation induced by ionizing radiation: p53 remains a central player. Molecular Oncology 2011; 5:336-48; PMID:21795128; http://dx.doi.org/10.1016/j.molonc.2011.06.004
- Rozenblatt-Rosen O, Deo RC, Padi M, Adelmant G, Calderwood MA, Rolland T, Grace M, Dricot A, Askenazi M, Tavares M, et al. Interpreting cancer genomes using systematic host network perturbations by tumour virus proteins. Nature 2012; 487:491-5; PMID:22810586; http://dx.doi.org/10.1038/nature11288
- Kracikova M, Akiri G, George A, Sachidanandam R, Aaronson SA. A threshold mechanism mediates p53 cell fate decision between growth arrest and apoptosis. Cell Death and Differentiation 2013; 20:576-88; PMID:23306555; http://dx.doi.org/10.1038/cdd.2012.155
- Goldstein I, Ezra O, Rivlin N, Molchadsky A, Madar S, Goldfinger N, Rotter V. p53 a novel regulator of lipid metabolism pathways. J Hepatol 2012; 56:656-62; PMID:22037227; http://dx.doi.org/10.1016/j.jhep.2011.08.022
- Smeenk L, van Heeringen SJ, Koeppel M, van Driel MA, Bartels SJ, Akkers RC, Denissov S, Stunnenberg HG, Lohrum M. Characterization of genome-wide p53-binding sites upon stress response. Nucleic Acids Res 2008; 36:3639-54; PMID:18474530; http://dx.doi.org/10.1093/nar/gkn232
- Botcheva K, McCorkle SR, McCombie WR, Dunn JJ, Anderson CW. Distinct p53 genomic binding patterns in normal and cancer-derived human cells. Cell Cycle 2011; 10:4237-49; PMID:22127205; http://dx.doi.org/10.4161/cc.10.24.18383
- Kenzelmann BD, Spano Mello S, Bieging KT, Jiang D, Dusek RL, Brady CA, Sidow A, Attardi LD. Global genomic profiling reveals an extensive p53-regulated autophagy program contributing to key p53 responses. Genes Dev 2013; 27:1016-31; PMID:23651856; http://dx.doi.org/10.1101/gad.212282.112
- Rashi-Elkeles S, Warnatz HJ, Elkon R>, Kupershtein A, Chobod Y, Paz A, Amstislavskiy V, Sultan M, Safer H, Nietfeld W, et al. Parallel profiling of the transcriptome cistrome and epigenome in the cellular response to ionizing radiation. Sci Signal 2014; 7:rs3; PMID:24825921; http://dx.doi.org/10.1126/scisignal.2005032
- (a) Allen MA, Andrysik Z, Dengler VL, Mellert HS, Guarnieri A, Freeman JA, Sullivan KD, Galbraith MD, Luo X, Kraus WL, et al. Global analysis of p53-regulated transcription identifies its direct targets and unexpected regulatory mechanisms. Elife 2014; 3:e02200; PMID:24867637; http://dx.doi.org/10.7554/eLife.02200; (b) Janky R, Verfaillie A, Imrichová H, Van de Sande B, Standaert L, et al. iRegulon: From a gene list to a gene regulatory network using large motif and track collections. PLoS Comput Biol 2014; 10(7):e1003731; PMID:25058159; http://dx.doi.org/10.1371/journal.pcbi.100373
- Mathieu MC, Lapierre I, Brault K, Raymond M. Aromatic hydrocarbon receptor (AhR)center dot AhR nuclear translocator- and p53-mediated induction of the murine multidrug resistance mdr1 gene by 3-methylcholanthrene and benzo(a)pyrene in hepatoma cells. J Biol Chem 2001; 276:4819-27; PMID:11096091
- Comer KA, Dennis PA, Armstrong L, Catino JJ, Kastan MB, Kumar CC. Human smooth muscle alpha-actin gene is a transcriptional target of the p53 tumor suppressor protein. Oncogene 1998; 16:1299-1308; PMID:9546431
- Kawase T, Ichikawa H, Ohta T, Nozaki N, Tashiro F, Ohki R, Taya Y. p53 target gene AEN is a nuclear exonuclease required for p53-dependent apoptosis. Oncogene 2008; 27:3797-810; PMID:18264133; http://dx.doi.org/10.1038/onc.2008.32
- Pierzchalski P, Reiss K, Cheng W, Cirielli C, Kajstura J, Nitahara JA, Rizk M, Capogrossi MC, Anversa P. p53 induces myocyte apoptosis via the activation of the renin-angiotensin system. Exp Cell Res 1997; 234:57-65; PMID:9223370
- Stambolsky P, Weisz L, Shats I, Klein Y, Goldfinger N, Oren M, Rotter V. Regulation of AIF expression by p53. Cell Death Differ 2006; 13:2140-49; PMID:16729031
- Wu M, Xu LG, Su T, Tian Y, Zhai Z, Shu HB. AMID is a p53-inducible gene downregulated in tumors. Oncogene 2004; 23:6815-9; PMID:15273740
- Lecona E, Barrasa JI, Olmo N, Llorente B, Turnay J, Lizarbe MA. Upregulation of annexin A1 expression by butyrate in human colon adenocarcinoma cells: Role of p53 NF-Y and p38 mitogen-activated protein kinase. Mol Cell Biol 2008; 28:4665-74; PMID:18541673; http://dx.doi.org/10.1128/MCB.00650-07
- Robles AI, Bemmels NA, Foraker AB, Harris CC. APAF-1 is a transcriptional target of p53 in DNA damage-induced apoptosis. Cancer Res 2001; 61:6660-4; PMID:11559530
- Fortin A, Cregan SP, MacLaurin JG, Kushwaha N, Hickman ES, Thompson CS, Hakim A, Albert PR, Cecconi F, Helin K, et al. APAF1 is a key transcriptional target for p53 in the regulation of neuronal cell death. J Cell Biol 2001; 155:207-16; PMID:11591730
- Rozenfeld-Granot G, Krishnamurthy J, Kannan K, Toren A, Amariglio N, Givol D, Rechavi G. A positive feedback mechanism in the transcriptional activation of Apaf-1 by p53 and the coactivator Zac-1. Oncogene 2002; 21:1469-76; PMID:11896574
- Vrba L, Junk DJ, Novak P, Futscher BW. p53 induces distinct epigenetic states at its direct target promoters. BMC Genomics 2008; 9:486; PMID:18922183; http://dx.doi.org/10.1186/1471-2164-9-486
- Wallace DM, Cotter TG. Histone Deacetylase Activity in Conjunction With E2F-1 and p53 Regulates Apaf-1 Expression in 661 W Cells and the Retina. J Neurosci Res 2009; 87 887-905; PMID:18951482; http://dx.doi.org/10.1002/jnr.21910
- Jaiswal AS, Narayan S. p53-dependent transcriptional regulation of the APC promoter in colon cancer cells treated with DNA alkylating agents. J Biol Chem 2001; 276:18193-9; PMID:11279192
- Ma KW, Araki K, Ichwan SJ, Suganuma T, Tamamori-Adachi M, Ikeda MA. E2FBP1DRIL1 an AT-rich interaction domain-family transcription factor is regulated by p53. Mol Cancer Res 2003; 1:438-44; PMID:12692263
- Zhang C, Gao C, Kawauchi J, Hashimoto Y, Tsuchida N, Kitajima S. Transcriptional activation of the human stress-inducible transcriptional repressor ATF3 gene promoter by p53. Biochem Biophys Res Commun 2002; 297:1302-10; PMID:12372430
- Graupner V, Alexander E, Overkamp T, Rothfuss O, De Laurenzi V, Gillissen BF, Daniel PT, Schulze-Osthoff K, Essmann F. Differential regulation of the proapoptotic multidomain protein Bak by p53 and p73 at the promoter level. Cell Death Differ 2011; 18:1130-9; PMID:21233848; http://dx.doi.org/10.1038/cdd.2010.179
- Miyashita T, Krajewski S, Krajewska M, Wang HG, Lin HK, Liebermann DA, Hoffman B, Reed JC. Tumor suppressor p53 is a regulator of bcl-2 and bax gene expression in vitro and in vivo. Oncogene 1994; 9:1799-805; PMID:8183579
- Thornborrow EC, Patel S, Mastropietro AE, Schwartzfarb EM, Manfredi JJ. A conserved intronic response element mediates direct p53-dependent transcriptional activation of both the human and murine bax genes. Oncogene 2002; 21:990-9; PMID:11850816
- Nakano K, Vousden KH. PUMA a novel proapoptotic gene is induced by p53. Mol Cell 2001; 7:683-94; PMID:11463392
- Miled C, Pontoglio M, Garbay S, Yaniv M, Weitzman JB. A genomic map of p53 binding sites identifies novel p53 targets involved in an apoptotic network. Cancer Res 2005; 65:5096-104; PMID:15958553
- Margalit O, Amram H, Amariglio N, Simon AJ, Shaklai S, Granot G, Minsky N, Shimoni A, Harmelin A, Givol D, et al. BCL6 is regulated by p53 through a response element frequently disrupted in B-cell non-Hodgkin lymphoma. Blood 2006; 107:1599-607; PMID:16249378
- Saifudeen Z, Du H, Dipp S, El-Dahr SS. The bradykinin type 2 receptor is a target for p53-mediated transcriptional activation. J Biol Chem 2000; 275:15557-62; PMID:10748162
- Saifudeen Z, Dipp S, Fan H, El-Dahr SS. Combinatorial control of the bradykinin B2 receptor promoter by p53 CREB KLF-4 and CBP: implications for terminal nephron differentiation. Am J Physiol-Renal Physiol 2005; 288:F899-F909; PMID:15632413
- Sax JK, Fei P, Murphy ME, Bernhard E, Korsmeyer SJ, El-Deiry WS. BID regulation by p53 contributes to chemosensitivity. Nat Cell Biol 2002; 4:842-9; PMID:12402042
- Fei PW, Wang W, Kim SH, Wang S, Burns TF, Sax JK, Buzzai M, Dicker DT, McKenna WG, Bernhard EJ, et al. Bnip3L is induced by p53 under hypoxia and its knockdown promotes tumor growth. Cancer Cell 2004; 6:597-609; PMID:15607964
- Rouault JP, Falette N, Guéhenneux F, Guillot C, Rimokh R, Wang Q, Berthet C, Moyret-Lalle C, Savatier P, Pain B, et al. Identification of BTG2 an antiproliferative p53-dependent component of the DNA damage cellular response pathway. Nat Genet 1996; 14:482-6; PMID:8944033
- Duriez C, Falette N, Audoynaud C, Moyret-Lalle C, Bensaad K, Courtois S, Wang Q, Soussi T, Puisieux A. The human BTG2TIS21PC3 gene: genomic structure transcriptional regulation and evaluation as a candidate tumor suppressor gene. Gene 2002; 282:207-14; PMID:11814693
- Ou YH, Chung PH, Hsu FF, Sun TP, Chang WY, Shieh SY. The candidate tumor suppressor BTG3 is a transcriptional target of p53 that inhibits E2F1. EMBO J 2007; 26:3968-80; PMID:17690688
- Jen KY, Cheung VG. Identification of novel p53 target genes in ionizing radiation response. Cancer Res 2005; 65:7666-73; PMID:16140933
- Bensaad K, Tsuruta A, Selak MA, Vidal MN, Nakano K, Bartrons R, Gottlieb E, Vousden KH. TIGAR a p53-inducible regulator of glycolysis and apoptosis. Cell 2006; 126:107-20; PMID:16839880
- Saigusa K, Imoto I, Tanikawa C, Aoyagi M, Ohno K, Nakamura Y, Inazawa J. RGC32 a novel p53-inducible gene is located on centrosomes during mitosis and results in G2M arrest. Oncogene 2007; 26:1110-21; PMID:17146433
- Brown L, Ongusaha PP, Kim HG, Nuti S, Mandinova A, Lee JW, Khosravi-Far R, Aaronson SA, Lee SW. CDIP a novel pro-apoptotic gene regulates TNF alpha-mediated apoptosis in a p53-dependent manner. EMBO J 2007; 26:3410-22; PMID:17599062
- Gupta S, Radha V, Furukawa Y, Swarup G. Direct transcriptional activation of human caspase-1 by tumor suppressor p53. J Biol Chem 2001; 276:10585-8; PMID:11278253
- Rikhof B, Corn PG, el-Deiry WS. Caspase 10 levels are increased following DNA damage in a p53-dependent manner. Cancer Biol Ther 2003; 2:707-12; PMID:14688482
- MacLachan TK, el-Deiry WS. Apoptotic threshold is lowered by p53 transactivation of caspase-6. Proc Natl Acad Sci USA 2002; 99:9492-7; PMID:12089322
- Bist A, Fielding CJ, Fielding PE. p53 regulates caveolin gene transcription cell cholesterol and growth by a novel mechanism. Biochem 2000; 39:1966-72; PMID:10684646
- Yu X, Riley T, Levine AJ. The regulation of the endosomal compartment by p53 the tumor suppressor gene. FEBS J 2009; 276:2201-12; PMID:19302216; http://dx.doi.org/10.1111/j.1742-4658.2009.06949.x
- Okamoto K, Beach D. Cyclin-G Is A transcriptional target of the P53 tumor-suppressor protein. EMBO J 1994; 13:4816-22; PMID:7957050
- Zauberman A, Lupo A, Oren M. Identification of P53 target genes through immune selection of genomic DNA - the cyclin-G gene contains 2 distinct P53 binding-sites. Oncogene 1995; 10:2361-6; PMID:7784084
- Mori T, Anazawa Y, Matsui K, Fukuda S, Nakamura Y, Arakawa H. Cyclin K as a direct transcriptional target of the p53 tumor suppressor. Neoplasia 2002; 4:268-74; PMID:11988847
- Mashimo T, Watabe M, Hirota S, Hosobe S, Miura K, Tegtmeyer PJ, Rinker-Shaeffer CW, Watabe K. The expression of the KAI1 gene a tumor metastasis suppressor is directly activated by p53. Proc Natl Acad Sci USA 1998; 95:11307-11; PMID:9736732
- Resnick-Silverman L, St Clair S, Maurer M, Zhao K, Manfredi JJ. Identification of a novel class of genomic DNA-binding sites suggests a mechanism for selectivity in target gene activation by the tumor suppressor protein p53. Gen Dev 1998; 12:2102-7; PMID:9679054
- el-Deiry WS, Kern SE, Pietenpol JA, Kinzler KW, Vogelstein B. Definition of a consensus binding site for p53. Nat. Genet 1992; 1:45-9; PMID:1301998
- el-Deiry WS, Tokino T, Velculescu VE, Levy DB, Parsons R, Trent JM, Lin D, Mercer WE, Kinzler KW, Vogelstein B. WAF1 a potential mediator of p53 tumor suppression. Cell 1993; 75:817-25; PMID:8242752
- Hearnes JM, Mays DJ, Schavolt KL, Tang L, Jiang X, Pietenpol JA. Chromatin immunoprecipitation-based screen to identify functional genomic binding sites for sequence-specific transactivators. Mol Cell Biol 2005; 25:10148-58; PMID:16260627
- Saramaki A, Banwell CM, Campbell MJ, Carlberg C. Regulation of the human p21((waf1cip1)) gene promoter via multiple binding sites for p53 and the vitamin D-3 receptor. Nucleic Acids Res 2006; 34:543-54; PMID:16434701
- Sohr S, Engeland K. The tumor suppressor p53 induces expression of the pregnancy-supporting human chorionic gonadotropin (hCG) CGB7 gene. Cell Cycle 2011; 10:3758-67; PMID:22032922; http://dx.doi.org/10.4161/cc.10.21.17946
- Jackson P, Shield M, Buskin J, Hawkes S, Reed M, Perrem K, Hauschka SD, Braithwaite A. P53-dependent activation of the mouse Mck gene promoter - identification of a novel P53-responsive sequence and evidence for cooperation between distinct P53 binding-sites. Gene Expr 1995; 5:19-33; PMID:7488858
- Zhao JQ, Schmieg FI, Logsdon N, Freedman D, Simmons DT, Molloy GR. p53 binds to a novel recognition sequence in the proximal promoter of the rat muscle creatine kinase gene and activates its transcription. Oncogene 1996; 13:293-302; PMID:8710368
- Wang LQ, Wu Q, Qiu P, Mirza A, McGuirk M, Kirschmeier P, Greene JR, Wang Y, Pickett CB, Liu S. Analyses of p53 target genes in the human genome by bioinformatic and microarray approaches. J Biol Chem 2001; 276:43604-10; PMID:11571296
- Wu GS, Saftig P, Peters C, el-Deiry WS. Potential role for cathepsin D in p53-dependent tumor suppression and chemosensitivity. Oncogene 1998; 16:2177-83; PMID:9619826
- Shiraishi K, Fukuda S, Mori T, Matsuda K, Yamaguchi T, Tanikawa C, Ogawa M, Nakamura Y, Arakawa H. Identification of fractalkine a CX3C-type chemokine as a direct target of p53. Cancer Res 2000; 60:3722-26; PMID:10919640
- Jackson RS, Cho YJ, Stein S, Liang P. CYFIP2 a direct p53 target is leptomycin-B sensitive. Cell Cycle 2007; 6:95-103; PMID:17245118
- Kudoh T, Kimura J, Lu ZG, Miki Y, Yoshida K. D4S234E a novel p53-responsive gene induces apoptosis in response to DNA damage. Exp Cell Res 2010; 316:2849-58; PMID:20599942; http://dx.doi.org/10.1016/j.yexcr.2010.06.025
- Ohnishi S, Futamura M, Kamino H, Nakamura Y, Kitamura N, Miyamoto Y, Miyamoto T, Shinogi D, Goda O, Arakawa H. Identification of NEEP21 encoding neuron-enriched endosomal protein of 21 kDa as a transcriptional target of tumor suppressor p53. Int J Oncol 2010; 37:1133-41; PMID:20878061
- Martoriati A, Doumont G, Alcalay M, Bellefroid E, Pelicci PG, Marine JC. dapk1 encoding an activator of a p19(ARF)-p53-mediated apoptotic checkpoint is a transcription target of p53. Oncogene 2005; 24:1461-6; PMID:15608685
- Tan T, Chu G. p53 binds and activates the xeroderma pigmentosum DDB2 gene in humans but not mice. Mol Cell Biol 2002; 22:3247-54; PMID:11971958
- Sakuma S, Saya H, Tada M, Nakao M, Fujiwara T, Roth JA, Sawamura Y, Shinohe Y, Abe H. Receptor protein tyrosine kinase DDR is up-regulated by p53 protein. FEBS Lett 1996; 398:165-9; PMID:8977099
- Qian YJ, Zhang J, Yan BF, Chen XB. DEC1 a basic helix-loop-helix transcription factor and a novel target gene of the p53 family mediates p53-dependent premature senescence. J Biol Chem 2008; 283:2896-905; PMID:18025081
- Kirschner RD, Rother K, Müller GA, Engeland K. The retinal dehydrogenasereductase retSDR1DHRS3 gene is activated by p53 and p63 but not by mutants derived from tumors or EECADULT malformation syndromes. Cell Cycle 2010; 9:2177-88; PMID:20543567
- Wang J, Shou J, Chen X. Dickkopf-1 an inhibitor of the Wnt signaling pathway is induced by p53. Oncogene 2000; 19:1843-8; PMID:10777218
- Crighton D, Wilkinson S, O'Prey J, Syed N, Smith P, Harrison PR, Gasco M, Garrone O, Crook T, Ryan KM. DRAM a p53-induced modulator of autophagy is critical for apoptosis. Cell 2006; 126:121-34; PMID:16839881
- Li MX, Zhou JY, Ge YB, Matherly LH, Wu GS. The phosphatase MKP1 is a transcriptional target of p53 involved in cell cycle regulation. J Biol Chem 2003; 278:41059-68; PMID:12890671
- Yang HJ, Wu GS. p53 transactivates the phosphatase MKP1 through both intronic and exonic p53 responsive elements. Cancer Biol Ther 2004; 3:1277-82; PMID:15611668
- Liu YX, Wang J, Guo J, Wu J, Lieberman HB, Yin Y. DUSP1 is controlled by p53 during the cellular response to oxidative stress. Mole Cancer Res 2008; 6:624-33; PMID:18403641; http://dx.doi.org/10.1158/1541-7786.MCR-07-2019
- Yin Y, Liu YX, Jin YJ, Hall EJ, Barrett JC. PAC1 phosphatase is a transcription target of p53 in signalling apoptosis and growth suppression. Nature 2003; 422:527-31; PMID:12673251
- Shen WH, Wang JL, Wu JJ, Zhurkin VB, Yin YX. Mitogen-activated protein kinase phosphatase 2: A novel transcription target of p53 in apoptosis. Cancer Res 2006; 66:6033-9; PMID:16778175
- Ueda K, Arakawa H, Nakamura Y. Dual-specificity phosphatase 5 (DUSP5) as a direct transcriptional target of tumor suppressor p53. Oncogene 2003; 22:5586-91; PMID:12944906
- Piya S, Kim JY, Bae J, Seol DW, Moon AR, Kim TH. DUSP6 is a novel transcriptional target of p53 and regulates p53-mediated apoptosis by modulating expression levels of Bcl-2 family proteins. FEBS Lett. 2012; 586:4233-40; PMID:23108049; http://dx.doi.org/10.1016/j.febslet.2012.10.031
- Carvajal LA, Hamard PJ, Tonnessen C, Manfredi JJ. E2F7 a novel target is up-regulated by p53 and mediates DNA damage-dependent transcriptional repression. Genes Dev 2012; 26:1533-45; PMID:22802528; http://dx.doi.org/10.1101/gad.184911.111
- Tanikawa C, Furukawa Y, Yoshida N, Arakawa H, Nakamura Y, Matsuda K. XEDAR as a putative colorectal tumor suppressor that mediates p53-regulated anoikis pathway. Oncogene 2009; 28:3081-92; PMID:19543321; http://dx.doi.org/10.1038/onc.2009.154
- LudesMeyers JH, Subler MA, Shivakumar CV, Munoz RM, Jiang P, Bigger JE, Brown DR, Deb SP, Deb S. Transcriptional activation of the human epidermal growth factor receptor promoter by human p53. Mol Cell Biol 1996; 16:6009-19; PMID:8887630
- Jin YJ, Wang J, Qiao C, Hei TK, Brandt-Rauf PW, Yin Y. A novel mechanism for p53 to regulate its target gene ECK in signaling apoptosis. Mole Cancer Res 2006; 4:769-8; PMID:17050670
- Wang B, Niu D, Lai L, Ren EC. p53 increases MHC class I expression by upregulating the endoplasmic reticulum aminopeptidase ERAP1. Nat Commun 2013; 4:2359; PMID:23965983; http://dx.doi.org/10.1038/ncomms3359
- Shirley SH, Rundhaug JE, Tian J, Cullinan-Ammann N, Lambertz I, Conti CJ, Fuchs-Young R. Transcriptional Regulation of Estrogen Receptor-alpha by p53 in Human Breast Cancer Cells. Cancer Res 2009; 69:3405-14; PMID:19351845; http://dx.doi.org/10.1158/0008-5472.CAN-08-3628
- Liebetrau W, Budde A, Savoia A, Grummt F, Hoehn H. p53 activates Fanconi anemia group C gene expression. Hu Mole Genet 1997; 6:277-83; PMID:9063748
- Muller M, Wilder S, Bannasch D, Israeli D, Lehlbach K, Li-Weber M, Friedman SL, Galle PR, Stremmel W, Oren M, et al. p53 activates the CD95 (APO-1Fas) gene in response to DNA damage by anticancer drugs. J Exp Med 1998; 188:2033-45; PMID:9841917
- Munsch D, Watanabe-Fukunaga R, Bourdon JC, Nagata S, May E, Yonish-Rouach E, Reisdorf P. Human and mouse Fas (APO-1CD95) death receptor genes each contain a p53-responsive element that is activated by p53 mutants unable to induce apoptosis. J Biol Chem 2000; 275:3867-72; PMID:10660538
- Schilling T, Schleithoff ES, Kairat A, Melino G, Stremmel W, Oren M, Krammer PH, Müller M. Active transcription of the human FASCD95TNFRSF6 gene involves the p53 family. Biochem Biophys Res Commun 2009; 387:399-404; PMID:19615968; http://dx.doi.org/10.1016/j.bbrc.2009.07.063
- Liu G, Chen XB. The ferredoxin reductase gene is regulated by the p53 family and sensitizes cells to oxidative stress-induced apoptosis. Oncogene 2002; 21:7195-204; PMID:12370809
- Ciribilli Y, Andreotti V, Menendez D, Langen JS, Schoenfelder G, Resnick MA, Inga A. The coordinated P53 and estrogen receptor cis-regulation at an FLT1 promoter SNP is specific to genotoxic stress and estrogenic compound. PLoS One 2010; 5(4):e10236; PMID:20422012; http://dx.doi.org/10.1371/journal.pone.0010236
- Elkeles A, Juven-Gershon T, Israeli D, Wilder S, Zalcenstein A, Oren M. The c-fos proto-oncogene is a target for transactivation by the p53 tumor suppressor. Mol Cell Biol 1999; 19:2594-600; PMID:10082525
- Singh S, Raina V, Chavali PL, Dubash T, Kadreppa S, Parab P, Chattopadhyay S. Regulation of GAD65 expression by SMAR1 and p53 upon Streptozotocin treatment. Bmc Molecular Biology 2012; 13:28; PMID:22978699
- Kastan MB, Zhan Q, el-Deiry WS, Carrier F, Jacks T, Walsh WV, Plunkett BS, Vogelstein B, Fornace AJ Jr. A mammalian cell cycle checkpoint pathway utilizing p53 and GADD45 is defective in ataxia-telangiectasia. Cell 1992; 71:587-97; PMID:1423616
- Ide T, Brown-Endres L, Chu K, Ongusaha PP, Ohtsuka T, El-Deiry WS, Aaronson SA, Lee SW. GAMT a p53-inducible modulator of apoptosis is critical for the adaptive response to nutrient stress (Retracted article. See vol. 51 pg. 552 2013). Mole Cell 2009; 36:379-92; PMID:19917247; http://dx.doi.org/10.1016/j.molcel.2009.09.031
- Zhu ZX, Wei J, Shi Z, Yang Y, Shao D, Li B, Wang X, Ma Z. Identification of human guanylate-binding protein 1 gene (hGBP1) as a direct transcriptional target gene of p53. Biochem Biophys Res Commun 2013; 436:204-11; PMID:23727578; http://dx.doi.org/10.1016/j.bbrc.2013.05.074
- Tan MJ, Wang YX, Guan KL, Sun Y. PTCF-beta a type beta transforming growth factor (TGF-beta) superfamily member is a p53 target gene that inhibits tumor cell growth via TGF-beta signaling pathway. Proc Natl Acad Sci USA 2000; 97:109-4; PMID:10618379
- Osada M, Park HL, Park MJ, Liu JW, Wu G, Trink B, Sidransky D. A p53-type response element in the GDF15 promoter confers high specificity for p53 activation. Biochem Biophys Res Commun 2007; 354:913-8; PMID:17276395
- Suzuki S, Tanaka T, Poyurovsky MV, Nagano H, Mayama T, Ohkubo S, Lokshin M, Hosokawa H, Nakayama T, Suzuki Y, et al. Phosphate-activated glutaminase (GLS2) a p53-inducible regulator of glutamine metabolism and reactive oxygen species. Proc Natl Acad Sci USA 2010; 107 7461-6; PMID:20351271; http://dx.doi.org/10.1073/pnas.1002459107
- Zhang YH, Qian YJ, Lu WF, Chen XB. The G protein-coupled receptor 87 is necessary for p53-dependent cell survival in response to genotoxic stress. Cancer Res 2009; 69:6049-56; PMID:19602589; http://dx.doi.org/10.1158/0008-5472.CAN-09-0621
- Tan MJ, Li S, Swaroop M, Guan K, Oberley LW, Sun Y. Transcriptional activation of the human glutathione peroxidase promoter by p53. J Biol Chem 1999; 274:12061-6; PMID:10207030
- Lo HW, Stephenson L, Cao X, Milas M, Pollock R, Ali-Osman F. Identification and functional characterization of the human glutathione S-transferase P1 gene as a novel transcriptional target of the p53 tumor suppressor gene. Mole Cancer Res 2008; 6 843-50; PMID:18505928; http://dx.doi.org/10.1158/1541-7786.MCR-07-2105
- Weizer-Stern O, Adamsky K, Margalit O, Ashur-Fabian O, Givol D, Amariglio N, Rechavi G. Hepcidin a key regulator of iron metabolism is transcriptionally activated by p53. Br J Haematol 2007; 138:253-62; PMID:17593032
- Wu XW, Bayle JH, Olson D, Levine AJ. The P53 Mdm-2 autoregulatory feedback loop. Genes Dev 1993; 7:1126-32; PMID:8319905
- Juven T, Barak Y, Zauberman A, George DL, Oren M. Wild-type P53 can mediate sequence-specific transactivation of an internal promoter within the Mdm2 gene. Oncogene 1993; 8:3411-6.
- Barak Y, Juven T, Haffner R, Oren M. mdm2 expression is induced by wild type p53 activity. EMBO J 1993; 12 461-8; PMID:8440237
- Zauberman A, Flusberg D, Haupt Y, Barak Y, Oren M. A functional P53-responsive intronic promoter is contained within the human Mdm2 gene. Nucleic Acids Res 1995; 23:2584-92; PMID:7651818
- Saucedo LJ, Carstens BP, Seavey SE, Albee LD, Perry ME. Regulation of transcriptional activation of mdm2 gene by p53 in response to UV radiation. Cell Growth Differ 1998; 9:119-30; PMID:9486848
- Britschgi C, Rizzi M, Grob TJ, Tschan MP, Hügli B, Reddy VA, Andres AC, Torbett BE, Tobler A, Fey MF. Identification of the p53 family-responsive element in the promoter region of the tumor suppressor gene hypermethylated in cancer 1. Oncogene 2006;l 25:2030-9; PMID:16301995
- Baraz L, Haupt Y, Elkin M, Peretz T, Vlodavsky I. Tumor suppressor p53 regulates heparanase gene expression. Oncogene 2006; 25 3939-47; PMID:16474844
- Deguin-Chambon V, Vacher M, Jullien M, May E, Bourdon JC, Direct transactivation of c-Ha-Ras gene by p53: evidence for its involvement in p53 transactivation activity and p53-mediated apoptosis. Oncogene 2000; 19:5831-41; PMID:11127813
- Feng Z, Jin S, Zupnick A, Hoh J, de Stanchina E, Lowe S, Prives C, Levine AJ. p53 tumor suppressor protein regulates the levels of huntingtin gene expression. Oncogene 2006; 25:1-7; PMID:16278683
- Schafer H, Trauzold A, Sebens T, Deppert W, Fölsch UR, Schmidt WE. The proliferation-associated early response gene p22PRG1 is a novel p53 target gene. Oncogene 1998; 16:2479-87; PMID:9627114
- Schafer H, Diebel J, Arlt A, Trauzold A, Schmidt WE. The promoter of human p22PACAP response gene 1 (PRG1) contains functional binding sites for the p53 tumor suppressor and for NF kappa B. FEBS Lett 1998; 436 139-43; PMID:9781666
- Huang YH, Wu JYJ, Zhang YJ, Wu MX. Synergistic and opposing regulation of the stress-responsive gene IEX-1 by p53 c-Myc and multiple NF-kappa Brel complexes. Oncogene 2002; 21:6819-28; PMID:12360408
- Song LL, Alimirah F, Panchanathan R, Xin H, Choubeyt D. Expression of an IFN-inducible cellular senescence gene IFI16 Is up-regulated by p53. Mole Cancer Res 2008; 6:1732-41; PMID:18974396; http://dx.doi.org/10.1158/1541-7786.MCR-08-0208
- Buckbinder L, Talbott R, Velasco-Miguel S, Takenaka I, Faha B, Seizinger BR, Kley N. Induction of the growth inhibitor Igf-binding protein-3 by P53. Nature 1995; 377:646-9; PMID:7566179
- Koeppel M, van Heeringen SJ, Smeenk L, Navis AC, Janssen-Megens EM, Lohrum M. The novel p53 target gene IRF2BP2 participates in cell survival during the p53 stress response. Nucleic Acids Res 2009; 37:322-5; PMID:19042971; http://dx.doi.org/10.1093/nar/gkn940
- Mori T, Anazawa Y, Iiizumi M, Fukuda S, Nakamura Y, Arakawa H. Identification of the interferon regulatory factor 5 gene (IRF-5) as a direct target for p53. Oncogene 2002; 21 2914-8; PMID:11973653
- Cui HY, Kamino H, Nakamura Y, Kitamura N, Miyamoto T, Shinogi D, Goda O, Arakara H, Futamura M. Regulation of apoptosis by p53-inducible transmembrane protein containing sushi domain. Oncol Rep 2010; 24:1193-200; PMID:20878110
- Mukhopadhyay T, Roth JA. p53 involvement in activation of the cytokeratin 8 gene in tumor cell lines. Anticancer Res 1996; 16:105-12; PMID:8615594
- Hu W, Feng Z, Teresky AK, Levine AJ. p53 regulates maternal reproduction through LIF. Nature 2007; 450:721-4; PMID:18046411
- Assaily W, Rubinger DA, Wheaton K, Lin Y, Ma W, Xuan W, Brown-Endres L, Tsuchihara K, Mak TW, Benchimol S. ROS-mediated p53 induction of Lpin1 regulates fatty acid oxidation in response to nutritional stress. Mol. Cell 2011; 44:491-501; PMID:22055193; http://dx.doi.org/10.1016/j.molcel.2011.08.038
- Seol DW, Chen QY, Smith ML, Zarnegar R. Regulation of the c-met proto-oncogene promoter by p53. J Biol Chem 1999; 274:3565-72; PMID:9920903
- Wang WL, Cheng X, Lu J, Wei J, Fu G, Zhu F, Jia C, Zhou L, Xie H, Zheng S. Mitofusin-2 is a novel direct target of p53. Biochem Biophys Res Commun 2010; 400:587-92; PMID:20804729; http://dx.doi.org/10.1016/j.bbrc.2010.08.108
- Chen JG, Sadowski I. Identification of the mismatch repair genes PMS2 and MLH1 as p53 target genes by using serial analysis of binding elements. Proc Natl Acad Sci USA 2005; 102:4813-8; PMID:15781865
- Bian JH, Sun Y, Transcriptional activation by p53 of the human type IV collagenase (gelatinase A or matrix metalloproteinase 2) promoter. Mol Cell Biol 1997; 17:6330-8; PMID:9343394
- Staun-Ram E, Goldman S, Shalev E. Ets-2 and p53 mediate cAMP-induced MMP-2 expression activity and trophoblast invasion. Reprod Biol Endocrinol 2009; 7:135; PMID:19939245; http://dx.doi.org/10.1186/1477-7827-7-135
- Stein S, Thomas EK, Herzog B, Westfall MD, Rocheleau JV, Jackson RS 2nd, Wang M, Liang P. NDRG1 is necessary for p53-dependent apoptosis. J Biol Chem 2004; 279:48930-40; PMID:15377670
- Cho SJ, Rossi A, Jung YS, Yan W, Liu G, Zhang J, Zhang M, Chen X. Ninjurin1 a target of p53 regulates p53 expression and p53-dependent cell survival senescence and radiation-induced mortality. Proc Natl Acad Sci USA 2013; 110:9362-7; PMID:23690620; http://dx.doi.org/10.1073/pnas.1221242110
- Sadasivam S, Gupta S, Radha V, Batta K, Kundu TK, Swarup G. Caspase-1 activator Ipaf is a p53-inducible gene involved in apoptosis. Oncogene 2005; 24 627-36; PMID:15580302
- Lefort K, Mandinova A, Ostano P, Kolev V, Calpini V, Kolfschoten I, Devgan V, Lieb J, Raffoul W, Hohl D, et al. Notch1 is a p53 target gene involved in human keratinocyte tumor suppression through negative regulation of ROCK12 and MRCK alpha kinases. Genes Dev 2007; 21:562-77; PMID:17344417
- Yugawa T, Handa K, Narisawa-Saito M, Ohno S, Fujita M, Kiyono T. Regulation of Notch1 gene expression by p53 in epithelial cells. Mol Cell Biol 2007; 27:3732-42; PMID:17353266
- Böhlig L, Metzger R, Rother K, Till H, Engeland K. The CCN3 gene coding for an extracellular adhesion-related protein is transcriptionally activated by the p53 tumor suppressor. Cell Cycle 2008; 7:1254-61; PMID:18418052
- Tasheva ES, Maki CG, Conrad AH, Conrad GW. Transcriptional activation of bovine mimecan by p53 through an intronic DNA-binding site. Biochim Biophys Acta 2001; 1517:333-8; PMID:11342211
- Saifudeen Z, Liu J, Dipp S, Yao X, Li Y, McLaughlin N, Aboudehen K, El-Dahr SS. A p53-pax2 pathway in kidney development: implications for nephrogenesis. PLoS One 2012; 7(9):e44869; PMID:22984579; http://dx.doi.org/10.1371/journal.pone.0044869
- Zhu JH, Chen XB. MCG10 a novel p53 target gene that encodes a KH domain RNA-binding protein is capable of inducing apoptosis and cell cycle arrest in G(2)-M. Mol Cell Biol 2000; 20:5602-18; PMID:10891498
- Shivakumar CV, Brown DR, Deb S, Deb SP. Wild-type human P53 transactivates the human proliferating cell nuclear antigen promoter. Mol Cell Biol 1995; 15:6785-93; PMID:8524244
- Morris GF, Bischoff JR, Mathews MB. Transcriptional activation of the human proliferating-cell nuclear antigen promoter by p53. Proc Natl Acad Sci USA 1996; 93:895-9; PMID:8570655
- Shan B, Xu J, Zhuo Y, Morris CA, Morris GF. Induction of p53-dependent activation of the human proliferating cell nuclear antigen gene in chromatin by ionizing radiation. J Biol Chem 2003; 278:44009-17; PMID:12947108
- Shan B, Morris GF. Binding sequence-dopendent regulation of the human proliferating cell nuclear antigen promoter by p53. Exp Cell Res 2005; 305:10-22; PMID:15777783
- Lee SJ, Lee SH, Yoon MH, Park BJ. A new p53 target gene RKIP is essential for DNA damage-induced cellular senescence and suppression of ERK activation. Neoplasia 2013; 15:727-37; PMID:23814485
- Reczek EE, Flores ER, Tsay AS, Attardi LD, Jacks T. Multiple response elements and differential p53 binding control perp expression during apoptosis. Mole Cancer Res 2003; 1:1048-57; PMID:14707288
- Lin YP, Ma WL, Benchimol S. Pidd a new death-domain-containing protein is induced by p53 and promotes apoptosis. Nat Genet 2000; 26:122-5; PMID:10973264
- Burns TF, Fei PW, Scata KA, Dicker DT, el-Deiry WS. Silencing of the novel p53 target gene SnkPlk2 leads to mitotic catastrophe in paclitaxel (Taxol)-exposed cells. Mol Cell Biol 2003; 23:5556-71; PMID:12897130
- Hudson CD, Morris PJ, Latchman DS, Budhram-Mahadeo VS. Brn-3a transcription factor blocks p53-mediated activation of proapoptotic target genes Noxa and Bax in vitro and in vivo to determine cell fate. J Biol Chem 2005; 280:11851-8; PMID:15598651
- de Stanchina E, Querido E, Narita M, Davuluri RV, Pandolfi PP, Ferbeyre G, Lowe SW. PML is a direct p53 target that modulates p53 effector functions. Mole Cell 2004; 13:523-35; PMID:14992722
- Rossi M, Demidov.ON, Anderson CW, Appella E, Mazur SJ. Induction of PPM1D following DNA-damaging treatments through a conserved p53 response element coincides with a shift in the use of transcription initiation sites. Nucleic Acids Res 2008; 36:7168-80; PMID:19015127; http://dx.doi.org/10.1093/nar/gkn888
- Yan JL, Jiang J, Lim CA, Wu Q, Ng HH, Chin KC. BLIMP1 regulates cell growth through repression of p53 transcription. Proc Natl Acad Sci USA 2007; 104:1841-6; PMID:17264218
- Stambolic V, MacPherson D, Sas D, Lin Y, Snow B, Jang Y, Benchimol S, Mak TW. Regulation of PTEN transcription by p53. Mole Cell 2001; 8:317-25; PMID:11545734
- Min SH, Kim DM, Heo YS, Kim YI, Kim HM, Kim J, Han YM>, Kim IC, Yoo OJ. New p53 target phosphatase of regenerating liver 1 (PRL-1) downregulates p53. Oncogene 2009; 28 545-54; PMID:18997816; http://dx.doi.org/10.1038/onc.2008.409
- Doumont G, Martoriati A, Marine JC. PTPRV is a key mediator of p53-induced cell cycle exit. Cell Cycle 2005; 4:1703-5; PMID:16258284
- Huang BH, Zhuo JL, Leung CH, Lu GD, Liu JJ, Yap CT, Hooi SC. PRAP1 is a novel executor of p53-dependent mechanisms in cell survival after DNA damage. Cell Death Disease 2012; 3:e442; PMID:23235459; http://dx.doi.org/10.1038/cddis.2012.180
- Feng ZH, Hu W, de Stanchina E, Teresky AK, Jin S, Lowe S, Levine AJ. The regulation of AMPK beta 1 TSC2 and PTEN expression by p53: Stress cell and tissue specificity and the role of these gene products in modulating the IGF-1-AKT-mTOR pathways. Cancer Res 2007; 67:3043-53; PMID:17409411
- Maxwell SA, Kochevar GJ. Identification of a p53-response element in the promoter of the proline oxidase gene. Biochem Biophys Res Commun 2008; 369:308-13; PMID:18279664; http://dx.doi.org/10.1016/j.bbrc.2008.01.171
- Raimondi I, Ciribilli Y, Monti P, Bisio A, Pollegioni L, Fronza G, Inga A, Campomenosi P. P53 Family members modulate the expression of PRODH but not PRODH2 via intronic p53 response elements. PLoS One 2013; 8(7):e69152; PMID:23861960; http://dx.doi.org/10.1371/journal.pone.0069152
- Ohtsuka T, Ryu H, Minamishima YA, Macip S, Sagara J, Nakayama KI, Aaronson SA, Lee SW. ASC is a Bax adaptor and regulates the p53-Bax mitochondrial apoptosis pathway. Nat Cell Biol 2004; 6(2):121-8; PMID:14730312
- Zhang XY, He Y, Lee KH, Dubois W, Li Z, Wu X, Kovalchuk A, Zhang W, Huang J. Rap2b a novel p53 target regulates p53-mediated pro-survival function. Cell Cycle 2013; 12:1279-91; PMID:23535297; http://dx.doi.org/10.4161/cc.24364
- Tian K, Wang Y, Xu H. WTH3 is a direct target of the p53 protein. Br J Cancer 2007; 96:1579-86; PMID:17426708
- Hsu TH, Chu CC, Jiang SY, Hung MW, Ni WC, Lin HE, Chang TC. Expression of the class II tumor suppressor gene RIG1 is directly regulated by p53 tumor suppressor in cancer cell lines. FEBS Lett 2012; 586 1287-93; PMID:22616991; http://dx.doi.org/10.1016/j.febslet.2012.03.020
- Shu LM, Yan WS, Chen XB. RNPC1 an RNA-binding protein and a target of the p53 family is required for maintaining the stability of the basal and stress-induced p21 transcript. Genes Dev 2006; 20:2961-72; PMID:17050675
- Ongusaha PP, Kim JI, Fang L, Wong TW, Yancopoulos GD, Aaronson SA, Lee SW. p53 induction and activation of DDR1 kinase counteract p53-mediated apoptosis and influence p53 regulation through a positive feedback loop. EMBO J 2003; 22:1289-301; PMID:12628922
- Ng CC, Arakawa H, Fukuda S, Kondoh H, Nakamura Y. p53RFP a p53-inducible RING-finger protein regulates the stability of p21(WAF1). Oncogene 2003; 22 4449-58; PMID:12853982
- Tanaka H, Arakawa H, Yamaguchi T, Shiraishi K, Fukuda S, Matsui K, Takei Y, Nakamura Y. A ribonucleotide reductase gene involved in a p53-dependent cell- cycle checkpoint for DNA damage. Nature 2000; 404:42-9; PMID:10716435
- Tan MJ, Heizmann CW, Guan KL, Schafer BW, Sun Y. Transcriptional activation of the human S100A2 promoter by wild-type p53. FEBS Lett 1999; 445:265-8; PMID:10094469
- Li CS, Chen H, Ding F, Zhang Y, Luo A, Wang M, Liu Z. A novel p53 target gene S100A9 induces p53-dependent cellular apoptosis and mediates the p53 apoptosis pathway. Biochem J 2009; 422:363-72; PMID:19534726; http://dx.doi.org/10.1042/BJ20090465
- Adachi K, Toyota M, Sasaki Y, Yamashita T, Ishida S, Ohe-Toyota M, Maruyama R, Hinoda Y, Saito T, Imai K, et al. Identification of SCN3B as a novel p53-inducible proapoptotic gene. Oncogene 2004; 23:7791-8; PMID:15334053
- Futamura M, Kamino H, Miyamoto Y, Kitamura N, Nakamura Y, Ohnishi S, Masuda Y, Arakawa H. Possible role of semaphorin 3F a candidate tumor suppressor gene at 3p2l1.3 in p53-regulated tumor angiogenesis suppression. Cancer Res 2007; 67 1451-60; PMID:17308083
- Zou Z, Gao C, Nagaich AK, Connell T, Saito S, Moul JW, Seth P, Appella E, Srivastava S. p53 regulates the expression of the tumor suppressor gene maspin. J Biol Chem 2000; 275:6051-4; PMID:10692390
- Wang SZE, Narasanna A, Whitell CW, Wu FY, Friedman DB, Arteaga CL. Convergence of p53 and transforming growth factor beta (TGF beta) signaling on activating expression of the tumor suppressor gene maspin in mammary epithelial cells. J Biol Chem 2007; 282:5661-9; PMID:17204482
- Kunz C, Pebler S, Otte J, Vonderahe D. Differential regulation of plasminogen-activator and inhibitor gene-transcription by the tumor-suppressor P53. Nucleic Acids Res 1995; 23:3710-7; PMID:7479001
- Velasco-Miguel S, Buckbinder L, Jean P, Gelbert L, Talbott R, Laidlaw J, Seizinger B, Kley N. PA26 a novel target of the p53 tumor suppressor and member of the GADD family of DNA damage and growth arrest inducible genes. Oncogene 1999; 18:127-37; PMID:9926927
- Maiyar AC, Huang AJ, Phu PT, Cha HH, Firestone GL. p53 stimulates promoter activity of the sgk serumglucocorticoid-inducible serinethreonine protein kinase gene in rodent mammary epithelial cells. J Biol Chem 1996; 271:12414-22; PMID:8647846
- Nagy N, Takahara M, Nishikawa J, Bourdon JC, Kis LL, Klein G, Klein E. Wild-type p53 activates SAP expression in lymphoid cells. Oncogene 2004; 23:8563-70; PMID:15378026
- Fiucci G, Beaucourt S, Duflaut D, Lespagnol A, Stumptner-Cuvelette P, Géant A, Buchwalter G, Tuynder M, Susini L, Lassalle JM, et al. Siah-1b is a direct transcriptional target of p53: Identification of the functional p53 responsive element in the siah-1b promoter. Proc Nat Acad Sci USA 2004; 101:3510-5; PMID:14985507
- Fortin A, MacLaurin JG, Arbour N, Cregan SP, Kushwaha N, Callaghan SM, Park DS, Albert PR, Slack RS. The proapoptotic gene SIVA is a direct transcriptional target for the tumor suppressors p53 and E2F1. J Biol Chem 2004; 279:28706-14; PMID:15105421
- Anazawa Y, Arakawa H, Nakagawa H, Nakamura Y. Identification of STAG1 as a key mediator of a p53-dependent apoptotic pathway. Oncogene 2004; 23:7621-7; PMID:15361841
- Zhang YH, Shu LM, Chen XB. Syntaxin 6 a regulator of the protein trafficking machinery and a target of the p53 family is required for cell adhesion and survival. J Biol Chem 2008; 283:30689-98; PMID:18779328; http://dx.doi.org/10.1074/jbc.M801711200
- Kishore AH, Batta K, Das C, Agarwal S, Kundu TK. p53 regulates its own activator: transcriptional co-activator PC4 a new p53-responsive gene. Biochem J 2007; 406:437-44; PMID:17555406
- da Costa NM, Hautefeuille A, Cros MP, Melendez ME, Waters T, Swann P, Hainaut P, Pinto LF. Transcriptional regulation of thymine DNA glycosylase (TDG) by the tumor suppressor protein p53. Cell Cycle 2012; 11:4570-8; PMID:23165212; http://dx.doi.org/10.4161/cc.22843
- Li H, Watts GS, Oshiro MM, Futscher BW, Domann FE. AP-2 alpha and AP-2 gamma are transcriptional targets of p53 in human breast carcinoma cells. Oncogene 2006; 25 5405-15; PMID:16636674
- Shin TH, Paterson AJ, Kudlow JE. P53 stimulates transcription from the human transforming growth-factor-alpha promoter - a potential growth-stimulatory role for P53. Mol Cell Biol 1995; 15:4694-701; PMID:7651386
- Menendez D, Shatz M, Azzam K, Garantziotis S, Fessler MB, Resnick MA. The toll-like receptor gene family is integrated into human DNA damage and p53 networks. Plos Genetics 2011; 7(3):e1001360; PMID:21483755; http://dx.doi.org/10.1371/journal.pgen.1001360
- Taura M, Eguma A, Suico MA, Shuto T, Koga T, Komatsu K, Komune T, Sato T, Saya H, Li JD, et al. p53 Regulates toll-like receptor 3 expression and function in human epithelial cell Lines. Mol Cell Biol 2008; 28:6557-67; PMID:18779317; http://dx.doi.org/10.1128/MCB.01202-08
- Liu XG, Yue P, Khuri FR, Sun SY. p53 upregulates death receptor 4 expression through an intronic p53 binding site. Cancer Res 2004; 64:5078-83; PMID:15289308
- Takimoto R, el-Deiry WS. Wild-type p53 transactivates the KILLERDR5 gene through an intronic sequence-specific DNA-binding site. Oncogene 2000; 19:1735-43; PMID:10777207
- Wang SL, el-Deiry WS. P73 or p53 directly regulates human p53 transcription to maintain cell cycle checkpoints. Cancer Res 2006; 66:6982-9; PMID:16849542
- Oda K, Arakawa H, Tanaka T, Matsuda K, Tanikawa C, Mori T, Nishimori H, Tamai K, Tokino T, Nakamura Y, et al. p53AlP1 a potential mediator of p53-dependent apoptosis and its regulation by Ser-46-phosphorylated p53. Cell 2000; 102:849-62; PMID:11030628
- Contente A, Dittmer A, Koch MC, Roth J, Dobbelstein M. A polymorphic microsatellite that mediates induction of PIG3 by p53. Nat Genet 2002; 30:315-20; PMID:11919562
- Okamura S, Arakawa H, Tanaka T, Nakanishi H, Ng CC, Taya Y, Monden M, Nakamura Y. p53DINP1 a p53-inducible gene regulates p53-dependent apoptosis. Molecular Cell 2001; 8:85-94; PMID:11511362
- Harmes DC, Bresnick E, Lubin EA, Watson JK, Heim KE, Curtin JC, Suskind AM, Lamb J, DiRenzo J. Positive and negative regulation of Delta N-p63 promoter activity by p53 and Delta N-p63-alpha contributes to differential regulation of p53 target genes. Oncogene 2003; 22:7607-16; PMID:14576823
- Chen XB, Zheng YM, Zhu JH, Jiang JY, Wang J. p73 is transcriptionally regulated by DNA damage p53 and p73. Oncogene 2001; 20:769-74; PMID:11314010
- Kartasheva NN, Contente A, Lenz-Stoppler C, Roth J, Dobbelstein M. p53 induces the expression of its antagonist p73 Delta N establishing an autoregulatory feedback loop. Oncogene 2002; 21:4715-27; PMID:12101410
- Wang JL, Liu YX, Hande MP, Wong AC, Jin YJ, Yin Y. TAp73 is a downstream target of p53 in controlling the cellular defense against stress. J Biol Chem 2007; 282:29152-62; PMID:17693405
- Park WR, Nakamura Y. p53CSV a novel p53-inducible gene involved in the p53-dependent cell-survival pathway. Cancer Res 2005; 65:1197-206; PMID:15735003
- Obad S, Brunnström H, Vallon-Christersson J, Borg A, Drott K, Gullberg U. Staf50 is a novel p53 target gene conferring reduced clonogenic growth of leukemic U-937 cells. Oncogene 2004; 23:4050-9; PMID:15064739
- Nylander K, Bourdon JC, Bray SE, Gibbs NK, Kay R, Hart I, Hall PA. Transcriptional activation of tyrosinase and TRP-I by p53 links UV irradiation to the protective tanning response. J Pathol 2000; 190:39-46; PMID:10640990
- Miyamoto Y, Futamura M, Kitamura N, Nakamura Y, Baba H, Arakawa H. Identification of UNC5A as a novel transcriptional target of tumor suppressor p53 and a regulator of apoptosis. Int J Oncol 2010; 36 1253-60; PMID:20372800
- Yoon H, Liyanarachchi S, Wright FA, Davuluri R, Lockman JC, de la Chapelle A, Pellegata NS. Gene expression profiling of isogenic cells with different TP53 gene dosage reveals numerous genes that are affected by TP53 dosage and identifies CSPG2 as a direct target of p53. Proc Natl Acad Sci USA 2002; 99:15632-7; PMID:12438652
- Maruyama R, Aoki F, Toyota M, Sasaki Y, Akashi H, Mita H, Suzuki H, Akino K, Ohe-Toyota M, Maruyama Y, et al. Comparative genome analysis identifies the vitamin D receptor gene as a direct target of p53-mediated transcriptional activation. Cancer Res 2006; 66:4574-83; PMID:16651407
- Adimoolam S, Ford JM. p53 and DNA damage-inducible expression of the xeroderma pigmentosum group C gene. Proc Natl Acad Sci USA 2002; 99:12985-90; PMID:12242345
- Lee JY, Kim HJ, Yoon NA, Lee WH, Min YJ, Ko BK, Lee BJ, Lee A, Cha HJ, Cho WJ, et al. Tumor suppressor p53 plays a key role in induction of both tristetraprolin and let-7 in human cancer cells. Nucleic Acids Res 2013; 41:5614-25; PMID:23595149; http://dx.doi.org/10.1093/nar/gkt222.
- Wilhelm MT, Mendez-Vidal C, Wiman KG. Identification of functional p53-binding motifs in the mouse wig-1 promoter. Febs Lett 2002; 524:69-72; PMID:12135743
- Nishimori H, Shiratsuchi T, Urano T, Kimura Y, Kiyono K, Tatsumi K, Yoshida S, Ono M, Kuwano M, Nakamura Y, et al. A novel brain-specific p53-target gene BAI1 containing thrombospondin type 1 repeats inhibits experimental angiogenesis. Oncogene 1997; 15:2145-50; PMID:9393972
- Ellisen LW, Ramsayer KD, Johannessen CM, Yang A, Beppu H, Minda K, Oliner JD, McKeon F, Haber DA. REDD1 a evelopmentally regulated transcriptional target of p63 and p53 links p63 to regulation of reactive oxygen species. Mol Cell 2002; 10:995-1005; PMID:12453409
- Kimura Y, Furuhata T, Urano T, Hirata K, Nakamura Y, Tokino T. Genomic structure and chromosomal localization of GML (GPI-anchored molecule-like protein) a gene induced by p53. Genomics 1997; 41:477-80; PMID:9169150
- Metcalfe AM, Dixon RM, Radda GK, Wild-type but not mutant p53 activates the hepatocyte growth factorscatter factor promoter. Nucleic Acids Res. 1997; 25:983-6; PMID:9023107
- Kato MV, Sato H, Nagayoshi M, Ikawa Y. Upregulation of the elongation factor-1alpha gene by p53 in association with death of an erythroleukemic cell line. Blood 1997; 90:1373-8; PMID:9269753
- Scherer SJ, Maier SM, Seifert M, Hanselmann RG, Zang KD, Muller-Hermelink HK, Angel P, Welter C, Schartl M. p53 and c-Jun functionally synergize in the regulation of the DNA repair gene hMSH2 in response to UV. J Biol Chem 2000; 275:37469-73; PMID:10984493
- Warnick CT, Dabbas B, Ford CD, Strait KA. Identification of a p53 response element in the promoter region of the hMSH2 gene required for expression in A2780 ovarian cancer cells. J Biol Chem 2001; 276:27363-70; PMID:11350971
- Urano T, Nishimori H, Han H, Furuhata T, Kimura Y, Nakamura Y, Tokino T. Cloning of P2XM a novel human P2X receptor gene regulated by p53. Cancer Res 1997; 57:3281-7; PMID:9242461
- Dornan D, Bheddah S, Newton K, Ince W, Frantz GD, Dowd P, Koeppen H, Dixit VM, French DM. COP1 the negative regulator of p53 is overexpressed in breast and ovarian adenocarcinomas. Cancer Res 2004; 64:7226-30; PMID:15492238
- Han HJ, Tokino T, Nakamura Y. CSR a scavenger receptor-like protein with a protective role against cellular damage causedby UV irradiation and oxidative stress. Hum Mol Genet 1998; 7:1039-46; PMID:9580669
- Hermeking H, Lengauer C, Polyak K, He TC, Zhang L, Thiagalingam S, Kinzler KW, Vogelstein B. 14-3-3 sigma is a p53-regulated inhibitor of G2M progression. Mol Cell 1997; 1:3-11; PMID:9659898
- Passer BJ, Nancy-Portebois V, Amzallag N, Prieur S, Cans C, Roborel de Climens A, Fiucci G, Bouvard V, Tuynder M, Susini L, et al. The p53-inducible TSAP6 gene product regulates apoptosis and the cell cycle and interacts with Nix and the Myt1 kinase. Proc Natl Acad Sci USA 2003; 100:2284-9; PMID:12606722
- Herzer K, Falk CS, Encke J, Eichhorst ST, Ulsenheimer A, Seliger B, Krammer PH. Upregulation of major histocompatibility complex class I on liver cells by hepatitis C virus core protein via p53 and TAP1 impairs natural killer cell cytotoxicity. J Virol 2003; 77:8299-309; PMID:12857899
- Johnson RA, Ince TA, Scotto KW. Transcriptional repression by p53 through direct binding to a novel DNA element. J Biol Chem 2001; 276:27716-20; PMID:11350951
- Lee KC, Crowe AJ, Barton MC. p53-mediated repression of alpha-fetoprotein gene expression by specific DNA binding. Mol. Cell Biol 1999; 19:1279-88; PMID:9891062
- Ogden SK, Lee KC, Wernke-Dollries K, Stratton SA, Aronow B, Barton MC. p53 targets chromatin structure alteration to repress alpha-fetoprotein gene expression. J Biol Chem 2001; 276:42057-62; PMID:11572852
- Nguyen TT, Cho K, Stratton SA, Barton MC. Transcription factor interactions and chromatin modifications associated with p53-mediated, developmental repression of the alpha-fetoprotein gene. Mol Cell Biol 2005; 25:2147-57; PMID:15743813
- Tsai WW, Nguyen TT, Shi Y, Barton MC. P53-targeted LSD1 functions in repression of chromatin structure and transcription in vivo. Mol Cell Biol 2008; 28:5139-46; PMID:18573881; http://dx.doi.org/10.1128/MCB.00287-08
- Mirza A, Wu Q, Wang L, McClanahan T, Bishop WR, Gheyas F, Ding W, Hutchins B, Hockenberry T, Kirschmeier P, et al. Global transcriptional program of p53 target genes during the process of apoptosis and cell cycle progression. Oncogene 2003; 22:3645-54; PMID:12789273
- Zaky A, Busso C, Izumi T, Chattopadhyay R, Bassiouny A, Mitra S, Bhakat KK. Regulation of the human AP-endonuclease (APE1Ref-1) expression by the tumor suppressor p53 in response to DNA damage. Nucleic Acids Res 2008; 36:1555-66; PMID:18208837; http://dx.doi.org/10.1093/nar/gkm1173
- Alimirah F, Panchanathan R, Chen J, Zhang X, Ho SM, Choubey D. Expression of androgen receptor is negatively regulated by p53. Neoplasia 2007; 9:1152-9; PMID:18084622
- Ceribelli M, Alcalay M, Vigano MA, Mantovani R. Repression of new p53 targets revealed by ChIP on chip experiments. Cell Cycle 2006; 5:1102-10; PMID:16721047
- Budhram-Mahadeo V, Morris PJ, Smith MD, Midgley CA, Boxer LM, Latchman DS. p53 suppresses the activation of the Bcl-2 promoter by the Brn-3a POU family transcription factor. J Biol Chem 1999; 274:15237-44; PMID:10329733
- Wu Y, Mehew JW, Heckman CA, Arcinas M, Boxer LM, Negative regulation of bcl-2 expression by p53 in hematopoietic cells. Oncogene 2001; 20:240-51; PMID:11313951
- Hoffman WH, Biade S, Zilfou JT, Chen J, Murphy M. Transcriptional repression of the anti-apoptotic survivin gene by wild type p53. J Biol Chem 2002; 277:3247-57; PMID:11714700
- Esteve PO, Chin HG, Pradhan S. Human maintenance DNA (cytosine-5)-methyltransferase and p53 modulate expression of p53-repressed promoters. Proc Natl Acad Sci USA 2005; 102:1000-5; PMID:15657147
- Raj D, Liu T, Samadashwily G, Li F, Grossman D. Survivin repression by p53 Rb and E2F2 in normal human melanocytes. Carcinogenesis 2008; 29:194-201; PMID:17916908
- Nabilsi NH, Broaddus RR, Loose DS. DNA methylation inhibits p53-mediated survivin repression. Oncogene 2009; 28:2046-50; PMID:19363521; http://dx.doi.org/10.1038/onc.2009.62
- Zalcenstein A, Weisz L, Stambolsky P, Bar J, Rotter V, Oren M. epression of the MSPMST-1 gene contributes to the antiapoptotic gain of function of mutant p53. Oncogene 2006; 25:359-69; PMID:16170349
- Feng X, Liu X, Zhang W, Xiao W.H. p53 directly suppresses BNIP3 expression to protect against hypoxia-induced cell death. Embo Journal 2011; 30:3397-415; PMID:21792176; http://dx.doi.org/10.1038/emboj.2011.248
- Jin W, Chen Y, Di GH, Miron P, Hou YF, Gao H, Shao ZM. Estrogen Receptor (ER)beta or p53 Attenuates ER alpha-mediated Transcriptional Activation on the BRCA2 Promoter. J Biol Chem 2008; 283:29671-80; PMID:18765668; http://dx.doi.org/10.1074/jbc.M802785200
- Innocente SA, Lee JM. p53 is a NF-Y- and p21-independent Sp1-dependent repressor of cyclin B1 transcription. Febs Lett. 2005; 579:1001-7; PMID:15710382
- Lipski R, Lippincott DJ, Durden BC, Kaplan AR, Keiser HE, Park JH, Levesque AA. p53 Dimers associate with a head-to-tail response element to repress cyclin B transcription. PLoS One 2012; 7(8):e42615; PMID:22905155; http://dx.doi.org/10.1371/journal.pone.0042615
- Imbriano C, Gurtner A, Cocchiarella F, Di Agostino S, Basile V, Gostissa M, Dobbelstein M, Del Sal G, Piaggio G, Mantovani R. Direct p53 transcriptional repression: in vivo analysis of CCAAT-containing G2M promoters. Mol Cell Biol 2005; 25:3737-51; PMID:15831478
- Dalvai M, Mondesert O, Bourdon JC, Ducommun B, Dozier C. Cdc25B is negatively regulated by p53 through Sp1 and NF-Y transcription factors. Oncogene 2011; 30:2282-8; PMID:21242964; http://dx.doi.org/10.1038/onc.2010.588
- St.Clair S, Giono L, Varmeh-Ziaie S, Resnick-Silverman L, Liu WJ, Padi A, Dastidar J, DaCosta A, Mattia M, Manfredi JJ. DNA damage-induced downregulation of Cdc25C is mediated by p53 via two independent mechanisms: one involves direct binding to the cdc25C promoter. Mol Cell 2004; 16:725-36; PMID:15574328
- Le Gac G, Esteve PO, Ferec C, Pradhan S. DNA damage-induced down-regulation of human Cdc25C and Cdc2 is mediated by cooperation between p53 and maintenance DNA (cytosine-5) methyltransferase 1. J Biol Chem 2006; 281:24161-70; PMID:16807237
- Zeng YX, Kotake Y, Pei XH, Smith MD, Xiong Y. p53 Binds to and Is Required for the Repression of Arf Tumor Suppressor by HDAC and Polycomb. Cancer Res 2011; 71:2781-92; PMID:21447739; http://dx.doi.org/10.1158/0008-5472.CAN-10-3483
- Bansal N, Kadamb R, Mittal S, Vig L, Sharma R, Dwarakanath BS, Saluja D. Tumor Suppressor Protein p53 Recruits Human Sin3BHDAC1 Complex for Down-Regulation of Its Target Promoters in Response to Genotoxic Stress. PLoS One 2011; 6(10):e26156; PMID:22028823; http://dx.doi.org/10.1371/journal.pone.0026156
- Banerjee T, Nath S, Roychoudhury S. DNA damage induced p53 downregulates Cdc20 by direct binding to its promoter causing chromatin remodeling. Nucleic Acids Res 2009; 37:2688-98; PMID:19273532; http://dx.doi.org/10.1093/nar/gkp110
- Godar S, Ince TA, Bell GW, Feldser D, Donaher JL, Bergh J, Liu A, Miu K, Watnick RS, Reinhardt F, et al. Growth-inhibitory and tumor- suppressive functions of p53 depend on its repression of CD44 expression. Cell 2008; 134:62-73; PMID:18614011; http://dx.doi.org/10.1016/j.cell.2008.06.006
- Yang MZ, Yuan F, Li P, Chen Z, Chen A, Li S, Hu C. Interferon regulatory factor 4 binding protein is a novel p53 target gene and suppresses cisplatininduced apoptosis of breast cancer cells. Mol Cancer 2012; 11:54; PMID:22888789
- Kho PS, Wang Z, Zhuang L, Li Y, Chew JL, Ng HH, Liu ET, Yu Q. p53-regulated transcriptional program associated with genotoxic stress-induced apoptosis. J Biol Chem 2004; 279:21183-92; PMID:15016801
- Peterson EJ, Bogler O, Taylor SM. p53-mediated repression of DNA methyltransferase 1 expression by specific DNA binding. Cancer Res 2003; 63:6579-82; PMID:14583449
- Lin RK, Wu CY, Chang JW, Juan LJ, Hsu HS, Chen CY, Lu YY, Tang YA, Yang YC, Yang PC, et al. Dysregulation of p53Sp1 Control Leads to DNA Methyltransferase-1 Overexpression in Lung Cancer. Cancer Res 2010; 70:5807-17; PMID:20570896; http://dx.doi.org/10.1158/0008-5472.CAN-09-4161
- Wilson PM, Fazzone W, LaBonte MJ, Lenz HJ, Ladner RD. Regulation of human dUTPase gene expression and p53-mediated transcriptional repression in response to oxaliplatin-induced DNA damage. Nucleic Acids Res 2009; 37:78-95; PMID:19015155; http://dx.doi.org/10.1093/nar/gkn910
- Scoumanne A, Chen XB, The epithelial cell transforming sequence 2, a guanine nucleotide exchange factor for Rho GTPases is repressed by p53 via protein methyltransferases and is required for G(1)-S transition. Cancer Res 2006; 66:6271-9; PMID:16778203
- Sankpal NV, Willman MW, Fleming TP, Mayfield JD, Gillanders WE. Transcriptional Repression of Epithelial Cell Adhesion Molecule Contributes to p53 Control of Breast Cancer Invasion. Cancer Res 2009; 69:753-7; PMID:19141643; http://dx.doi.org/10.1158/0008-5472.CAN-08-2708
- Salah Z, Haupt S, Maoz M, Baraz L, Rotter V, Peretz T, Haupt Y, Bar-Shavit R. p53 controls hPar1 function and expression. Oncogene 2008; 27:6866-74; PMID:18820708; http://dx.doi.org/10.1038/onc.2008.324
- Wong J, Li PX, Klamut HJ. A novel p53 transcriptional repressor element (p53TRE) and the asymmetrical contribution of two p53 binding sites modulate the response of the placental transforming growth factor-beta promoter to p53. J Biol Chem 2002; 277:26699-707; PMID:12011055
- Du P, Tang FQ, Qiu YL, Dong F. GFI1 is repressed by p53 and inhibits DNA damage-induced apoptosis. PLoS One 2013; 8(9):e73542; PMID:24023884; http://dx.doi.org/10.1371/journal.pone.0073542
- Maeda Y, Hwang-Verslues WW, Wei G, Fukazawa T, Durbin ML, Owen LB, Liu X, Sladek FM. Tumour suppressor p53 down-regulates the expression of the human hepatocyte nuclear factor 4alpha (HNF4alpha) gene. Biochem J 2006; 400:303-13; PMID:16895524
- Zhang Y, Wang JS, Chen LL, Zhang Y, Cheng XK, Heng FY, Wu NH, Shen YF. Repression of hsp90 beta gene by p53 in UV irradiation-induced apoptosis of Jurkat cells. J Biol Chem 2004; 279:42545-51; PMID:15284248
- Paolella BR, Havrda MC, Mantani A, Wray CM, Zhang Z, Israel MA. p53 Directly represses id2 to inhibit the proliferation of neural progenitor cells. Stem Cells 2011; 29:1090-101; PMID:21608079; http://dx.doi.org/10.1002/stem.660
- Im HJ, Pittelkow MR, Kumar R. Divergent regulation of the growth-promoting gene IEX-1 by the p53 tumor suppressor and Sp1. J Biol Chem 2002; 277:14612-21; PMID:11844788
- Werner H, Karnieli E, Rauscher FJ, LeRoith D. Wild-type and mutant p53 differentially regulate transcription of the insulin-like growth factor I receptor gene. Proc Natl Acad Sci USA 1996; 93:8318-23; PMID:8710868
- Kavurma MM, Figg N, Bennett MR, Mercer J, Khachigian LM, Littlewood TD. Oxidative stress regulates IGF1R expression in vascular smooth-muscle cells via p53 and HDAC recruitment. Biochem J 2007; 407:79-87; PMID:17600529
- Zhang LJ, Kashanchi F, Zhan Q, Zhan S, Brady JN, Fornace AJ, Seth P, Helman LJ. Regulation of insulin-like growth factor II p3 promoter by p53: A potential mechanism for tumorigenesis. Cancer Res 1996; 56 1367-73; PMID:8640827
- Cai BH, Hsu PC, Hsin IL, Chao CF, Lu MH, Lin HC, Chiou SH, Tao PL, Chen JY. p53 Acts as a Co-Repressor to Regulate Keratin 14 Expression during Epidermal Cell Differentiation. PLoS One 2012; 7(7):e41742; PMID:22911849; http://dx.doi.org/10.1371/journal.pone.0041742
- Wang B, Feng P, Xiao ZW, Ren EC. LIM and SH3 protein 1 (Lasp1) is a novel p53 transcriptional target involved in hepatocellular carcinoma. J Hepatol 2009; 50:528-37; PMID:19155088; http://dx.doi.org/10.1016/j.jhep.2008.10.025
- Cecchinelli B, Lavra L, Rinaldo C, Iacovelli S, Gurtner A, Gasbarri A, Ulivieri A, Del Prete F, Trovato M, Piaggio G, et al. Repression of the antiapoptotic molecule galectin-3 by homeodomain-interacting protein kinase 2-activated p53 is required for p53-induced apoptosis. Mol Cell Biol 2006; 26:4746-57; PMID:16738336
- Chun AC, Jin DY. Transcriptional regulation of mitotic checkpoint gene MAD1 by p53. J. Biol. Chem 2003; 278:37439-50; PMID:12876282
- Murphy M, Ahn J, Walker KK, Hoffman WH, Evans RM, Levine AJ, George DL. Transcriptional repression by wild-type p53 utilizes histone deacetylases mediated by interaction with mSin3a. Genes Dev 1999; 13:2490-501; PMID:10521394
- Jiang P, Du WJ, Mancuso A, Wellen KE, Yang XL. Reciprocal regulation of p53 and malic enzymes modulates metabolism and senescence. Nature 2013; 493(7434):689-93; PMID:23334421; http://dx.doi.org/10.1038/nature11776
- Lee MH, Na H, Kim EJ, Lee HW, Lee MO. Poly(ADP-ribosyl)ation of p53 induces gene-specific transcriptional repression of MTA1. Oncogene 2012; 31:5099-107; PMID:22286760; http://dx.doi.org/10.1038/onc.2012.2
- Ho JS, Ma W, Mao DY, Benchimol S. p53-Dependent transcriptional repression of c-myc is required for G1 cell cycle arrest. Mol. Cell Biol 2005; 25:7423-31; PMID:16107691
- Lin TX, Chao C, Saito S, Mazur SJ, Murphy ME, Appella E, Xu Y. P53 induces differentiation of mouse embryonic stem cells by suppressing Nanog expression. Nat Cell Biol 2005; 7:165-U80; PMID:15619621
- Nabilsi NH, Ryder DJ, Peraza-Penton AC, Poudyal R, Loose DS, Kladde MP. Local Depletion of DNA Methylation Identifies a Repressive p53 Regulatory Region in the NEK2 Promoter. J Biol Chem 2013; 288:35940-51; PMID:24163369; http://dx.doi.org/10.1074/jbc.M113.523837
- Li YZ, Lu DY, Tan WQ, Wang JX, Li PF. p53 initiates apoptosis by transcriptionally targeting the antiapoptotic protein ARC. Mol. Cell Biol 2008; 28:564-74; PMID:17998337
- Mortensen K, Skouv J, Hougaard DM, Larsson LI. Endogenous endothelial cell nitric-oxide synthase modulates apoptosis in cultured breast cancer cells and is transcriptionally regulated by p53. J Biol Chem 1999; 274:37679-84; PMID:10608825
- Saifudeen Z, Marks J, Du H, El-Dahr SS. Spatial repression of PCNA by p53 during kidney development. Am J Physiol Renal Physiol 2002; 283:F727-33; PMID:12217864
- Astanehe A, Arenillas D, Wasserman WW, Leung PC, Dunn SE, Davies BR, Mills GB, Auersperg N. Mechanisms underlying p53 regulation of PIK3CA transcription in ovarian surface epithelium and in ovarian cancer. J Cell Sci 2008; 121:664-74; PMID:18270270; http://dx.doi.org/10.1242/jcs.013029
- McKenzie L, King S, Marcar L, Nicol S, Dias SS, Schumm K, Robertson P, Bourdon JC, Perkins N, Fuller-Pace F, et al. p53-dependent repression of polo-like kinase-1 (PLK1;. Cell Cycle 2010; 9:4200-12; PMID:20962589
- Zhou Z, Cao JX, Li SY, An GS, Ni JH, Jia HT. p53 Suppresses E2F1-dependent PLK1 expression upon DNA damage by forming p53-E2F1-DNA complex. Exp Cell Res 2013; 319:3104-15; PMID:24076372; http://dx.doi.org/10.1016/j.yexcr.2013.09.012
- Van Bodegom D, Saifudeen Z, Dipp S, Puri S, Magenheimer BS, Calvet JP, El-Dahr SS. The polycystic kidney disease-1 gene is a target for p53-mediated transcriptional repression. J Biol Chem 2006; 281:31234-44; PMID:16931520
- Van Bodegom D, Roessingh W, Pridjian A, El Dahr SS. Mechanisms of p53-mediated repression of the human polycystic kidney disease-1 promoter. Biochim Biophys Acta 2010; 1799:502-09; PMID:20388565
- Islam MR, Jimenez T, Pelham C, Rodova M, Puri S, Magenheimer BS, Maser RL, Widmann C, Calvet JP. MAPERK Kinase Kinase 1 (MEKK1) Mediates Transcriptional Repression by Interacting with Polycystic Kidney Disease-1 (PKD1) Promoter-bound p53 Tumor Suppressor Protein. J Biol Chem 2010; 285:38818-31.
- Li BQ ,Lee MYW Transcriptional regulation of the human DNA polymerase delta catalytic subunit gene POLD1 by p53 tumor suppressor and Sp1. J Biol Chem 2001; 276:29729-39; PMID:11375983
- Sahin E, Colla S, Liesa M, Moslehi J, Müller FL, Guo M, Cooper M, Kotton D, Fabian AJ, Walkey C, et al. Telomere dysfunction induces metabolic and mitochondrial compromise. Nature 2011; 470:359-65; PMID:21307849; http://dx.doi.org/10.1038/nature09787
- Li C, Lin M, Liu J. Identification of PRC1 as the p53 target gene uncovers a novel function of p53 in the regulation of cytokinesis. Oncogene 2004; 23:9336-47; PMID:15531928; http://dx.doi.org/10.1038/sj.onc.1208114
- Hsieh WJ, Hsieh SC, Chen CC, Wang FF. Human DDA3 is an oncoprotein down-regulated by p53 and DNA damage. Biochem Biophys Res Commun 2008; 369:567-72; PMID:10196169; http://dx.doi.org/10.1016/j.bbrc.2008.02.047
- Subbaramaiah K, Altorki N, Chung WJ, Mestre JR, Sampat A, Dannenberg AJ. Inhibition of cyclooxygenase-2 gene expression by p53. J. Biol. Chem 1999; 274:10911-5; PMID:10196169; http://dx.doi.org/10.1074/jbc.274.16.10911
- Montano X. Repression of SHP-1 expression by p53 leads to trkA tyrosine phosphorylation and suppression of breast cancer cell proliferation. Oncogene 2009; 28:3787-800; PMID:19749791; http://dx.doi.org/10.1038/onc.2009.143
- Golubovskaya V, Kaur A, Cance W. Cloning and characterization of the promoter region of human focal adhesion kinase gene: nuclear factor kappa B and p53 binding sites. B B A-Gene Struct Expr 2004; 1678:111-25; http://dx.doi.org/10.1016/j.bbaexp.2004.03.002
- Golubovskaya VM, Finch R, Kweh F, Massoll NA, Campbell-Thompson M, Wallace MR, Cance WG. P53 regulates FAK expression in human tumor cells. Mol Carcinogen 2008; 47:373-82; PMID:17999388; http://dx.doi.org/10.1002/mc.20395
- Arias-Lopez C, Lazaro-Trueba I, Kerr P, Lord CJ, Dexter T, Iravani M, Ashworth A, Silva A. p53 modulates homologous recombination by transcriptional regulation of the RAD51 gene. EMBO Rep 2006; 7:219-24; PMID:16322760; http://dx.doi.org/10.1038/sj.embor.7400587
- Osifchin NE, Jiang D, Ohtani-Fujita N, Fujita T, Carroza M, Kim SJ, Sakai T, Robbins PD. Identification of A P53 Binding-Site in the Human Retinoblastoma Susceptibility Gene Promoter. J Biol Chem 1994; 269:6383-9.380; PMID:8119988
- Sengupta S, Shimamoto A, Koshiji M, Pedeux R, Rusin M, Spillare EA, Shen JC, Huang LE, Lindor NM, Furuichi Y, et al. Tumor suppressor p53 represses transcription of RECQ4 helicase. Oncogene 2005; 24:1738-48; PMID:15674334; http://dx.doi.org/10.1038/sj.onc.1208380
- Farkas C, Martins CP, Escobar D, Hepp MI, Donner DB, Castro AF, Evan G, Gutiérrez JL, Warren R, Pincheira R. Wild Type p53 Transcriptionally Represses the SALL2 Transcription Factor under Genotoxic Stress. PLoS One 2013; 8:e73817; PMID:24040083; http://dx.doi.org/10.1371/journal.pone.0073817
- Schwartzenberg-Bar-Yoseph F, Armoni M, Karnieli E. The tumor suppressor p53 down-regulates glucose transporters GLUT1 and GLUT4 gene expression. Cancer Res 2004; 64:2627-33; PMID:15059920; http://dx.doi.org/10.1158/0008-5472.CAN-03-0846
- Han XB, Patters AB, Chesney RW. Transcriptional repression of taurine transporter gene (TauT) by p53 in renal cells. J Biol Chem 2002; 277:39266-73; PMID:12163498; http://dx.doi.org/10.1074/jbc.M205939200
- Faraonio R, Vergara P, Di Marzo D, Pierantoni MG, Napolitano M, Russo T, Cimino F. p53 suppresses the Nrf2-dependent transcription of antioxidant response genes. J Biol Chem 2006; 281:39776-84; PMID:17077087; http://dx.doi.org/10.1074/jbc.M605707200
- Dhar SK, Xu Y, Chen YM, Clair DKS. Specificity protein 1-dependent p53-mediated suppression of human manganese superoxide dismutase gene expression. J Biol Chem 2006; 281:21698-709; PMID:16740634; http://dx.doi.org/10.1074/jbc.M601083200
- Dhar SK, Xu Y, St Clair DK. Nuclear Factor kappa B- and Specificity Protein 1-dependent p53-mediated Bi-directional Regulation of the Human Manganese Superoxide Dismutase Gene. J Biol Chem 2010; 285:9835-46; PMID:20061391; http://dx.doi.org/10.1074/jbc.M109.060715
- Dhar SK, Tangpong J, Chaiswing L, Oberley TD, St Clair DK. Manganese Superoxide Dismutase Is a p53-Regulated Gene That Switches Cancers between Early and Advanced Stages. Cancer Res 2011; 71:6684-95; PMID:22009531; http://dx.doi.org/10.1158/0008-5472.CAN-11-1233
- Kim E, Günther W, Yoshizato K, Meissner H, Zapf S, Nüsing RM, Yamamoto H, Van Meir EG, Deppert W, Giese A. Tumor suppressor p53 inhibits transcriptional activation of invasion gene thromboxane synthase mediated by the proto-oncogenic factor ets-1. Oncogene 2003; 22:7716-27; PMID:14586398; http://dx.doi.org/10.1038/sj.onc.1207155
- Tu SP, Chi AL, Ai W, Takaishi S, Dubeykovskaya Z, Quante M, Fox JG, Wang TC. p53 inhibition of AP1-dependent TFF2 expression induces apoptosis and inhibits cell migration in gastric cancer cells. A J Physiol-Gast L Phys 2009; 297:G385-96; PMID:19541923
- D’Souza S, Xin H, Walter S, Choubey D. The gene encoding p202 an interferon-inducible negative regulator of the p53 tumor suppressor is a target of p53-mediated transcriptional repression. J. Biol. Chem 2001; 276:298-305; PMID:11013253; http://dx.doi.org/10.1074/jbc.M007155200
- Amson R, Pece S, Lespagnol A, Vyas R, Mazzarol G, Tosoni D, Colaluca I, Viale G, Rodrigues-Ferreira S, Wynendaele J, et al. Reciprocal repression between P53 and TCTP. Nat. Med 2011; 18:91-9; PMID:22157679; http://dx.doi.org/10.1038/nm.2546
- Zhang DW, Jeang KT, Lee CGL. p53 negatively regulates the expression of FAT10 a gene upregulated in various cancers. Oncogene 2006; 25:2318-27; PMID:16501612; http://dx.doi.org/10.1038/sj.onc.1209220
- van der Watt PJ, Leaner VD. The nuclear exporter Crm1 is regulated by NFY and Sp1 in cancer cells and repressed by p53 in response to DNA damage. BiochimBioph Acta-Gene Regul Mech 2011; 1809:316-26.
- Radhakrishnan VM, Putnam CW, Qi WQ, Martinez JD. P53 suppresses expression of the 14-3-3gamma oncogene. Bmc Cancer 2011; 11; PMID:21791091; http://dx.doi.org/10.1186/1471-2407-11-378
- Yan HL, Xue G, Mei Q, Wang YZ, Ding FX, Liu MF, Lu MH, Tang Y, Yu HY, Sun SH. Repression of the miR-17-92 cluster by p53 has an important function in hypoxia-induced apoptosis. EMBO J 2009; 28:2719-32; PMID:19696742; http://dx.doi.org/10.1038/emboj.2009.214
- Johnson RA, Ince TA, Scotto KW. Transcriptional repression by p53 through direct binding a novel DNA element. J Biol Chem 2001; 276:27716-20; PMID:11350951; http://dx.doi.org/10.1074/jbc.C100121200
- Budhram-Mahadeo V, Morris PJ, Smith MD, Midgley CA, Boxer LM, Latchman DS. p53 suppresses the activation of the Bcl-2 promoter by the Brn-3a POU family transcription factor. J Biol Chem 1999; 274:15237-44; PMID:10329733; http://dx.doi.org/10.1074/jbc.274.21.15237
- Innocente SA, Lee JM. P53 is a NF-Y- and p21-independent Sp1-dependent repressor of cyclin B1 transcription. Febs Lett 2005; 579:1001-7; PMID:15710382; http://dx.doi.org/10.1016/j.febslet.2004.12.073
- Banerjee T, Nath S, Roychoudhury S. DNA damage induced p53 downregulates Cdc20 by direct binding to its promoter causing chromatin remodeling. Nucleic Acids Res 2009; 37:2688-98; PMID:19273532; http://dx.doi.org/10.1093/nar/gkp110
- Chun ACS, Jin DY. Transcriptional regulation of mitotic checkpoint gene MAD1 by p53. J Biol Chem 2003; 278:37439-50; PMID:12876282; http://dx.doi.org/10.1074/jbc.M307185200
- de Toledo SM, Azzam EI, Keng P, Laffrenier S, Little JB. Regulation by ionizing radiation of CDC2 cyclin A cyclin B thymidine kinase topoisomerase IIalpha and RAD51 expression in normal human diploid fibroblasts is dependent on p53p21Waf1. Cell Growth Differ 1998; 9:887-96; PMID:9831241
- Chang BD, Watanabe K, Broude EV, Fang J, Poole JC, Kalinichenko TV, Roninson IB. Effects of p21(Waf1Cip1Sdi1) on cellular gene expression: Implications for carcinogenesis senescence and age-related diseases. Proc Natl Acad Sci USA 2000; 97:4291-6.
- Flatt PM, Tang LJ, Scatena CD, Szak ST, Pietenpol JA. p53 regulation of G(2) checkpoint is retinoblastoma protein dependent. Mol Cell Biol 2000; 20:4210-23; PMID:10825186; http://dx.doi.org/10.1128/MCB.20.12.4210-4223.2000
- Löhr K, Möritz C, Contente A, Dobbelstein M. p21CDKN1A mediates negative regulation of transcription by p53. J Biol Chem 2003; 278:32507-16; PMID:12748190
- Tabach Y, Milyavsky M, Shats I, Brosh R, Zuk O, Yitzhaky A, Mantovani R, Domany E, Rotter V, Pilpel Y. The promoters of human cell cycle genes integrate signals from two tumor suppressive pathways during cellular transformation. Mol Syst Biol 2005; 1:2005; PMID:16729057
- Jackson MW, Agarwal MK, Yang J, Bruss P, Uchiumi T, Agarwal ML, Stark GR, Taylor WR. p130p107p105Rb-dependent transcriptional repression during DNA-damage-induced cell-cycle exit at G2. J Cell Sci 2005; 118:1821-32; PMID:15827088; http://dx.doi.org/10.1242/jcs.02307
- Ginsberg D, Mechta F, Yaniv M, Oren M, Wild-type p53 can down-modulate the activity of various promoters. Proc Natl Acad Sci USA 1991; 88:9979-83; PMID:1946467; http://dx.doi.org/10.1073/pnas.88.22.9979
- Wang B, Xiao ZW, Ren EC. Redefining the p53 response element. Proc Natl Acad Sci USA 2009; 106:14373-8; PMID:19597154
- Imbriano C, Gnesutta N, Mantovani R. The NF-Yp53 liaison: Well beyond repression. Biochim Biophys Acta 2012; 1825:131-9; PMID:22138487
- Chen X, Müller GA, Quaas M, Fischer M, Han N, Stutchbury B, Sharrocks AD, Engeland K. The forkhead transcription factor FOXM1 controls cell cycle-dependent gene expression through an atypical chromatin binding mechanism. Mol Cell Biol 2013; 33:227-36; PMID:23109430; http://dx.doi.org/10.1128/MCB.00881-12
- Chicas A, Wang X, Zhang C, McCurrach M, Zhao Z, Mert O, Dickins RA, Narita M, Zhang M, Lowe SW. Dissecting the unique role of the retinoblastoma tumor suppressor during cellular senescence. Cancer Cell 2010; 17:376-87; PMID:20385362; http://dx.doi.org/10.1016/j.ccr.2010.01.023
- Litovchick L, Sadasivam S, Florens L, Zhu X, Swanson SK, Velmurugan S, Chen R, Washburn MP, Liu XS, DeCaprio JA. Evolutionarily conserved multisubunit RBL2p130 and E2F4 protein complex represses human cell cycle-dependent genes in quiescence. Mol. Cell 2007; 26:539-51; PMID:17531812; http://dx.doi.org/10.1016/j.molcel.2007.04.015
- Sadasivam S, Duan S, DeCaprio JA. The MuvB complex sequentially recruits B-Myb and FoxM1 to promote mitotic gene expression. Genes Dev 2012; 26:474-89; PMID:22391450; http://dx.doi.org/10.1101/gad.181933.111
- Resnick-Silverman L, St Clair S, Maurer M, Zhao K, Manfredi JJ. Identification of a novel class of genomic DNA-binding sites suggests a mechanism for selectivity in target gene activation by the tumor suppressor protein p53. Genes Dev 1998; 12:2102-7; PMID:9679054; http://dx.doi.org/10.1101/gad.12.14.2102
- Krause K, Haugwitz U, Wasner M, Wiedmann M, Mössner J, Engeland K. Expression of the cell cycle phosphatase cdc25C is down-regulated by the tumour suppressor protein p53 but not by p73. Biochem Biophys Res Commun 2001; 284:743-50; PMID:11396965; http://dx.doi.org/10.1006/bbrc.2001.5040
- Benson EK, et al. p53-dependent gene repression through p21 is mediated by recruitment of E2F4 repression complexes. Oncogene 2014; 33:3959-69;http://dx.doi.org/10.1038/onc.2013.378
- Spitkovsky D, Schulze A, Boye B, Jansen-Durr P, Down-regulation of cyclin A gene expression upon genotoxic stress correlates with reduced binding of free E2F to the promoter. Cell Growth Differ 1997; 8:699-710; PMID:9186003
- Azzam EI, deToledo SM, Pykett MJ, Nagasawa H, Little JB. CDC2 is down-regulated by ionizing radiation in a p53-dependent manner. Cell Growth Differ 1997; 8:1161-9.
- Gottifredi V, Karni-Schmidt O, Shieh SS, Prives C. p53 down-regulates CHK1 through p21 and the retinoblastoma protein. Mol Cell Biol 2001; 21:1066-76; PMID:11158294; http://dx.doi.org/10.1128/MCB.21.4.1066-1076.2001
- Zhu H, Chang BD, Uchiumi T, Roninson IB. Identification of promoter elements responsible for transcriptional inhibition of polo-like kinase 1 and topoisomerase IIalpha genes by p21(WAF1CIP1SDI1. Cell Cycle 2002; 1:59-66; PMID:12429910
- Shats I, Milyavsky M, Tang X, Stambolsky P, Erez N, Brosh R, Kogan I, Braunstein I, Tzukerman M, Ginsberg D, et al. p53-dependent down-regulation of telomerase is mediated by p21waf1. J Biol Chem 2004; 279:50976-85; PMID:15371422; http://dx.doi.org/10.1074/jbc.M402502200
- Jackson JG, Pereira-Smith OM. Primary and compensatory roles for RB family members at cell cycle gene promoters that are deacetylated and downregulated in doxorubicin-induced senescence of breast cancer cells. Mol Cell Biol 2006; 26:2501-10; PMID:16537896; http://dx.doi.org/10.1128/MCB.26.7.2501-2510.2006
- Barsotti AM, Prives C. Pro-proliferative FoxM1 is a target of p53-mediated repression. Oncogene 2009; 28:4295-305; PMID:19749794; http://dx.doi.org/10.1038/onc.2009.282
- Schvartzman JM, Duijf PH, Sotillo R, Coker C, Benezra R. Mad2 is a critical mediator of the chromosome instability observed upon Rb and p53 pathway inhibition. Cancer Cell 2011; 19:701-14; PMID:21665145; http://dx.doi.org/10.1016/j.ccr.2011.04.017
- Mannefeld M, Klassen E, Gaubatz S. B-MYB is required for recovery from the DNA damage-induced G2 checkpoint in p53 mutant cells. Cancer Res 2009; 69:4073-80; PMID:19383908; http://dx.doi.org/10.1158/0008-5472.CAN-08-4156
- Quaas M, Müller GA, Engeland K. p53 can repress transcription of cell cycle genes through a p21(WAF1CIP1)-dependent switch from MMB to DREAM protein complex binding at CHR promoter elements. Cell Cycle 2012; 11:4661-72; PMID:23187802; http://dx.doi.org/10.4161/cc.22917
- Fischer M, Grundke I, Sohr S, Quaas M, Hoffmann S, Knörck A, Gumhold C, Rother K. p53 and cell cycle dependent transcription of kinesin family member 23 (KIF23) is controlled via a CHR promoter element bound by DREAM and MMB complexes. PLoS One 2013; 8:e63187; PMID:23650552; http://dx.doi.org/10.1371/journal.pone.0063187
- Fischer M, Quaas M, Wintsche A, Müller GA, Engeland K. Polo-like kinase 4 transcription is activated via CRE and NRF1 elements repressed by DREAM through CDECHR sites and deregulated by HPV E7 protein. Nucleic Acids Res 2014; 42:163-80; PMID:24071582; http://dx.doi.org/10.1093/nar/gkt849
- Schmit F, Korenjak M, Mannefeld M, Schmitt K, Franke C, von Eyss B, Gagrica S, Hänel F, Brehm A, Gaubatz S. LINC a human complex that is related to pRB-containing complexes in invertebrates regulates the expression of G2M genes. Cell Cycle 2007; 6:1903-13; PMID:17671431; http://dx.doi.org/10.4161/cc.6.15.4512
- Sadasivam S, DeCaprio JA. The DREAM complex: master coordinator of cell cycle-dependent gene expression. Nat. Rev. Cancer 2013; 13:585-95; PMID:23842645; http://dx.doi.org/10.1038/nrc3556
- Taylor WR, Schonthal AH, Galante J, Stark GR. p130E2F4 binds to and represses the cdc2 promoter in response to p53. J Biol Chem 2001; 276:1998-2006; PMID:11032828; http://dx.doi.org/10.1074/jbc.M005101200
- Scian MJ, Carchman EH, Mohanraj L, Stagliano KE, Anderson MA, Deb D, Crane BM, Kiyono T, Windle B, Deb SP, et al. Wild-type p53 and p73 negatively regulate expression of proliferation related genes. Oncogene 2008; 27:2583-93; PMID:17982488; http://dx.doi.org/10.1038/sj.onc.1210898
- Desdouets C, Ory C, Matesic G, Soussi T, Bréchot C, Sobczak-Thépot J. ATF/CREB site mediated transcriptional activation and p53 dependent repression of the cyclin A promoter. Febs Lett 1996; 385:34-8; PMID:8641461; http://dx.doi.org/10.1016/0014-5793(96)00330-4
- Kidokoro T, Tanikawa C, Furukawa Y, Katagiri T, Nakamura Y, Matsuda K. CDC20 a potential cancer therapeutic target is negatively regulated by p53. Oncogene 2008; 27:1562-71; PMID:17873905; http://dx.doi.org/10.1038/sj.onc.1210799
- Badie C, Bourhis J, Sobczak-Thépot J, Haddada H, Chiron M, Janicot M, Janot F, Tursz T, Vassal G. p53-dependent G2 arrest associated with a decrease in cyclins A2 and B1 levels in a human carcinoma cell line. Br J Cancer 2000; 82:642-50; PMID:10682678
- Tang X, Milyavsky M, Shats I, Erez N, Goldfinger N, Rotter V. Activated p53 suppresses the histone methyltransferase EZH2 gene. Oncogene 2004; PMID:15208672; 23:5759-69; http://dx.doi.org/10.1038/sj.onc.1207706
- Moberg KH, Tyndall WA, Hall DJ. Wild-type murine p53 represses transcription from the murine c-myc promoter in a human glial cell line. J Cell Biochem 1992; 49:208-15; PMID:1400626; http://dx.doi.org/10.1002/jcb.240490213
- Mungamuri SK, Benson EK, Wang S, Gu W, Lee SW, Aaronson SA. p53-mediated heterochromatin reorganization regulates its cell fate decisions. Nat Struct Mol Biol 2012; 19:478-84 S1; PMID:22466965; http://dx.doi.org/10.1038/nsmb.2271
- Krause K, Wasner M, Reinhard W, Haugwitz U, Dohna CL, Mössner J, Engeland K. The tumour suppressor protein p53 can repress transcription of cyclin B. Nucleic Acids Res 2000; 28:4410-8; PMID:11071927; http://dx.doi.org/10.1093/nar/28.22.4410
- Rother K, Kirschner R, Sänger K, Böhlig L, Mössner J, Engeland K. p53 downregulates expression of the G(1)S cell cycle phosphatase Cdc25A. Oncogene 2007; 26:1949-53; PMID:17001315; http://dx.doi.org/10.1038/sj.onc.1209989
- Rother K, Dengl M, Lorenz J, Tschöp K, Kirschner R, Mössner J, Engeland K. Gene expression of cyclin-dependent kinase subunit Cks2 is repressed by the tumor suppressor p53 but not by the related proteins p63 or p73. Febs Lett 2007; 581:1166-72; PMID:17336302; http://dx.doi.org/10.1016/j.febslet.2007.02.028
- Rother K, Li YY, Tschöp K, Kirschner R, Müller GA, Mössner J, Engeland K. Expression of cyclin-dependent kinase subunit 1 (Cks1) is regulated during the cell cycle by a CDECHR Tandem element and is downregulated by p53 but not by p63 or p73. Cell Cycle 2007; 6:853-62; PMID:17377499; http://dx.doi.org/10.4161/cc.6.7.4017
- Dobbelstein M. Interchanging heads: p53 re-composes the DREAMMMB complex to repress transcription. Cell Cycle 2013; 12:11; PMID:23255095; http://dx.doi.org/10.4161/cc.23169
- Westendorp B, Mokry M, Groot Koerkamp MJ, Holstege FC, Cuppen E, de Bruin A. E2F7 represses a network of oscillating cell cycle genes to control S-phase progression. Nucleic Acids Res 2012; 40:3511-23; PMID:22180533; http://dx.doi.org/10.1093/nar/gkr1203
- Moody CA, Laimins LA. Human papillomavirus oncoproteins: pathways to transformation. Nat Rev Cancer 2010; 10:550-60; PMID:20592731; http://dx.doi.org/10.1038/nrc2886
- Dyson N, Howley PM, Munger K, Harlow E. The human papilloma virus-16 E7 oncoprotein is able to bind to the retinoblastoma gene product. Science 1989; 243:934-7; PMID:2537532; http://dx.doi.org/10.1126/science.2537532
- Nor Rashid N, Yusof R, Watson RJ. Disruption of repressive p130-DREAM complexes by human papillomavirus 16 E6E7 oncoproteins is required for cell-cycle progression in cervical cancer cells. J Gen Virol 2011; 92:2620-7; PMID:21813705; http://dx.doi.org/10.1099/vir.0.035352-0
- Mack DH, Vartikar J, Pipas JM, Laimins L. A. Specific repression of TATA-mediated but not initiator-mediated transcription by wild-type p53. Nature 1993; 363:281-3; PMID:8387645; http://dx.doi.org/10.1038/363281a0
- Shaulian E, Haviv I, Shaul Y, Oren M. Transcriptional repression by the C-terminal domain of p53. Oncogene 1995; 10:671-80; PMID:7862444
- Agoff SN, Hou J, Linzer DI, Wu B. Regulation of the human hsp70 promoter by p53. Science 1993; 259:84-7; PMID:8418500; http://dx.doi.org/10.1126/science.8418500
- Kubicka S, Kühnel F, Zender L, Rudolph KL, Plümpe J, Manns M, Trautwein C. p53 represses CAAT enhancer-binding protein (CEBP)-dependent transcription of the albumin gene. A molecular mechanism involved in viral liver infection with implications for hepatocarcinogenesis. J Biol Chem 1999; 274:32137-44; PMID:10542249; http://dx.doi.org/10.1074/jbc.274.45.32137
- Esteve PO, Chin HG, Pradhan S. Molecular mechanisms of transactivation and doxorubicin-mediated repression of survivin gene in cancer cells. J Biol Chem 2007; 282:2615-25; PMID:17124180; http://dx.doi.org/10.1074/jbc.M606203200
- Chun JY, Hu Y, Pinder E, Wu J, Li F, Gao AC. Selenium inhibition of survivin expression by preventing Sp1 binding to its promoter. Mol Cancer Ther 2007; 6:2572-80; PMID:17876054; http://dx.doi.org/10.1158/1535-7163.MCT-07-0172
- Wu K, Jiang SW, Couch FJ. p53 mediates repression of the BRCA2 promoter and down-regulation of BRCA2 mRNA and protein levels in response to DNA damage. J Biol Chem 2003; 278:15652-60; PMID:12591928; http://dx.doi.org/10.1074/jbc.M211297200
- Manni I, Mazzaro G, Gurtner A, Mantovani R, Haugwitz U, Krause K, Engeland K, Sacchi A, Soddu S, Piaggio G. NF-Y mediates the transcriptional inhibition of the cyclin B1 cyclin B2 and cdc25C promoters upon induced G2 arrest. J Biol Chem 2001; 276:5570-6; PMID:11096075; http://dx.doi.org/10.1074/jbc.M006052200
- Matsui T, Katsuno Y, Inoue T, Fujita F, Joh T, Niida H, Murakami H, Itoh M, Nakanishi M. Negative regulation of Chk2 expression by p53 is dependent on the CCAAT-binding transcription factor NF-Y. J Biol Chem 2004; 279:25093-100; PMID:15044452; http://dx.doi.org/10.1074/jbc.M403232200
- Gu L, Zhu N, Findley HW, Woods WG, Zhou M. Identification and characterization of the IKKalpha promoter: positive and negative regulation by ETS-1 and p53 respectively. J Biol Chem 2004; 279:52141-9; PMID:15469934; http://dx.doi.org/10.1074/jbc.M407915200
- Elias A, Wu J, Chen T. Tumor suppressor protein p53 negatively regulates human pregnane X receptor activity. Mol Pharmacol 2013; 83:1229-36; PMID:23536728; http://dx.doi.org/10.1124/mol.113.085092
- Zhu N, Gu L, Findley HW, Zhou M. Transcriptional repression of the eukaryotic initiation factor 4E gene by wild type p53. Biochem Biophys Res Commun 2005; 335:1272-9; PMID:16112647; http://dx.doi.org/10.1016/j.bbrc.2005.08.026
- Faniello MC, Di Sanzo M, Quaresima B, Baudi F, Di Caro V, Cuda G, Morrone G, Del Sal G, Spinelli G, Venuta S, et al. p53-mediated downregulation of H ferritin promoter transcriptional efficiency via NF-Y. Int J Biochem Cell Biol 2008; 40:2110-9; PMID:18372207; http://dx.doi.org/10.1016/j.biocel.2008.02.010
- Webster NJ, Resnik JL, Reichart DB, Strauss B, Haas M, Seely BL. Repression of the insulin receptor promoter by the tumor suppressor gene product p53: a possible mechanism for receptor overexpression in breast cancer. Cancer Res 1996; 56:2781-8; PMID:8665514
- Lin YC, Chen YN, Lin KF, Wang FF, Chou TY, Chen MY. Association of p21 with NF-YA suppresses the expression of Polo-like kinase 1 and prevents mitotic death in response to DNA damage. Cell Death Dis 2014; 5:e987; PMID:24407240; http://dx.doi.org/10.1038/cddis.2013.527
- Zhan M, Yu D, Liu J, Glazer RI, Hannay J, Pollock RE. Transcriptional repression of protein kinase Calpha via Sp1 by wild type p53 is involved in inhibition of multidrug resistance 1 P-glycoprotein phosphorylation. J Biol Chem 2005; 280:4825-33; PMID:15563462; http://dx.doi.org/10.1074/jbc.M407450200
- Zhou Y, Mehta KR, Choi AP, Scolavino S, Zhang X. DNA damage-induced inhibition of securin expression is mediated by p53. J Biol Chem 2003; 278:462-70; PMID:12403781
- Modugno M, Tagliabue E, Ardini E, Berno V, Galmozzi E, De Bortoli M, Castronovo V, Ménard S. p53-dependent downregulation of metastasis-associated laminin receptor. Oncogene 2002; 21:7478-87; PMID:12386810; http://dx.doi.org/10.1038/sj.onc.1205957
- Xu D, Wang Q, Gruber A, Björkholm M, Chen Z, Zaid A, Selivanova G, Peterson C, Wiman KG, Pisa P. Downregulation of telomerase reverse transcriptase mRNA expression by wild type p53 in human tumor cells. Oncogene 2000; 19:5123-33; PMID:11064449; http://dx.doi.org/10.1038/sj.onc.1203890
- Kanaya T, Kyo S, Hamada K, Takakura M, Kitagawa Y, Harada H, Inoue M. Adenoviral expression of p53 represses telomerase activity through down-regulation of human telomerase reverse transcriptase transcription. Clin Cancer Res 2000; 6:1239-47; PMID:10778946
- Joshi AA, Wu Z, Reed RF, Suttle DP. Nuclear factor-Y binding to the topoisomerase IIalpha promoter is inhibited by both the p53 tumor suppressor and anticancer drugs. Mol Pharmaco 2003;. 63:359-67; PMID:12527807; http://dx.doi.org/10.1124/mol.63.2.359
- Pal S, Datta K, Mukhopadhyay D. Central role of p53 on regulation of vascular permeability factorvascular endothelial growth factor (VPFVEGF) expression in mammary carcinoma. Cancer Res 2001; 61:6952-7; PMID:11559575
- Yamabe Y, Shimamoto A, Goto M, Yokota J, Sugawara M, Furuichi Y. Sp1-mediated transcription of the Werner helicase gene is modulated by Rb and p53. Mol Cell Biol 1998; 18:6191-200.PMID:9774636
- Yap N, Yu CL, Cheng SY Modulation of the transcriptional activity of thyroid hormone receptors by the tumor suppressor p53. Proc Natl Acad Sci USA 1996; 93:4273-7; PMID:8633054; http://dx.doi.org/10.1073/pnas.93.9.4273
- Taura M, Suico MA, Fukuda R, Koga T, Shuto T, Sato T, Morino-Koga S, Okada S, Kai H. MEFELF4 transactivation by E2F1 is inhibited by p53. Nucleic Acids Res 2011; 39:76-88; PMID:20805247; http://dx.doi.org/10.1093/nar/gkq762
- Tophkhane C, Yang SH, Jiang Y, Ma Z, Subramaniam D, Anant S, Yogosawa S, Sakai T, Liu WG, Edgerton S, et al. p53 inactivation upregulates p73 expression through E2F-1 mediated transcription. PLoS One 2012; 7:e43564; PMID:22952705; http://dx.doi.org/10.1371/journal.pone.0043564
- Yun J, Chae HD, Choy HE, Chung J, Yoo HS, Han MH, Shin DY. p53 negatively regulates cdc2 transcription via the CCAAT- binding NF-Y transcription factor. J Biol Chem 1999; 274:29677-82; PMID:10514438; http://dx.doi.org/10.1074/jbc.274.42.29677
- Salsi V, Caretti G, Wasner M, Reinhard W, Haugwitz U, Engeland K, Mantovani R. Interactions between p300 and multiple NF-Y trimers govern Cyclin B2 promoter function. J Biol Chem 2003; 278:6642-50; PMID:12482752; http://dx.doi.org/10.1074/jbc.M210065200
- Müller GA, Engeland K. The central role of CDECHR promoter elements in the regulation of cell cycle-dependent gene transcription. FEBS J 2010; 277:877-93; PMID:20015071
- Kasowski M, Grubert F, Heffelfinger C, Hariharan M, Asabere A, Waszak SM, Habegger L, Rozowsky J, Shi M, Urban AE, et al. Variation in transcription factor binding among humans. Science 2010; 328:232-5; PMID:20299548; http://dx.doi.org/10.1126/science.1183621
- (a) Zwicker J, Lucibello FC, Wolfraim LA, Gross C, Truss M, Engeland K, Müller R. Cell cycle regulation of the cyclin A cdc25C and cdc2 genes is based on a common mechanism of transcriptional repression. EMBO J 1995; 14:4514-22; PMID:7556094; (b) Müller GA, Wintsche A, Stangner K, Prohaska SJ, Stadler PF, Engeland K. The CHR site: Definition and genome-wide identification of a cell cycle transcriptional element. Nucl Acids Res. 2014; PMID:25106871; http://dx.doi.org/10.1093/nar/gku696
- Müller GA, Quaas M, Schümann M, Krause E, Padi M, Fischer M, Litovchick L, DeCaprio JA, Engeland K. The CHR promoter element controls cell cycle-dependent gene transcription and binds the DREAM and MMB complexes. Nucleic Acids Res 2012; 40:1561-78; PMID:22064854; http://dx.doi.org/10.1093/nar/gkr793
- Yang J, Song K, Krebs TL, Jackson MW, Danielpour D. RbE2F4 and Smad23 link survivin to TGF-beta-induced apoptosis and tumor progression. Oncogene 2008; 27:5326-38; PMID:18504435; http://dx.doi.org/10.1038/onc.2008.165
- Yee AS, Reichel R, Kovesdi I, Nevins JR. Promoter interaction of the E1A-inducible factor E2F and its potential role in the formation of a multi-component complex. EMBO J 1987; 6:2061-8; PMID:2820719
- Chellappan SP, Hiebert S, Mudryj M, Horowitz JM, Nevins JR. The E2F transcription factor is a cellular target for the RB protein. Cell 1991; 65:1053-61; PMID:1828392; http://dx.doi.org/10.1016/0092-8674(91)90557-F
- Wells J, Yan PS, Cechvala M, Huang T, Farnham PJ. Identification of novel pRb binding sites using CpG microarrays suggests that E2F recruits pRb to specific genomic sites during S phase. Oncogene 2003; 22:1445-60; PMID:12629508; http://dx.doi.org/10.1038/sj.onc.1206264
- Elkon R, Linhart C, Sharan R, Shamir R, Shiloh Y. Genome-wide in silico identification of transcriptional regulators controlling the cell cycle in human cells. Genome Res 2003; 13:773-80; PMID:12727897; http://dx.doi.org/10.1101/gr.947203
- Linhart C, Elkon R, Shiloh Y, Shamir R, Deciphering transcriptional regulatory elements that encode specific cell cycle phasing by comparative genomics analysis. Cell Cycle 2005; 4:1788-97; PMID:16294034; http://dx.doi.org/10.4161/cc.4.12.2173
- Suzuki H, Forrest AR, van Nimwegen E, Daub CO, Balwierz PJ, Irvine KM, Lassmann T, Ravasi T, Hasegawa Y, de Hoon MJ, et al. The transcriptional network that controls growth arrest and differentiation in a human myeloid leukemia cell line. Nat Genet 2009; 41:553-62; PMID:19377474; http://dx.doi.org/10.1038/ng.375
- He L, He X, Lim LP, de Stanchina E, Xuan Z, Liang Y, Xue W, Zender L, Magnus J, Ridzon D, et al. A microRNA component of the p53 tumour suppressor network. Nature 2007; 447:1130-4; PMID:17554337; http://dx.doi.org/10.1038/nature05939
- Huarte M, Guttman M, Feldser D, Garber M, Koziol MJ, Kenzelmann-Broz D, Khalil AM, Zuk O, Amit I, Rabani M, et al. A large intergenic noncoding RNA induced by p53 mediates global gene repression in the p53 response. Cell 2010; 142:409-19; PMID:20673990; http://dx.doi.org/10.1016/j.cell.2010.06.040
- Hung T, Wang Y, Lin MF, Koegel AK, Kotake Y, Grant GD, Horlings HM, Shah N, Umbricht C, Wang P, et al. Extensive and coordinated transcription of noncoding RNAs within cell-cycle promoters. Nat Genet 2011; 43:621-9; PMID:21642992; http://dx.doi.org/10.1038/ng.848
- Hwang CI, Matoso A, Corney DC, Flesken-Nikitin A, Körner S, Wang W, Boccaccio C, Thorgeirsson SS, Comoglio PM, Hermeking H, et al. Wild-type p53 controls cell motility and invasion by dual regulation of MET expression. Proc Natl Acad Sci USA 2011; 108:14240-5; PMID:21831840; http://dx.doi.org/10.1073/pnas.1017536108
- Concepcion CP, Han YC, Mu P, Bonetti C, Yao E, D'Andrea A, Vidigal JA, Maughan WP, Ogrodowski P, Ventura A. Intact p53-dependent responses in miR-34-deficient mice. PLoS Genet 2012; 8:e1002797; PMID:22844244; http://dx.doi.org/10.1371/journal.pgen.1002797
- Dimitrova N, Zamudio JR, Jong RM, Soukup D, Resnick R, Sarma K, Ward AJ, Raj A, Lee JT, Sharp PA, et al. LincRNA-p21 Activates p21 In cis to Promote Polycomb Target Gene Expression and to Enforce the G1S Checkpoint. Mol Cell 2014; 54:777-90; PMID:24857549; http://dx.doi.org/10.1016/j.molcel.2014.04.025
- Benatti P, Dolfini D, Viganò A, Ravo M, Weisz A, Imbriano C. Specific inhibition of NF-Y subunits triggers different cell proliferation defects. Nucleic Acids Res 2011; 39:5356-68; PMID:21415014; http://dx.doi.org/10.1093/nar/gkr128
- Meyer LR, Zweig AS, Hinrichs AS, Karolchik D, Kuhn RM, Wong M, Sloan CA, Rosenbloom KR, Roe G, Rhead B, et al. The UCSC Genome Browser database: extensions and updates 2013. Nucleic Acids Res 2013; 41:D64-9; PMID:23155063; http://dx.doi.org/10.1093/nar/gks1048
- Flicek P, Ahmed I, Amode MR, Barrell D, Beal K, Brent S, Carvalho-Silva D, Clapham P, Coates G, Fairley S, et al. Ensembl 2013. Nucleic Acids Res 2013; 41:D48-55; PMID:23203987; http://dx.doi.org/10.1093/nar/gks1236
- Quinlan AR, Hall IM, BEDTools: a flexible suite of utilities for comparing genomic features. Bioinformatics 2010; 26:841-2; PMID:20110278; http://dx.doi.org/10.1093/bioinformatics/btq033
- Siepel A, Bejerano G, Pedersen JS, Hinrichs AS, Hou M, Rosenbloom K, Clawson H, Spieth J, Hillier LW, Richards S, et al. Evolutionarily conserved elements in vertebrate insect worm and yeast genomes. Genome Res 2005; 15:1034-50; PMID:16024819; http://dx.doi.org/10.1101/gr.3715005
- Blanchette M, Kent WJ, Riemer C, Elnitski L, Smit AF, Roskin KM, Baertsch R, Rosenbloom K, Clawson H, Green ED, et al. Aligning multiple genomic sequences with the threaded blockset aligner. Genome Res 2004; 14:708-15; PMID:15060014; http://dx.doi.org/10.1101/gr.1933104
- Wang B, Xiao Z, Ko HL, Ren EC. The p53 response element and transcriptional repression. Cell Cycle 2010; 9:870-9; PMID:20160511; http://dx.doi.org/10.4161/cc.9.5.10825
