Abstract
Eribulin mesylate is a synthetic analog of halichondrin B known to bind tubulin and microtubules, specifically at their protein rich plus-ends, thereby dampening microtubule (MT) dynamics, arresting cells in mitosis, and inducing apoptosis. The proteins which bind to the MT plus-end are known as microtubule plus-end tracking proteins (+TIPs) and have been shown to promote MT growth and stabilization. Eribulin's plus-end binding suggests it may compete for binding sites with known +TIP proteins such as End-binding 1 (EB1). To better understand the impact of eribulin plus-end binding in regard to the proteins which normally bind there, cells expressing GFP-EB1 were treated with various concentrations of eribulin. In a concentration dependent manner, GFP-EB1 became dissociated from the MT plus-ends following drug addition. Similar results were found with immuno-stained fixed cells. Cells treated with low concentrations of eribulin also showed decreased ability to migrate, suggesting the decrease in MT dynamics may have a downstream effect. Extended exposure of eribulin to cells leads to total depolymerization of the MT array. Taken together, these data show eribulin effectively disrupts EB1 +TIP complex formation, providing mechanistic insights into the impact of eribulin on MT dynamics.
Keywords:
Abbreviations
| MT | = | microtubule |
| EB | = | end binding |
| +TIP | = | plus-end tracking protein |
Introduction
Eribulin mesylate (Halaven®) is FDA approved for the treatment of pretreated, advanced, and metastatic breast cancer.Citation1 Eribulin is a truncated synthetic analog of halichondrin B, a natural product isolated from the sea sponge Halichondria okadai.Citation2 Like many other natural products, eribulin interacts with the tubulin/microtubule (MT) system.Citation3-5 Specifically, eribulin binds with high affinity at the interface between tubulin heterodimers on their dynamic plus-ends.Citation4,6
Several lines of work have shown eribulin to have a potent impact on plus-end MT dynamics.Citation4-6 In vitro work with purified MTs has demonstrated that eribulin dramatically reduces MT growth without impacting shrinkage rates. Analysis of eribulin treated mitotic U2OS cells revealed a clear reduction in centromeric motility, likely through the suppression of spindle MT dynamics, leading to dysfunctional spindles.Citation5 In interphase MCF-7 cells, eribulin treatment led to dampened MT growth dynamics, visualized through GFP-tubulin.Citation4 However, the impact of eribulin on MT plus-end binding proteins which regulate MT dynamics has not been explored.
The MT-plus end is a particularly active region of the MT polymer with many families of proteins continually binding and dissociating from it. One notable family of proteins called end binding (EB) proteins bind to the +TIP and stabilize MTs by preventing depolymerization and promoting rescue. Such proteins can be used to hitchhike cargo to the cell cortex.Citation7,8 While eribulin binds in this same region, to our knowledge no work to date has shown its impact on the localization of cytosolic proteins competing for that binding site. Here we investigate the effect of eribulin on EB1 localization and dynamics in cells as well as its impact on cell migration.
Results
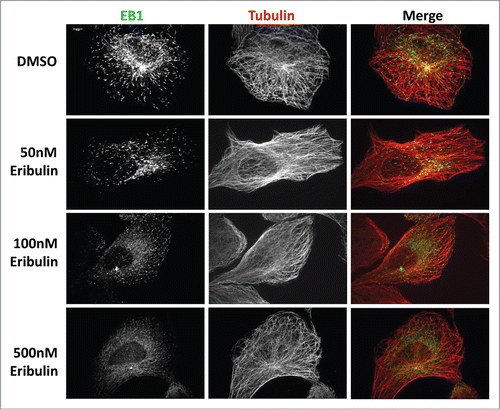
We studied the impact of eribulin on the localization and dynamics of the +TIP protein EB1 in the human osteosarcoma cell line U2OS. A short term dose response study in which cells were incubated in either 50, 100, or 500 nM eribulin for 2 minutes and then fixed and immunostained for tubulin and EB1 was conducted. While the MT arrays of eribulin treated cells appeared to be organized normally relative to controls, the EB1 comet staining patterns were significantly altered. At concentrations of 50 and 100 nM, EB1 comets were present but noticeably smaller, suggesting MT growth was diminished (). More dramatically, with a dose of 500 nM eribulin, comets appeared to become completely disassociated from MT plus-ends. Eribulin is known to bind with high affinity to the interface between the tubulin subunits, thereby disrupting EB1 localization.
Figure 1. EB1 dissociates from MTs after exposure to eribulin. U2OS cells were treated with 50, 100 and 500 nM eribulin for 2 min and EB1 comets were visualized by immuno-staining using anti-α-tubulin, and anti-EB1 antibodies, as described in Materials and Methods. The steady state MT array shows no obvious defects. Scale bar = 10μm.
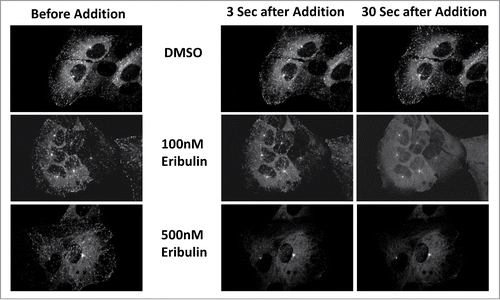
To directly visualize eribulin's impact in living cells, a U2OS cell line expressing EB1-GFP was used. This cell line was ideal as it emphasized only growing +TIPs without the background from a dense MT array that had complicated previous work. Cells were imaged before and throughout the addition of eribulin. Similar to the immunofluorescence studies discussed above, cells showed a dose dependent response. While controls continued to produce EB1 comets, eribulin treated cells showed dramatically reduced comet size and comet number almost instantaneously (). At 100 nM eribulin, EB1-GFP began dispersing within 3 seconds of drug addition and the amount and size of comets was dramatically reduced after 30 seconds. With a 500 nM dosage of eribulin, EB1 was immediately dispersed. These fixed and real time findings show that the high affinity binding eribulin displays in vitro effectively displaces all plus-tip EB1 in a concentration dependent manner. It is likely that other +TIP complex proteins also will be displaced, further reducing the stability of the MT polymer.
Figure 2. EB1-GFP is disrupted in living U2OS cells treated with eribulin. Living U2OS cells expressing EB1-GFP were treated with 100 and 500 nM eribulin. Images shown were taken before the addition of eribulin and 3 sec and 30 sec following drug treatment.
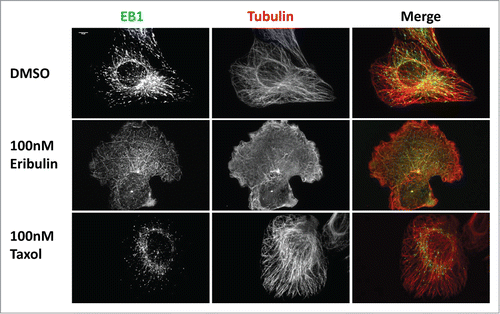
Previous in vitro studies have shown that long term exposure to eribulin leads to depolymerized MTs.Citation4 To test this in cells, we incubated U2OS cells in 100 nM eribulin for 16 hours. Cellular MTs were near completely lost () and EB1 comets were absent. Without the ability to stabilize and promote growth from their plus-ends by +TIP proteins, microtubules in eribulin treated cells were depolymerized over time. As expected, 16 hours of treatment with 100nM Taxol, a drug known to stabilize microtubules,Citation9 led to increased bundling of MTs and more pointed and smaller EB1 tip binding.
Figure 3. MTs are depolymerized after extended eribulin exposure. U2OS cells were treated with 100 nM eribulin or 100 nM Taxol for 16 h. EB1 and MTs were visualized by immunofluorescence as described in . Scale bar = 10μm.
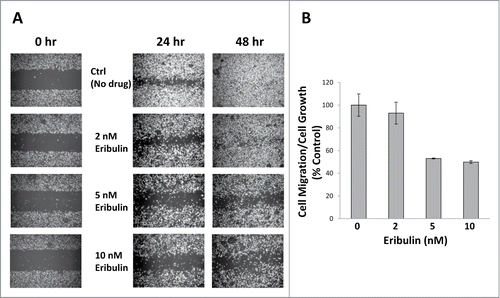
Dynamic MTs are essential for effective cell migration. Depletion of dynamic MTs with low concentrations of MT disrupting drugs has been shown to drastically reduce cell migration in NRK fibroblasts.Citation10,11 Since we have shown through our EB1 work, that eribulin altered MT growth dynamics, we studied the effect of eribulin on migration of the human breast cancer cell line MDA-MB-231. The IC50 values of eribulin for U2OS and MDA-MB-231 cells are very different (Table S1). MDA-MB-231 cells were 25-fold more sensitive to eribulin, compared to U2OS cells. Wound healing assays were performed using approximately 2X, 5X and 10X IC50 concentrations of eribulin, and the effects of eribulin on cell migration were normalized to those of cell growth. Concentrations of 2, 5 and 10 nM eribulin inhibited cell migration by 7%, 47% and 50%, respectively ( and Table S2).
Figure 4. Eribulin inhibits cell migration. (A) Wound healing assays were performed using approximately 90% confluent MDA-MB-231 cells treated with 2, 5 and 10 nM eribulin. Four× phase contrast images were taken 24 hr and 48 hr after drug treatment. The open space areas were measured using ImageJ. (B) Cell migration after 24 hr eribulin treatment was normalized to cell growth, n = 2.
Discussion
Taken together these data show a clear impact on interphase MT growth dynamics induced by eribulin in fixed and living U2OS cells. Eribulin effectively disrupts plus-tip growth and binding, preventing EB1 from localizing to +TIPs, reducing MT stability, and leading to MT depolymerization after prolonged exposure. These results suggest that the decrease in cell migration as shown in the in vitro wound assay results from decreased MT dynamics due to displaced EB1 binding. These data are consistent with the hypothesis that eribulin's mechanism of altering MT growth dynamics is through the direct disruption of plus-tip complex formation.
Materials and Methods
Drugs
Eribulin was synthesized at Eisai Research Institute and Taxol® was obtained from the Drug Development Branch, National Cancer Institute.
Antibodies
Commercially available primary antibodies were rabbit polyclonal anti-α-tubulin (1:750, Abcam, ab18251) and mouse monoclonal anti-EB1 (1:500, BD Biosciences, 610534). Secondary antibodies conjugated with fluorophores (Jackson ImmunoResearch Laboratories) were used at a final concentration of 7.5 μg/ml.
Cell culture
U2OS (ATCC) and U2OS EB1-GFP cells (a gift from Dr. Pavel Draber) were cultured at 37°C in DMEM (Invitrogen, 11965–092) containing 10% fetal-bovine serum (FBS), 1% Glutamax, and 2% penicillin/streptomycin in the presence of 5% CO2. The human breast cancer cell line MDA-MB-231 (ATCC) was grown in RPMI with 10% FBS.
Immunofluorescence
Cells were grown on coverslips and fixed in methanol at −20°C for 20 minutes and then fixed with 4% paraformaldehyde and 4.2% sucrose in phosphate buffered saline (PBS) at room temperature for 20 min. After washing with PBS-Tween (PBS-T), cells were blocked in buffer (5% normal goat serum in PBS with 0.1% Triton X-100) for 30 min. Following incubation with the indicated primary antibody for 1.5 hours, cells were washed with PBS-T, and incubated with fluorophore conjugated secondary antibody for 45 minutes. Cells were washed again and mounted on a slide with mounting media.
Microscopy
Fixed and live cells were imaged using a 4-D spinning-disk confocal microscope (PerkinElmer) with either a 60 × (1.4 NA) or a 100 × (1.4 NA) objective attached to a digital camera (Orca ER; Hamamatsu). Confocal images are displayed as the maximum intensity projections of all captured Z planes.
To visualize MT dynamics before, during and after the addition of eribulin, fluorescent images were acquired every 3 seconds using a heated chamber and 5% CO2. Images were exported and visualized using ImageJ (NIH).
Wound healing assays
Wound healing assays were performed using approximately 90% confluent MDA-MB-231 cells. 4X phase contrast images were taken 24 hr and 48 hr after eribulin treatment. The open space areas were measured using ImageJ. Cell growth following drug treatment was measured by counting cells with a Beckman Coulter Counter.
IC50 measurements
2000 cells were added to each well of a 96-well plate. Increasing concentrations of the drugs were added 18 h after plating. IC50 values were determined after 96hr incubation at 37°C, using the SRB method.Citation12
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
950143_Supplementary_Tables.zip
Download Zip (43 KB)Acknowledgments
The work was supported by funding from Eisai Inc, and by NIH CA077263, the National Foundation for Cancer Research and the Breast Cancer Research Foundation.
The authors thank Dr. Bruce Littlefield for helpful discussions.
References
- Verdaguer H, Morilla I, Urruticoechea A. Eribulin mesylate in breast cancer. Womens Health (Lond Engl) 2013; 9:517-26; PMID:24161305; http://dx.doi.org/10.2217/whe.13.61
- Uemura D, Takahashi K, Yamamoto T, Katayama C, Tanaka J, et al. Norhalichondrin A: an antitumor polyether macrolide from a marine sponge. J Am Chem Soc 1985; 107:4796-8; http://dx.doi.org/10.1021/ja00302a042
- Bai RL, Paull KD, Herald CL, Malspeis L, Pettit GR, Hamel E. Halichondrin B and homohalichondrin B, marine natural products binding in the vinca domain of tubulin. Discovery of tubulin-based mechanism of action by analysis of differential cytotoxicity data. J Biol Chem 1991; 266:15882-9; PMID:1874739
- Jordan MA, Kamath K, Manna T, Okouneva T, Miller HP, Davis C, Littlefield BA, Wilson L. The primary antimitotic mechanism of action of the synthetic halichondrin E7389 is suppression of microtubule growth. Mol Cancer Ther 2005; 4:1086-95; PMID:16020666; http://dx.doi.org/10.1158/1535-7163.MCT-04-0345
- Okouneva T, Azarenko O, Wilson L, Littlefield BA, Jordan MA. Inhibition of centromere dynamics by eribulin (E7389) during mitotic metaphase. Mol Cancer Ther 2008; 7:2003-11; PMID:18645010; http://dx.doi.org/10.1158/1535-7163.MCT-08-0095
- Smith JA, Wilson L, Azarenko O, Zhu X, Lewis BM, Littlefield BA, Jordan MA. Eribulin binds at microtubule ends to a single site on tubulin to suppress dynamic instability. Biochemistry 2010; 49:1331-7; PMID:20030375; http://dx.doi.org/10.1021/bi901810u
- Akhmanova A, Steinmetz MO. Tracking the ends: a dynamic protein network controls the fate of microtubule tips. Nat Rev Mol Cell Biol 2008; 9:309-22; PMID:18322465; http://dx.doi.org/10.1038/nrm2369
- Vitre B, Coquelle FM, Heichette C, Garnier C, Chretien D, Arnal I. EB1 regulates microtubule dynamics and tubulin sheet closure in vitro. Nat Cell Biol 2008; 10:415-21; PMID:18364701; http://dx.doi.org/10.1038/ncb1703]
- Schiff PB, Horwitz SB. Taxol stabilizes microtubules in mouse fibroblast cells. Proc Natl Acad Sci U S A 1980; 77: 1561-5; PMID:6103535; http://dx.doi.org/10.1073/pnas.77.3.1561
- Liao G, Nagasaki T, Gundersen GG. Low concentrations of nocodazole interfere with fibroblast locomotion without significantly affecting microtubule level: implications for the role of dynamic microtubules in cell locomotion. J Cell Sci 1995; 108(Pt 11):3473-83.
- Mikhailov A, Gundersen GG. Relationship between microtubule dynamics and lamellipodium formation revealed by direct imaging of microtubules in cells treated with nocodazole or taxol. Cell Motil Cytoskeleton 1998; 41:325-40; PMID:9858157; http://dx.doi.org/10.1002/(SICI)1097-0169(1998)41
- Skehan P, Storeng R, Scudiero D, Monks A, McMahon J, Vistica D, Warren JT, Bokesch H, Kenney S, Boyd MR. New colorimetric cytotoxicity assay for anticancer-drug screening. J Natl Cancer Inst 1990; 82:1107-12; PMID:2359136; http://dx.doi.org/10.1093/jnci/82.13.1107
