Abstract
Polo-like kinases are important regulators of multiple mitotic events; however, how Polo-like kinases are spatially and temporally regulated to perform their many tasks is not well understood. Here, we examined the subcellular localization of the budding yeast Polo-like kinase Cdc5 using a functional Cdc5-GFP protein expressed from the endogenous locus. In addition to the well-described localization of Cdc5 at the spindle pole bodies (SPBs) and the bud neck, we found that Cdc5-GFP accumulates in the nucleus in early mitosis but is released to the cytoplasm in late mitosis in a manner dependent on the Cdc14 phosphatase. This Cdc5 release from the nucleus is important for mitotic exit because artificial sequestration of Cdc5 in the nucleus by addition of a strong nuclear localization signal (NLS) resulted in mitotic exit defects. We identified a key cytoplasmic target of Cdc5 as Bfa1, an inhibitor of mitotic exit. Our study revealed a novel layer of Cdc5 regulation and suggests the existence of a possible coordination between Cdc5 and Cdc14 activity.
Abbreviations
| FEAR | = | Cdc Fourteen Early Anaphase Release |
| MEN | = | Mitotic Exit Network |
Introduction
Polo-like kinases are essential mitotic regulators that are highly conserved from yeast to humans. Since the first discovery of the polo gene in D. melanogaster,Citation1 Polo-like kinases have been identified to play many important roles in mitosis, including mitotic entry, spindle assembly, chromosome segregation, mitotic exit and cytokinesis.Citation2 How Polo-like kinases perform so many different functions coordinated both in space and in time is not fully understood.
In budding yeast, the Polo-like kinase Cdc5 plays important roles in many aspects of mitosis.Citation3 First, Cdc5 promotes mitotic entry by promoting degradation of Swe1, an inhibitor of the mitotic Cdk.Citation4-6 In metaphase, Cdc5 facilitates chromosome segregation. Cdc5 phosphorylates the cohesin subunit Scc1 to allow its separase-dependent cleavage, which is essential for sister chromatid separation.Citation7-9 In early anaphase, Cdc5 promotes Cdc14 early anaphase release (FEAR) by phosphorylating the Cdc14 sequestering protein Net1/Cfi1 in the nucleolus.Citation10-13 The nucleolar release of Cdc14 phosphatase is essential for stabilizing the anaphase spindle,Citation14 nucleolar segregationCitation15 and primes mitotic exit by removing Cdk-dependent phosphorylation.Citation16-18 In addition to FEAR, Cdc5 also regulates chromosome condensation by phosphorylating condensin subunits Brn1, Ycg1, and Ycs4 in anaphase.Citation19 Cdc5 is also an essential regulator of mitotic exit. A canonical signaling module called the mitotic exit network (MEN) promotes full activation of Cdc14 and thus is essential for mitotic exit in budding yeast.Citation20 Cdc5 activates the MEN by at least two pathways. First, Cdc5 phosphorylates and inactivates Bfa1, an inhibitor of MEN.Citation21-24 Cdc5 also phosphorylates Cdc15, a kinase that is essential for MEN.Citation23 Finally, Cdc5 is important for cytokinesis. Cdc5 phosphorylates Rho1 GEFs in anaphase and promotes the local activation of Rho1 at the cell division site for contractile actin ring assembly.Citation25 Thus, Cdc5 activity and functions need to be tightly regulated both in space and in time.
An important question is how Cdc5 can phosphorylate each substrate in a temporal order. Because kinase activity of Cdc5 is high from G2 phase to late mitosis,Citation26 it is likely that the interactions between Cdc5 and its substrates are spatially controlled. Supporting this idea, it has previously been reported that Cdc5 localizes to various subcellular structures/compartments during the cell cycle.Citation5,26-31 Cdc5 can localize to the SPBs, at the bud neck, and in the nucleus,Citation27 but the timing of Cdc5 localization to the specific subcellular compartments remains little explored. Cdc5 binding to its substrates depends on its polo-box domain (PBD)Citation32,33 and although previous studies have characterized Cdc5 PBD mutant function,Citation27,28 the role that Cdc5 substrate binding plays on its localization during mitosis has not been fully explored.
In this paper, we analyzed the spatial distribution of Cdc5 during the cell cycle. By taking advantage of both newly generated and previously published cdc5 alleles, we show that Cdc5 is exported from the nucleus to the cytoplasm in anaphase through an active mechanism. We further identify Cdc14 as a regulator of Cdc5 nuclear export and demonstrate that Cdc5 nuclear export is required for MEN activation and for timely mitotic exit. Our data suggest the existence of a possible positive feedback loop between Cdc5 and Cdc14 to regulate timely exit from mitosis.
Results
Cdc5 changes its localization during the cell cycle
To carefully investigate subcellular localization of Cdc5 during the cell cycle, we added a GFP tag to the carboxyl terminus of Cdc5 and expressed it from the endogenous locus. This CDC5-GFP construct is functional because the cells expressing CDC5-GFP as sole copy of CDC5 exhibited no obvious growth defects. In a log phase culture, Cdc5-GFP signal is absent in unbudded G1 cells () consistent with the fact that Cdc5 is targeted for degradation after mitosis by Cdh1.Citation34,35 In small to medium budded cells, which correspond to S or G2 phase, Cdc5-GFP starts to localize to the SPBs () as reported previously.Citation27,28,31 In large budded metaphase cells, we saw accumulation of Cdc5-GFP in the nucleus in addition to the SPBs. This is consistent with previous work showing that Cdc5 is transported into the nucleus upon mitotic entry.Citation36 In anaphase cells with fully segregated nuclei, Cdc5-GFP was localized at both SPBs and at the bud neck. The SPB signal of Cdc5-GFP was generally brighter in the daughter cell than mother cell in anaphase. We also noticed that nuclear Cdc5-GFP signal was significantly reduced in late mitotic cells compared to metaphase cells ().
Figure 1. Cdc5-GFP localization during the cell cycle. (A) Representative images of live cells showing the localization of Cdc5-GFP expressed from the endogenous locus (SY1653) during the cell cycle. The arrow points to the elongated nucleus which has reduced Cdc5-GFP signal compared to metaphase. (B) Cdc5-GFP localization in cdh1Δ (SY1202) cells. The numbers indicate metaphase (1), anaphase (2), and post-mitotic (3) cells.
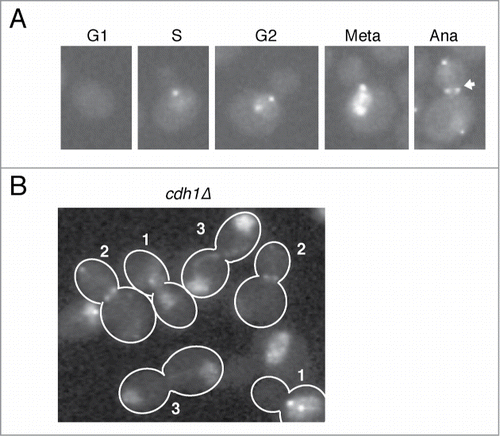
Because Cdc5 is degraded upon mitotic exit by an APC-Cdh1 dependent manner,Citation31,34 it is possible that the reduction of nuclear Cdc5-GFP signal in anaphase was due to degradation. In cdh1Δ, Cdc5-GFP is accumulated in the nucleus in metaphase (Cells 1 in ) and released from the nucleus and localized to the the SPBs and at the bud neck (Cells 2 in ) similar to wild type. Shortly after mitotic exit, as judged by loss of SPB and bud neck signals and segregated nuclei (Cells 3 in ), Cdc5-GFP accumulates back in the nucleus. The localization pattern of Cdc5-GFP in cdh1Δ suggests that there is an active nuclear transport mechanism for Cdc5 specifically during anaphase.
FEAR and Cdc14 activity are required for nuclear release of Cdc5 in anaphase
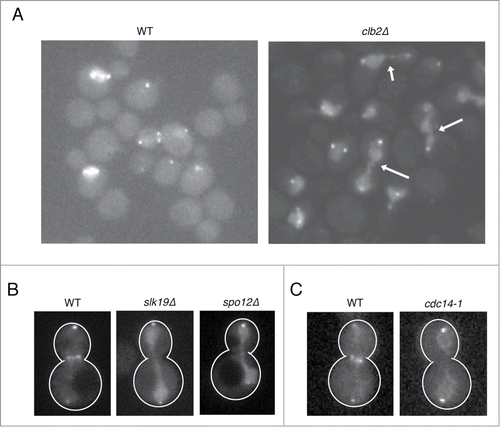
To identify the possible regulators that contribute for nuclear release of Cdc5 in anaphase, we examined the effects of gene deletion mutants implicated in late mitotic events (the gene deletions tested are listed in the ). First, we examined the effect of the major mitotic cyclin Clb2. We found that Cdc5-GFP remains accumulated in the nucleus in anaphase in clb2Δ (). This effect is specific to Clb2 because deletion of an S-phase cyclin CLB5 did not affect Cdc5 localization (). We also found a strong nuclear signal of Cdc5 in anaphase in slk19Δ and spo12Δ cells (), which are defective in Cdc14 release.Citation20 Because Clb2 is also implicated in Cdc14 release,Citation37 we suspected that Cdc14 has a key role in Cdc5 localization in late mitosis. Indeed, in cdc14-1, we found that Cdc5 was enriched in the nucleus in anaphase ().
Table 1. Analysis of Cdc5-GFP nuclear accumulation in anaphase cells in various mitotic mutants. Nuclear localization of Cdc5-GFP was judged ranging from no detectable nuclear Cdc5-GFP signal (−) to strong Cdc5-GFP nuclear signal (+++)
Figure 2. Nuclear release of Cdc5 in anaphase depends on the mitotic cyclin Clb2 and the Cdc14 phosphatase. (A and B) Representative images of Cdc5-GFP in clb2Δ (SY689), slk19Δ (SY1387), and spo12Δ (SY1579) live cells cultured at 25°C. The arrows indicate nuclear accumulation of Cdc5-GFP in anaphase. (C) Representative image of Cdc5-GFP in WT (VB467) and cdc14-1 (SY1382) cultured at 30°C.
The mechanism by which Cdc14 regulates Cdc5 localization
Cdc14 is a phosphatase that dephosphorylates Cdk1 substrates.Citation17 To understand the mechanism by which Cdc14 regulates the localization of Cdc5 in anaphase, we considered three possibilities. First, Cdc14 could directly dephosphorylate Cdc5 because Cdc5 is a substrate of Cdk1.Citation38 Second, Cdc14 could dephosphorylate Cdc5 anchoring proteins in the nucleus and thereby promote the liberation of Cdc5 from the nucleus, because Cdc5 localization often depends on docking of the PBD to phospho-proteins.Citation32 Third, Cdc14 could indirectly regulate Cdc5 localization by dephosphorylating regulators of Cdc5 nucleocytoplasmic transport.
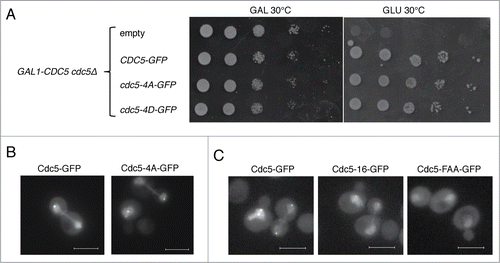
To test the first hypothesis, we have introduced mutations into the Cdk-dependent phosphorylation sites on Cdc5. Cdc5 is known to be phosphorylated by Cdk1/Clb2 at 5 residues: T23, T70, T242, S419, and S657.Citation38 Because phosphorylation of T242 is essential for Cdc5 kinase activity, it is unlikely that T242 is dephosphorylated in vivo in anaphase because Cdc5 kinase activity remains high until mitotic exit.Citation26,38 Therefore, we mutated the remaining 4 residues to unphosphorylatable alanine (cdc5-4A-GFP) or phosphomimetic glutamate (cdc5-4D-GFP). We found that both cdc5-4A-GFP and cdc5-4D-GFP expressed from a low-copy centromeric plasmid were able to complement the viability of a strain which had endogenous Cdc5 depleted (). If Cdc14-dependent dephosphorylation of Cdc5 promotes its cytoplasmic localization, Cdc5-4A-GFP should be readily excluded from the nucleus. For this experiment, we slightly overexpressed CDC5-GFP from a low copy number plasmid. In this condition, both Cdc5-GFP and Cdc5-4A-GFP were enriched in the nucleus even in anaphase (), possibly due to over-dosage. Because there was no obvious change in Cdc5-4A-GFP nuclear signal compared to Cdc5-GFP in this experiment, it is unlikely that Cdc14 is regulating Cdc5 localization by its direct dephosphorylation.
Figure 3. Involvement of Cdc5 phosphorylation by Cdk1 or the PBD of Cdc5 in nuclear release of Cdc5. (A) cdc5-4A-GFP and cdc5-4D-GFP are functional and complement depletion of CDC5. SY1619 (cdc5Δ::GAL1-CDC5), whose sole CDC5 gene is under glucose-repressible GAL1 promoter, was transformed with a low copy number plasmid containing either cdc5-4A-GFP (pVR35) or cdc5-4D-GFP (pVR36) and was tested for growth in galactose (GAL) or glucose (GLU) containing media for 2 days at 30°C. (B) Localization of Cdc5-4A-GFP expressed from a low copy number plasmid in a wild type strain (PY3295) at room temperature. Note that expression of Cdc5-GFP from a plasmid (pVR13) resulted in nuclear accumulation in anaphase most likely because of overexpression. Scale bar: 5 μm. (C) Localization of Cdc5 (pVR13) and of PBD mutants Cdc5-16-GFP (pVB38) and Cdc5-FAA-GFP (pVB39) expressed from a low copy number plasmid in a wild type strain (PY3295) at room temperature. Although the SPB and the bud neck signals of Cdc5-16-GFP and Cdc5-FAA-GFP were significantly impaired, their nuclear localization pattern was indistinguishable from wild type Cdc5-GFP. Scale bar: 5 μm.
We then tested our next hypothesis: the involvement of the PBD in nuclear localization. Because Cdk1 creates a phosphorylation-dependent docking site for the PBD of Cdc5,Citation32 Cdc14 could release Cdc5 from the nucleus via removing these docking sites by dephosphorylation. To test if Cdc5 localizes to the nucleus due to anchoring via the PBD, we used previously described PBD mutants of Cdc5. First, we used cdc5-16,Citation28,32,39 which is defective in binding to the proteins primed by phosphorylation and cdc5-FAA,Citation27 which is completely defective in PBD function. If Cdc5 is anchored by PBD-dependent binding, we expect to see a reduction of nuclear Cdc5 signal in the PBD mutants. Consistent with previous reports,Citation27,28 both Cdc5-16-GFP and Cdc5-FAA-GFP failed to localize to the SPBs and bud neck even when slightly overexpressed as an exogenous copy (). However, nucleocytoplasmic distribution of these mutants was comparable to wild type in this condition. Therefore, it is unlikely that removal of Cdc5 docking sites by Cdc14 is solely responsible for Cdc5 nuclear delocalization.
Taken together, the molecular mechanism by which Cdc14 promotes Cdc5 delocalization from the nucleus remains elusive. However, our analysis suggests that Cdc14-dependent nuclear release of Cdc5 is unlikely through direct regulation of Cdc5 but rather yet to be identified nuclear export or nuclear import factors are involved.
Release of Cdc5 from the nucleus in anaphase is important for mitotic exit
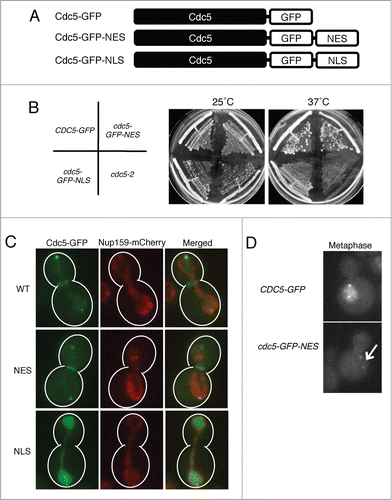
To understand the physiological relevance of Cdc5 delocalization from the nucleus in anaphase, we artificially enriched Cdc5 in the nucleus by adding a strong nuclear localization signal (NLS) of SV40 antigen to the carboxyl terminus of CDC5-GFP to generate CDC5-GFP-NLS (). For comparison, we also created a CDC5-GFP-NES in a similar manner. Cells expressing cdc5-GFP-NLS or cdc5-GFP-NES from the endogenous CDC5 locus as the only copy of Cdc5 were viable, however, the cdc5-GFP-NLS strain showed mild growth defects at 37°C (). As predicted, Cdc5-GFP-NLS remained accumulated in the nucleus throughout the cell cycle including in anaphase (). Consistent with this, no bud neck signal was observed in cdc5-GFP-NLS in anaphase. Cdc5-GFP-NES was excluded from the nucleus throughout the cell cycle including in metaphase when wild-type Cdc5-GFP is accumulated in the nucleus ().
Figure 4. Generation of Cdc5 mutants with altered nucleocytoplasmic distributions. (A) Schematic representation of Cdc5 mutant constructs. (B) cdc5Δ::CDC5-GFP (VB467), cdc5Δ::cdc5-GFP-NES (VB468), and cdc5Δ::cdc5-GFP-NLS (VB543) are viable both at 25° and 37°C. (C) Localization of Cdc5-GFP (VB813), Cdc5-GFP-NES (VB918), and Cdc5-GFP-NLS (VB815) in metaphase and anaphase cells. The nuclear pore protein Nup159-mCherry was used as a nuclear marker. (D) Localization of Cdc5-GFP-NES (VB918) in metaphase compared to Cdc5-GFP (VB813). The arrow points to the nucleus of Cdc5-GFP-NES, which has no detectable GFP signal.
We monitored the cell cycle kinetics of CDC5-GFP, cdc5-GFP-NLS, and cdc5-GFP-NES cells by staining their mitotic spindle and nuclei after release from G1 (). In cdc5-GFP-NES strain, we noticed no or little defect in the timing of spindle assembly (); however, there was a slight delay in the timing of spindle elongation suggesting that nuclear accumulation of Cdc5 is important for mitotic entry and/or anaphase onset.Citation8,36 cdc5-GFP-NES strain exists from mitosis with a 10 minutes delay, but this is likely reflected by the delay in the initiation of mitosis ().
Figure 5. Release of Cdc5 from the nucleus in anaphase is important for timely mitotic exit. Cell cycle progression of CDC5-GFP (VB467) (A), cdc5-GFP-NES (VB468) (B), and CDC5-GFP-NLS (VB543) (C) were analyzed after G1 release at 25°C. Cell cycle progression was judged by the budding index and by the length of mitotic spindle immunostained by anti-alpha tubulin antibody. (D) A representative image of tubulin and DAPI staining at the time point 100 min of (A) is shown. Scale bar: 5 μm.
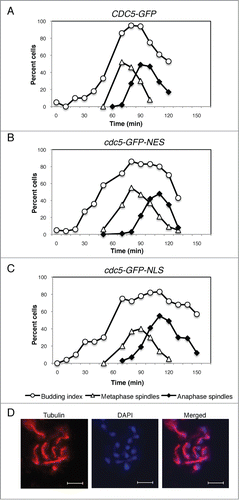
The cdc5-GFP-NLS strain has a slight delay (maybe 10-minutes) in early mitosis. This is possibly due to delay in Swe1 degradation, which occurs in the cytoplasm.Citation6 Strikingly, we found a 20–30 min delay in mitotic exit (). This result strongly suggests that Cdc5 release from the nucleus in anaphase is required for timely mitotic exit.
cdc5-GFP-NLS is synthetic lethal with FEAR mutants slk19Δ and spo12Δ
The mitotic exit defects of cdc5-GFP-NLS prompted us to test if this mutant has problems in a specific mitotic exit pathway. Cdc5 promotes mitotic exit through 2 distinct pathways. First, Cdc5 promotes Cdc14 release from the nucleolus as a part of the FEAR network.Citation16 Second, Cdc5 activates the cytoplasmic MEN by inactivating the Bfa1/Bub2 complexCitation22,24,30 and by activating the Cdc15 kinase.Citation23 Because Cdc5-GFP-NLS is accumulated in the nucleus in anaphase, we hypothesized that cdc5-GFP-NLS is defective in the MEN pathway.
To test this hypothesis, we tested the genetic interaction between cdc5-GFP-NLS and FEAR and MEN mutants. We found that cdc5-GFP-NLS was synthetic lethal with the FEAR mutants slk19Δ and spo12Δ (). This genetic interaction strongly suggests that cdc5-GFP-NLS is impaired in activation of MEN.
Figure 6. Genetic interaction of cdc5-GFP-NLS with mutants defective in mitotic exit. cdc5-GFP-NLS cells harboring a complementing CDC5 plasmid (with a URA3 selective marker) were crossed with slk19Δ, spo12Δ, and lte1Δ. After tetrad dissection, relevant double mutants were spotted onto SC-URA or SC with 5-fluororotic acid (5-FOA which counter selects for URA3+) plates together with each single mutants (VB677, VB683, VB657, VB659, VB661) at 25°C. cdc5-GFP-NLS slk19Δ (VB597, VB598), cdc5-GFP-NLS spo12Δ (VB622, VB624) and cdc5-GFP-NLS lte1Δ (VB645, VB646) all failed to lose the complementing plasmid indicating synthetic lethality.
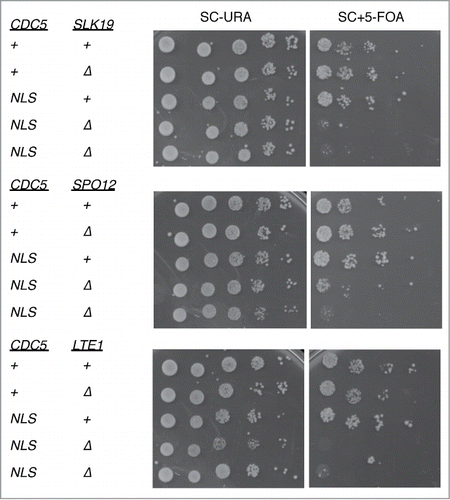
We also found that cdc5-GFP-NLS is synthetic lethal with deletion of LTE1 (). Because Lte1 is a non-essential component of MEN,Citation40-43 synthetic lethality between lte1Δ and cdc5-GFP-NLS further suggests reduced activity of MEN in the cdc5-GFP-NLS mutant.
Cdc5-dependent Inhibition of Bfa1 is Required for Timely Mitotic Exit
Bfa1 is the critical target of Cdc5 in mitotic exit.Citation22 In anaphase, Cdc5 phosphorylates and thereby inactivates Bfa1, allowing MEN to be switched on.Citation21,22,24 Because our cell cycle and genetic analysis strongly suggest that cdc5-GFP-NLS is defective in activation of MEN, and because Bfa1 localizes to the cytoplasmic surface of the SPB, we hypothesized that cdc5-GFP-NLS is impaired in Bfa1 phosphorylation.
To monitor the phosphorylation status of Bfa1-3HA, we arrested the cells in late anaphase by the cdc14-1 mutation for 3 hours. Consistent with the previous reports,Citation22,30 in the wild-type control (CDC5-GFP), slower migrating bands of Bfa1-3HA were dominant by Western blotting (). This slow migration is a result of Cdc5-dependent phosphorylation because it is largely reduced in the cdc5-2 mutant (). Consistent with our hypothesis, Bfa1-3HA was migrating faster in cdc5-GFP-NLS compared to wild type. This reduced phosphorylation of Bfa1-3HA was specific to cdc5-GFP-NLS because cdc5-GFP-NES did not show this pattern.
Figure 7. Bfa1 is the major cytoplasmic target of Cdc5. (A) Phosphorylation status of Bfa1-3HA in cdc5-GFP-NLS or cdc5-GFP-NES mutant cells. These strains (VB807, VB800, VB915) were arrested in telophase by cdc14-1 mutation at 37° for 3 hours and cell lysates were subjected for the Western blotting of Bfa1-3HA. cdc5-2 BFA1-3HA (VB725) was used as a negative control. Tubulin was used as a loading control. (B) Scc1 phosphorylation is not defective in cdc5-GFP-NLS in metaphase. CDC5-GFP (VB749), cdc5-GFP-NLS (VB753) and cdc5-2 (VB808) cells were arrested in G2/M phase with 15 μg/ml nocodazole for 3 hours at 25°C and shifted to 37°C for 2 hours (to inactivate cdc5-2). The cell lysates were subjected for the Western blotting of Scc1-3HA. Tubulin was used as a loading control. (C) Deletion of BFA1 (VB808) suppresses the temperature sensitivity of cdc5-GFP-NLS (VB572) at 37°C. Indicated strains were streaked to single colonies on YEPD plates at 25°C and 37°C for 2 days. (D) Deletion of BFA1 (VB920) rescues the synthetic lethality of cdc5-GFP-NLS spo12Δ (VB622). Indicated strains harboring a complementing CDC5-URA3 plasmid were spotted on plates either containing 5-FOA or SC-URA at room temperature for 4 days.
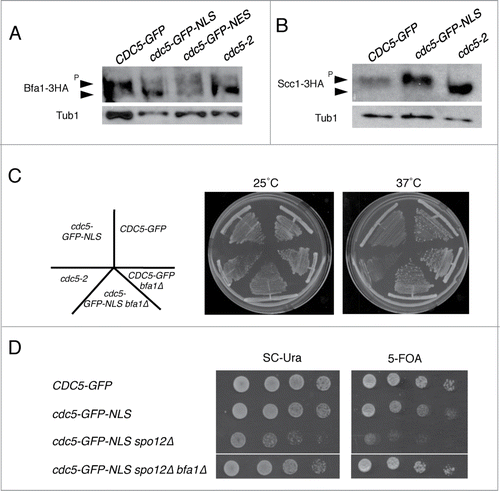
It is a formal possibility that cdc5-GFP-NLS has reduced kinase activity. To confirm this that cdc5-GFP-NLS retains kinase activity in vivo, we monitored the phosphorylation status of nuclear Cdc5 substrate Scc1. In nocodazole-arrested metaphase cells, Scc1-3HA was hyper-phosphorylated in both CDC5-GFP and in cdc5-GFP-NLS strains but not in cdc5-2 (). This result confirms that cdc5-GFP-NLS is able to efficiently phosphorylate its nuclear substrates. Thus, impaired phosphorylation of Bfa1 is due to its spatial separation from cdc5-GFP-NLS and not due to reduced kinase activity.
Lastly, to confirm the hypothesis that cdc5-GFP-NLS is specifically defective in activation of MEN, we asked if deletion of BFA1 was sufficient to rescue the defects associated with cdc5-GFP-NLS. We found that deletion of BFA1 rescued the slow growth phenotype of cdc5-GFP-NLS at the high temperature at 37°C (). Furthermore, synthetic lethality of cdc5-GFP-NLS spo12Δ was rescued by the deletion of BFA1 (). These data confirm that cdc5-GFP-NLS is defective in MEN activation and that Bfa1 is a critical cytoplasmic target of Cdc5 important for timely mitotic exit.
Discussion
Cdc5 localization during the cell cycle
In this study, we revisited Cdc5 localization during the cell cycle using a functional CDC5-GFP construct expressed at the endogenous locus. Consistent with previous reports,Citation5,27-31,36 we found that Cdc5-GFP localizes to the SPBs, nucleus, and bud neck during mitosis. Cdc5-GFP accumulates in the nucleus at the G2/M phase as has been previously described.Citation28,36 However, we found that Cdc5-GFP signal is lost from the nucleus in anaphase. Because this loss of nuclear Cdc5-GFP signal is not a result of Cdh1-dependent degradation, we suspect that Cdc5 is exported from the nucleus in anaphase. Cdc5-GFP localizes to the bud neck only in anaphase when nuclear Cdc5-GFP signal is reduced. It has been reported that Cdc5 tagged c-terminally with 5 copies of GFP and expressed from the endogenous locus can localize to the bud neck after prolonged nocodazole arrest.Citation27 However, our Cdc5-GFP construct, which contains just one copy of GFP, when expressed at the endogenous level localizes to the bud neck in an unperturbed cell cycle only during anaphase. Our data suggests that a robust bud neck localization of Cdc5 requires nuclear release. Because Cdc5 is a key regulator of cytokinesis, it is of interest to study the correlation of the timing of Cdc5 localization to the bud neck and cytokinetic actin ring assembly.
Possible mechanism of Cdc5 nuclear export
It is widely accepted that Polo-like kinase localization is dependent on its PBD. By binding to phospho-proteins primed by mitotic kinases, the PBD dictates the subcellular localization of Polo-like kinases.Citation44 Because FEAR and Cdc14 phosphatase are required for Cdc5 nuclear release in anaphase, we hypothesized that removal of Cdk1-dependent phosphorylation from Cdc5 docking proteins in the nucleus triggers Cdc5 release. However, because Cdc5 PBD mutants defective in substrate binding were not readily released from the nucleus, the PBD is not essential for nuclear retention of Cdc5. Because Cdc5 is known to be phosphorylated by Cdk1 during mitosis,Citation38 an alternative explanation for Cdc5 nuclear release in anaphase could be direct dephosphorylation by Cdc14. However, because the unphosphorylatable cdc5-4A-GFP localized similarly to wild type Cdc5-GFP, it is unlikely that Cdc14 controls Cdc5 localization directly. The caveat of these analysis was that we analyzed the localization of Cdc5-GFP exogenously expresses from a low-copy number plasmid. As has been previously reported,Citation27 when expressed ectopically, Cdc5-GFP can accumulate in the nucleus even in anaphase (). We did not observe any noticeable difference in nuclear localization among Cdc5-GFP, Cdc5-16-GFP, Cdc5-FAA-GFP, and Cdc5-4A-GFP in anaphase.
One of the alternative mechanisms by which Cdc14 can regulate Cdc5 nuclear release is via regulating exportins or importins for Cdc5. The identification of the exportins for Cdc5 is an important future question but because Cdc5 is released from the nucleus only during anaphase when Cdc14 is active, Cdc14 might be activating the exportin for Cdc5 in a cell cycle stage specific manner.
Physiological relevance for Cdc5 nuclear release in anaphase
We revealed that Cdc5 release from the nucleus in anaphase is required for timely mitotic exit. Cdc5-GFP-NLS, which is artificially trapped in the nucleus in anaphase (), showed a delay in spindle disassembly () and exhibited synthetic lethality with mutants defective in mitotic exit (). We further showed that Cdc5-GFP-NLS is defective in phosphorylation of MEN inhibitor Bfa1 (). Indeed, the mitotic exit defect of Cdc5-GFP-NLS can be explained by Bfa1 activity because deleting BFA1 rescued the temperature sensitive growth defect of cdc5-GFP-NLS () and synthetic lethality of cdc5-GFP-NLS spo12Δ ().
It is well established that Cdc5 triggers Cdc14 release from the nucleolus and thereby activates Cdc14.Citation10-12,20 The requirement of Cdc14 in Cdc5 release suggests a potential positive feedback loop between Cdc14 and Cdc5. The precise mechanism by which Cdc5 is regulated awaits future investigation. However, our study uncovered a novel spatial layer of Cdc5 regulation that is important for timely mitotic exit.
Materials and Methods
Yeast genetics and plasmid construction
All yeast strains in this study were isogenic or congenic to BY4741 (MATa leu2Δ0 his3Δ1 met15Δ0 ura3Δ0). The strains and plasmids are listed in and , respectively. Standard yeast genetics was used to generate the strains. PY and SY strains were gifts from D. Pellman (Dana-Farber Cancer Institute, Boston, MA). PJH plasmids were gifts from J. Haber (Brandeis University, Waltham, MA). Gene deletions or modifications were performed using pFA6a vectors provided by J. Pringle (Stanford University, Stanford, CA).Citation45 The NLS sequence taken from the SV40 antigen (AAAPKKKRKVG) and the NES sequence (AAALALKLAGLNI) were previously described.Citation46,47 The NLS or NES sequences were introduced directly after the GFP sequence of CDC5-GFP by PCR-mediated one-step gene replacement.Citation45 Alpha-factor was used at 2 μg/ml. Nocodazole was used at 15 μg/ml. All cultures were grown in YPED unless otherwise noted.
Table 2. Strains used in this study
Table 3. Plasmids used in this study
For construction of the pSY222 plasmid, CDC5-GFP fragment containing 500 bps of 5’UTR was PCR amplified using genomic DNA of SY1653 as a template and was inserted into pRS415 vector using KpnI and HindIII. pVR35, pVR36, pVB38, and pVB39 were derived from pSY222 by site-directed mutagenesis using QuikChange II XL Site-Directed Mutagenesis Kit (Agilent; 20051).
Biochemistry
For Western blotting, the cell lysates were prepared by trichloroacetic acid (TCA) method.Citation48 Mouse monoclonal anti-HA antibody (16B12, Covance) and rat anti-α tubulin monoclonal antibody (MCA78G, AbD Serotec) were used at 1: 3,000 dilution in TBST containing 5% skim milk. Horseradish peroxidase (HRP)-conjugated mouse secondary antibody (Millipore) was used at 1:10,000 dilution in TBST containing 5% skim milk. The signal was detected by Amersham ECL Prime Western Blotting Detection Reagent (GE Healthcare Life Sciences; RPN2232).
Fluorescence microscopy
Images were acquired using the Eclipse E600 (Nikon) epifluorescence microscope equipped with a charge-coupled device camera (DC350F; Andor) with a 60×, NA 1.4× oil objective. The images were captured and analyzed with NIS-Elements software (Nikon).
Cells for live imaging were cultured in synthetic complete (SC) or SC-Leu medium. Cell preparation for immunofluorescence was performed as previously described.Citation47 The mitotic spindle was visualized with a rat anti-α tubulin rat monoclonal antibody (MCA78G, AbD Serotec) conjugated with an Alexa Fluor 568 goat anti-rat antibody (Molecular Probes; A-11077). Nuclei were visualized with DAPI.
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Acknowledgments
We thank Drs. Jim Haber, Kyung Lee, John Pringle, David Pellman, Yoshiaki Kamada, and Brenda Andrews for the gifts of reagents. We are grateful to members of the Yoshida and Haber labs for discussion and sharing reagents, especially Yana Ostrovsky for her technical help.
References
- Sunkel CE, Glover DM. polo, a mitotic mutant of Drosophila displaying abnormal spindle poles. J Cell Sci 1988; 89 (Pt 1):25-38; PMID:3417791
- Archambault V, Glover DM. Polo-like kinases: conservation and divergence in their functions and regulation. Nat Rev Mol Cell Biol 2009; 10:265-75; PMID:19305416; http://dx.doi.org/10.1038/nrm2653
- Lee KS, Park JE, Asano S, Park CJ. Yeast polo-like kinases: functionally conserved multitask mitotic regulators. Oncogene 2005; 24:217-9; PMID:15640837; http://dx.doi.org/10.1038/sj.onc.1208271
- Bartholomew CR, Woo SH, Chung YS, Jones C, Hardy CF. Cdc5 interacts with the Wee1 kinase in budding yeast. Mol Cell Biol 2001; 21:4949-59; PMID:11438652; http://dx.doi.org/10.1128/MCB.21.15.4949-4959.2001
- Park JE, Park CJ, Sakchaisri K, Karpova T, Asano S, McNally J, Sunwoo Y, Leem SH, Lee KS. Novel functional dissection of the localization-specific roles of budding yeast polo kinase Cdc5p. Mol Cell Biol 2004; 24:9873-86; PMID:15509790; http://dx.doi.org/10.1128/MCB.24.22.9873-9886.2004
- Asano S, Park JE, Sakchaisri K, Yu LR, Song S, Supavilai P, Veenstra TD, Lee KS. Concerted mechanism of Swe1Wee1 regulation by multiple kinases in budding yeast. EMBO J 2005; 24:2194-04; PMID:15920482; http://dx.doi.org/10.1038/sj.emboj.7600683
- Uhlmann F, Lottspeich F, Nasmyth K. Sister-chromatid separation at anaphase onset is promoted by cleavage of the cohesin subunit Scc1. Nature 1999; 400:37-42; PMID:10403247; http://dx.doi.org/10.1038/21831
- Alexandru G, Uhlmann F, Mechtler K, Poupart MA, Nasmyth K. Phosphorylation of the cohesin subunit Scc1 by PoloCdc5 kinase regulates sister chromatid separation in yeast. Cell 2001; 105:459-72; PMID:11371343; http://dx.doi.org/10.1016/S0092-8674(01)00362-2
- Hornig NC, Uhlmann F. Preferential cleavage of chromatin-bound cohesin after targeted phosphorylation by Polo-like kinase. EMBO J 2004; 23:3144-53; PMID:15241476; http://dx.doi.org/10.1038/sj.emboj.7600303
- Shou W, Azzam R, Chen SL, Huddleston MJ, Baskerville C, Charbonneau H, Annan RS, Carr SA, Deshaies RJ. Cdc5 influences phosphorylation of Net1 and disassembly of the RENT complex. BMC Mol Biol 2002; 3:3; PMID:11960554; http://dx.doi.org/10.1186/1471-2199-3-3
- Yoshida S, Toh-e A. Budding yeast Cdc5 phosphorylates Net1 and assists Cdc14 release from the nucleolus. Biochem Biophys Res Commun 2002; 294:687-91; PMID:12056824; http://dx.doi.org/10.1016/S0006-291X(02)00544-2
- Visintin R, Stegmeier F, Amon A. The role of the polo kinase Cdc5 in controlling Cdc14 localization. Mol Biol Cell 2003; 14:4486-98; PMID:14551257; http://dx.doi.org/10.1091/mbc.E03-02-0095
- Yoshida S, Asakawa K, Toh-e A. Mitotic exit network controls the localization of Cdc14 to the spindle pole body in saccharomyces cerevisiae. Curr Biol 2002; 12:944-50; PMID:12062061; http://dx.doi.org/10.1016/S0960-9822(02)00870-9
- Higuchi T, Uhlmann F. Stabilization of microtubule dynamics at anaphase onset promotes chromosome segregation. Nature 2005; 433:171-6; PMID:15650742; http://dx.doi.org/10.1038/nature03240
- Sullivan M, Higuchi T, Katis VL, Uhlmann F. Cdc14 phosphatase induces rDNA condensation and resolves cohesin-independent cohesion during budding yeast anaphase. Cell 2004; 117:471-82; PMID:15137940; http://dx.doi.org/10.1016/S0092-8674(04)00415-5
- Stegmeier F, Amon A. Closing mitosis: the functions of the Cdc14 phosphatase and its regulation. Annu Rev Genet 2004; 38:203-32; PMID:15568976; http://dx.doi.org/10.1146/annurev.genet.38.072902.093051
- Visintin R, Craig K, Hwang ES, Prinz S, Tyers M, Amon A. The phosphatase Cdc14 triggers mitotic exit by reversal of Cdk-dependent phosphorylation. Mol Cell 1998; 2:709-18; PMID:9885559; http://dx.doi.org/10.1016/S1097-2765(00)80286-5
- Bremmer SC, Hall H, Martinez JS, Eissler CL, Hinrichsen TH, Rossie S, Parker LL, Hall MC, Charbonneau H. Cdc14 phosphatases preferentially dephosphorylate a subset of cyclin-dependent kinase (Cdk) sites containing phosphoserine. J Biol Chem 2012; 287:1662-9; PMID:22117071; http://dx.doi.org/10.1074/jbc.M111.281105
- St-Pierre J, Douziech M, Bazile F, Pascariu M, Bonneil E, Sauve V, Ratsima H, D’Amours D. Polo kinase regulates mitotic chromosome condensation by hyperactivation of condensin DNA supercoiling activity. Mol Cell 2009; 34:416-26; PMID:19481522; http://dx.doi.org/10.1016/j.molcel.2009.04.013
- Stegmeier F, Visintin R, Amon A. Separase, polo kinase, the kinetochore protein Slk19, and Spo12 function in a network that controls Cdc14 localization during early anaphase. Cell 2002; 108:207-20; PMID:11832211; http://dx.doi.org/10.1016/S0092-8674(02)00618-9
- Lee SE, Jensen S, Frenz LM, Johnson AL, Fesquet D, Johnston LH. The Bub2-dependent mitotic pathway in yeast acts every cell cycle and regulates cytokinesis. J Cell Sci 2001; 114:2345-54; PMID:11493673
- Hu F, Wang Y, Liu D, Li Y, Qin J, Elledge SJ. Regulation of the Bub2Bfa1 GAP complex by Cdc5 and cell cycle checkpoints. Cell 2001; 107:655-65; PMID:11733064; http://dx.doi.org/10.1016/S0092-8674(01)00580-3
- Rock JM, Amon A. Cdc15 integrates Tem1 GTPase-mediated spatial signals with Polo kinase-mediated temporal cues to activate mitotic exit. Genes Dev 2011; 25:1943-54; PMID:21937712; http://dx.doi.org/10.1101/gad.17257711
- Geymonat M, Spanos A, Walker PA, Johnston LH, Sedgwick SG. In vitro regulation of budding yeast Bfa1Bub2 GAP activity by Cdc5. J Biol Chem 2003; 278:14591-4; PMID:12637549; http://dx.doi.org/10.1074/jbc.C300059200
- Yoshida S, Kono K, Lowery DM, Bartolini S, Yaffe MB, Ohya Y, Pellman D. Polo-like kinase Cdc5 controls the local activation of Rho1 to promote cytokinesis. Science 2006; 313:108-11; PMID:16763112; http://dx.doi.org/10.1126/science.1126747
- Cheng L, Hunke L, Hardy CF. Cell cycle regulation of the Saccharomyces cerevisiae polo-like kinase cdc5p. Mol Cell Biol 1998; 18:7360-70; PMID:9819423
- Song S, Grenfell TZ, Garfield S, Erikson RL, Lee KS. Essential function of the polo box of Cdc5 in subcellular localization and induction of cytokinetic structures. Mol Cell Biol 2000; 20:286-98; PMID:10594031; http://dx.doi.org/10.1128/MCB.20.1.286-298.2000
- Ratsima H, Ladouceur AM, Pascariu M, Sauve V, Salloum Z, Maddox PS, D’Amours D. Independent modulation of the kinase and polo-box activities of Cdc5 protein unravels unique roles in the maintenance of genome stability. Proc Natl Acad Sci U S A 2011; 108:E914-23; PMID:21987786; http://dx.doi.org/10.1073/pnas.1106448108
- Valerio-Santiago M, de Los Santos-Velazquez AI, Monje-Casas F. Inhibition of the mitotic exit network in response to damaged telomeres. PLoS Genet 2013; 9:e1003859; PMID:24130507
- Maekawa H, Priest C, Lechner J, Pereira G, Schiebel E. The yeast centrosome translates the positional information of the anaphase spindle into a cell cycle signal. J Cell Biol 2007; 179:423-36; PMID:17967947; http://dx.doi.org/10.1083/jcb.200705197
- Shirayama M, Zachariae W, Ciosk R, Nasmyth K. The Polo-like kinase Cdc5p and the WD-repeat protein Cdc20pfizzy are regulators and substrates of the anaphase promoting complex in saccharomyces cerevisiae. EMBO J 1998; 17:1336-49; PMID:9482731; http://dx.doi.org/10.1093/emboj/17.5.1336
- Elia AE, Rellos P, Haire LF, Chao JW, Ivins FJ, Hoepker K, Mohammad D, Cantley LC, Smerdon SJ, Yaffe MB. The molecular basis for phosphodependent substrate targeting and regulation of Plks by the Polo-box domain. Cell 2003; 115:83-95; PMID:14532005; http://dx.doi.org/10.1016/S0092-8674(03)00725-6
- Elia AE, Cantley LC, Yaffe MB. Proteomic screen finds pSerpThr-binding domain localizing Plk1 to mitotic substrates. Science 2003; 299:1228-31; PMID:12595692; http://dx.doi.org/10.1126/science.1079079
- Charles JF, Jaspersen SL, Tinker-Kulberg RL, Hwang L, Szidon A, Morgan DO. The polo-related kinase Cdc5 activates and is destroyed by the mitotic cyclin destruction machinery in S. cerevisiae. Curr Biol 1998; 8:497-507; PMID:9560342; http://dx.doi.org/10.1016/S0960-9822(98)70201-5
- Shirayama M, Toth A, Galova M, Nasmyth K. APC(Cdc20) promotes exit from mitosis by destroying the anaphase inhibitor Pds1 and cyclin Clb5. Nature 1999; 402:203-7; PMID:10647015; http://dx.doi.org/10.1038/46080
- Nakashima A, Maruki Y, Imamura Y, Kondo C, Kawamata T, Kawanishi I, Takata H, Matsuura A, Lee KS, Kikkawa U, et al. The yeast Tor signaling pathway is involved in G2M transition via polo-kinase. PLoS One 2008; 3:e2223; PMID:18493323; http://dx.doi.org/10.1371/journal.pone.0002223
- Azzam R, Chen SL, Shou W, Mah AS, Alexandru G, Nasmyth K, Annan RS, Carr SA, Deshaies RJ. Phosphorylation by cyclin B-Cdk underlies release of mitotic exit activator Cdc14 from the nucleolus. Science 2004; 305:516-9; PMID:15273393; http://dx.doi.org/10.1126/science.1099402
- Mortensen EM, Haas W, Gygi M, Gygi SP, Kellogg DR. Cdc28-dependent regulation of the Cdc5Polo kinase. Curr Biol 2005; 15:2033-7; PMID:16303563; http://dx.doi.org/10.1016/j.cub.2005.10.046
- Miller CT, Gabrielse C, Chen YC, Weinreich M. Cdc7p-Dbf4p regulates mitotic exit by inhibiting polo kinase. PLoS Genet 2009; 5:e1000498; PMID:19478884; http://dx.doi.org/10.1371/journal.pgen.1000498
- Shirayama M, Matsui Y, Tanaka K, Toh-e A. Isolation of a CDC25 family gene, MSI2LTE1, as a multicopy suppressor of ira1. Yeast 1994; 10:451-61; PMID:7941731; http://dx.doi.org/10.1002/yea.320100404
- Shirayama M, Matsui Y, Toh-e A. Dominant mutant alleles of yeast protein kinase gene CDC15 suppress the lte1 defect in termination of M phase and genetically interact with CDC14. Mol Gen Genet 1996; 251:176-85; PMID:8668128
- Bardin AJ, Visintin R, Amon A. A mechanism for coupling exit from mitosis to partitioning of the nucleus. Cell 2000; 102:21-31; PMID:10929710; http://dx.doi.org/10.1016/S0092-8674(00)00007-6
- Pereira G, Hofken T, Grindlay J, Manson C, Schiebel E. The Bub2p spindle checkpoint links nuclear migration with mitotic exit. Mol Cell 2000; 6:1-10; PMID:10949022; http://dx.doi.org/10.1016/S1097-2765(00)00002-2
- Neef R, Gruneberg U, Kopajtich R, Li X, Nigg EA, Sillje H, Barr FA. Choice of Plk1 docking partners during mitosis and cytokinesis is controlled by the activation state of Cdk1. Nat Cell Biol 2007; 9:436-44; PMID:17351640; http://dx.doi.org/10.1038/ncb1557
- Longtine MS, McKenzie A, 3rd, Demarini DJ, Shah NG, Wach A, Brachat A, Philippsen P, Pringle JR. Additional modules for versatile and economical PCR-based gene deletion and modification in Saccharomyces cerevisiae. Yeast 1998; 14:953-61; PMID:9717241; http://dx.doi.org/10.1002/(SICI)1097-0061(199807)14 :10%3c953::AID-YEA293%3e3.0.CO;2-U
- Hodel AE, Harreman MT, Pulliam KF, Harben ME, Holmes JS, Hodel MR, Berland KM, Corbett AH. Nuclear localization signal receptor affinity correlates with in vivo localization in saccharomyces cerevisiae. J Biol Chem 2006; 281:23545-56; PMID:16785238; http://dx.doi.org/10.1074/jbc.M601718200
- Rossio V, Yoshida S. Spatial regulation of Cdc55-PP2A by Zds1Zds2 controls mitotic entry and mitotic exit in budding yeast. J Cell Biol 2011; 193:445-54; PMID:21536748; http://dx.doi.org/10.1083/jcb.201101134
- Rossio V, Michimoto T, Sasaki T, Ohbayashi I, Kikuchi Y, Yoshida S. Nuclear PP2A-Cdc55 prevents APC-Cdc20 activation during the spindle assembly checkpoint. J Cell Sci 2013; 126:4396-405; PMID:23886942; http://dx.doi.org/10.1242/jcs.127365
