Abstract
Fanconi anemia (FA) is a rare recessive genetic disease characterized by congenital abnormalities, bone marrow failure and heightened cancer susceptibility in early adulthood. FA is caused by biallelic germ-line mutation of any one of 16 genes. While several functions for the FA proteins have been ascribed, the prevailing hypothesis is that the FA proteins function cooperatively in the FA-BRCA pathway to repair damaged DNA. A pivotal step in the activation of the FA-BRCA pathway is the monoubiquitination of the FANCD2 and FANCI proteins. Despite their importance for DNA repair, the domain structure, regulation, and function of FANCD2 and FANCI remain poorly understood. In this review, we provide an overview of our current understanding of FANCD2 and FANCI, with an emphasis on their posttranslational modification and common and unique functions.
Abbreviations
| AML | = | acute myeloid leukemia |
| APC/C | = | anaphase-promoting complex/cyclosome |
| APH | = | aphidicolin |
| ARM | = | armadillo repeat domain |
| AT | = | ataxia-telangiectasia |
| ATM | = | ataxia-telangiectasia mutated |
| ATR | = | ATM and Rad3-related |
| BAC | = | bacterial-artificial-chromosome |
| BS | = | Bloom syndrome |
| ChIP-seq | = | CHIP sequencing |
| CtBP | = | C-terminal binding protein |
| CtIP | = | CtBP-interacting protein |
| CUE | = | coupling of ubiquitin conjugation to endoplasmic reticulum degradation |
| EPS15 | = | epidermal growth factor receptor pathway substrate 15 |
| FA | = | Fanconi anemia |
| FAN1 | = | FANCD2-associated nuclease1 |
| FISH | = | fluorescence in situ hybridization |
| HECT | = | homologous to E6-AP Carboxy Terminus |
| HJ | = | Holliday junction |
| HR | = | homologous recombination |
| iPOND | = | isolation of proteins on nascent DNA |
| MCM2-MCM7 | = | minichromosome maintenance 2–7 |
| MEFs | = | mouse embryonic fibroblasts |
| MMC | = | mitomycin C |
| MRN | = | MRE11/RAD50/NBS1 |
| NLS | = | nuclear localization signal |
| PCNA | = | proliferating cell nuclear antigen |
| PIKK | = | phosphatidylinositol-3-OH-kinase-like family of protein kinases |
| PIP-box | = | PCNA-interacting protein motif |
| POL κ | = | DNA polymerase κ |
| RACE | = | rapid amplification of cDNA ends |
| RING | = | really interesting new gene |
| RTK | = | receptor tyrosine kinase |
| SCF | = | Skp1/Cullin/F-box protein complex |
| SCKL1 | = | seckel syndrome |
| SILAC | = | stable isotope labeling with amino acids in cell culture |
| SLD1/SLD2 | = | SUMO-like domains |
| SLIM | = | SUMO-like domain interacting motif |
| TIP60 | = | 60 kDa Tat-interactive protein |
| TLS | = | Translesion DNA synthesis |
| UAF1 | = | USP1-associated factor 1 |
| UBD | = | ubiquitin-binding domain |
| UBZ | = | ubiquitin-binding zinc finger |
| UFB | = | ultra-fine DNA bridges |
| UIM | = | ubiquitin-interacting motif |
| ULD | = | ubiquitin-like domain |
| USP1 | = | ubiquitin-specific protease 1 |
| VRR-nuc | = | virus-type replication repair nuclease |
The Discovery of FANCD2 and FANCI
FANCD2
The FANCD2 gene was identified in 2001 by the Grompe, D’Andrea, and Moses Laboratories using a positional gene cloning approach, following many years of collaborative efforts.Citation1-4 The PD20 FA patient line was originally assigned to the FA-D complementation group because of its inability to complement the mitomycin C (MMC) hypersensitivity of the FA-D reference line HSC62.Citation4-6 Using a microcell-mediated chromosome transfer approach, the FANCD gene was initially mapped to 3p22-26.Citation4 Hejna and colleagues subsequently narrowed the critical region to ∼200 kb, a region which harbored 3 candidate genes: TIGR-A004X28, SGC34603, and AA609512.Citation2 Timmers and colleagues then used rapid amplification of cDNA ends (RACE) to obtain the respective full-length cDNAs and sequenced these 3 genes in the PD20 cells.Citation1 No sequence changes were uncovered in A004X28 and AA609512. However, 5 sequence changes were found in SGC34603; 3 of which represented common polymorphisms and 2 represented bona fide mutations. One sequence alteration was a maternally inherited A to G transition at nucleotide 376, which resulted in a S to G missense change. This transition was also associated with missplicing and insertion of 13 nt from intron 5 into the mRNA, causing a frameshift and production of a severely truncated protein. A second paternally inherited sequence change in SGC34603 resulted in an R to H missense and hypomorphic change at position 1236 (R1236H),Citation1 strongly suggesting that SGC34603 was indeed the FANCD gene. Confirming this, transduction of PD20 cells with full-length wild-type FANCD rescued the MMC hypersensitivity of these cells.Citation1 No sequence changes in SGC34603 however were uncovered in HSC62. These findings led to the delineation of 2 FA-D complementation groups, FA-D1 and FA-D2, with biallelic mutations in SGC34603/FANCD2 causative for the FA-D2 complementation group.Citation1 Early biochemical studies of the FANCD2 protein uncovered 2 isoforms: an unmodified form and a higher molecular mass isoform posttranslationally modified through the covalent attachment of a single ubiquitin polypeptide, referred to as monoubiquitination.Citation3
FANCI
The existence of the FA-I complementation group was first established by Levitus and colleagues, on the basis of somatic cell hybrid complementation analyses,Citation7 and the FANCI gene was subsequently identified by 3 groups in 2007.Citation8-10 Using a 2-step genome-wide linkage approach with 4 genetically informative families, Dorsman and colleagues identified 4 candidate regions encompassing 39.4 Mb and comprising 351 genes. The sequencing of several candidate DNA repair genes failed to reveal sequence alterations. However, 2 strong alternative candidate genes - KIAA1794/NP_060663 and C15orf42/NP_689472 - were interrogated due to their resemblance to known FA genes in their extent of evolutionary conservation and the presence of nuclear localization signals, as many FA proteins, including FANCD2, were known to be nuclear.Citation3,Citation11-14 Sequencing of the KIAA1794 gene in all 8 FA-I patients uncovered pathogenic mutations.Citation8
Employing a phosphoproteomics screen to identify substrates of the DNA damage response kinases ATM (ataxia-telangiectasia mutated) and ATR (ATM and Rad3-related), the Elledge group identified KIAA1794 as an ionizing radiation-inducible phosphoprotein.Citation15 An amino acid sequence alignment of KIAA1794/FANCI and FANCD2 uncovered a modest overall 13% identity and 20% primary sequence similarity. However, a higher degree of sequence conservation was revealed in the region encompassing KIAA1794/FANCI K523 and that flanking FANCD2 K561, the site of FANCD2 monoubiquitination.Citation3,10 Subsequent experiments established that, like FANCD2, FANCI is monoubiquitinated following treatment of cells with DNA damaging agents and during S-phase of the cell cycle.Citation10 While full-length KIAA1794/FANCI protein was detected by immunoblotting in BD0952, an Epstein-Barr virus-transformed lymphocyte line from an FA patient of unassigned complementation group, monoubiquitinated FANCI was not detected in these cells. DNA sequencing revealed homozygosity for 2 base substitutions in KIAA1794/FANCI in BD0952 cells. These included a C to T transition leading to a P55L amino acid change, and a G to A transversion leading to a R1285Q change, the latter representing the pathogenic mutation.Citation10
In an alternative strategy, Sims and colleagues performed a BLAST search using the conserved amino acid sequence flanking the site of FANCD2 monoubiquitination K561, LVIRK, to identify proteins with similar monoubiquitination ‘consensus’ sequences.Citation9 A highly similar sequence - LVLRK - was uncovered in the uncharacterized protein KIAA1794, which, upon further examination, displayed approximately 40% sequence similarity within the regions surrounding KIAA1794 K523 and FANCD2 K561.Citation9 KIAA1794/FANCI was shown to undergo DNA damage-inducible monoubiquitination, and, importantly, this monoubiquitination was dependent on both the presence and monoubiquitination of FANCD2.Citation9 Sims and colleagues subsequently sequenced all of the exons and intron-exon boundaries of the FANCI gene for several FA patients of unassigned complementation group with defective FANCD2 monoubiquitination. Biallelic mutations in FANCI were uncovered in 4 patients. Patient EUFA592 expressed an amino-terminal truncated FANCI as a result of a homozygous T to A transition in exon 2 leading to the creation of a cryptic start site at M94.Citation9 Patient F010191 was compound heterozygous for FANCI mutations, with one allele encoding a R614X variant and the other encoding FANCI R1299X. Only the latter variant was detectable by immunoblotting. F010191 cells were defective for both FANCI and FANCD2 monoubiquitination indicating an important role for the extreme carboxy-terminus of FANCI in the regulation of this posttranslational modification. Importantly, all FA-I patients examined expressed some FANCI protein suggesting that FANCI null mutations may lead to embryonic lethality.Citation9
FA-D2 and FA-I clinical aspects
FA is a clinically heterogeneous disease characterized by a wide spectrum of congenital abnormalities, a predisposition to develop early-onset progressive bone marrow failure (BMF), and an increased risk of developing solid tumors in early adulthood. The carrier frequency of FA was previously estimated at 1:300 with an expected incidence rate of approximately 1:360,000.Citation16 However, a more recent report cites a higher carrier frequency of 1:156–1:209, leading to an expected incidence rate of 1:130,000.Citation17 A much higher carrier frequency of approximately 1:100 is found in certain populations, including Spanish Romani, Ashkenazi Jews, and Afrikaaners; this increase being attributed to the founder effect.Citation17
It had previously been estimated that the FA-D2 complementation group accounts for ∼1–3% of all FA patients.Citation18,19 However, more recently it has been reported that this complementation group accounts for approximately 6% of all FA patients.Citation20 Patients belonging to the FA-D2 complementation group display the common FA congenital malformations including skin hypo- or hyper-pigmentation, short stature, and microcephaly ().Citation20-22 Importantly, these physical abnormalities, along with skeletal irregularities such as radial ray deformities, often allow for early clinical diagnosis. One notable difference between the FA-D2 complementation group and others is the penetrance of congenital abnormalities: while it is estimated that ∼30% of individuals with FA do not display any congenital abnormality, every FA-D2 patient in one study exhibited a congenital defect.Citation20 Distinct phenotypes have also been observed in Fancd2 knockout mice that are not observed in Fanca, Fancc, or Fancg mutant mice.Citation23 Houghtaling and colleagues discovered that Fancd2-/- mice have a high prevalence of micropthalmia, an increased incidence of epithelial tumors, severe hypogonadism, as well as perinatal lethality.Citation23 In addition to the congenital abnormalities, an earlier mean age of onset of BMF is observed for FA-D2 patients, compared to patients from all other FA complementation groups - 4.5 y compared to 8.0 y.Citation20,24 Similar to that mentioned above for FA-I patients, residual FANCD2 protein expression is detected in the vast majority of FA-D2 patient cells, suggesting that only hypomorphic FANCD2 mutations are compatible with viability. The majority of FANCD2 mutations detected among FA-D2 patients are homozygous missense or compound heterozygous for a missense mutation and nucleotide change affecting splicing, often leading to monoallelic expression of low levels of both non- and monoubiquitinated FANCD2.Citation1,20
Table 1. Summary of clinical data available for FA-D2 patients
To date, only 12 patients from the FA-I complementation group have been described, compared to 30 FA-D2 patients. Much less is known about FA-I patients’ clinical manifestations, and this complementation group only accounts for an estimated less than 1% of all FA cases.Citation18 Of the few FA-I patients with available clinical data, all have growth retardation ().Citation8,9 In addition, approximately 50% of FA-I patients exhibit radial ray defects as well as both kidney and heart anomalies. Furthermore, the age of onset of hematological manifestations among FA-I patients varies widely, ranging from 1 - 8 y, and the average age of onset of BMF for the small number of FA-I patients documented is 5.4 y.Citation8 Of the limited FA-I population size examined to date, to our knowledge no unique clinical manifestations have been observed.
Table 2. Summary of clinical data available for FA-I patients
Domain Architecture and Structure of FANCD2 and FANCI
FANCD2 is a 1,451 amino acid protein with a molecular mass of 164 kDa. When originally identified, FANCD2 was described as an orphan protein, as no recognizable domains or enzymatic motifs were evident.Citation1 To date, only 4 FANCD2 domains/motifs have been experimentally characterized (). The C-terminal EDGE motif, named for the single letter amino acid code, is not required for FANCD2 monoubiquitination or nuclear foci formation. However, this motif is necessary for complementation of the ICL-sensitivity of FA-D2 patient cells.Citation25 An interaction between FANCD2 and the major DNA processivity factor PCNA has also been described.Citation26 FANCD2 contains a highly conserved PIP-motif (PCNA-interacting protein motif) which is necessary for the association of FANCD2 with PCNA (proliferating cell nuclear antigen), and for efficient FANCD2 monoubiquitination, nuclear foci formation and ICL repair, suggesting that PCNA may function as a molecular scaffold to facilitate FANCD2 function.Citation26 FANCD2 also harbors an amino-terminal CUE (coupling of ubiquitin conjugation to endoplasmic reticulum degradation) ubiquitin-binding domain (UBD).Citation27 The CUE domain adopts a triple α-helical configuration and mediates noncovalent ubiquitin binding. The CUE domain is important for interaction with FANCI, for the retention of monoubiquitinated FANCD2 and FANCI in chromatin, and for efficient ICL repair.Citation27 In addition, the N-terminal 58 amino acids of FANCD2 harbors a nuclear localization signal (NLS).Citation28 This sequence is required for the efficient nuclear localization of both FANCD2 and FANCI, and for their monoubiquitination and function in ICL repair.Citation28
Figure 1. Domain architecture and structure of FANCD2 and FANCI. (A) Schematic of the FANCD2 protein indicating the amino-terminal NLS (nuclear localization signal) domain (green), CUE (coupling of ubiquitin conjugation to endoplasmic reticulum degradation) domain (maroon), PIP-box (PCNA-interacting protein motif) (orange), and the C-terminal EDGE motif (purple). Functionally-characterized phosphorylation sites (teal) and K561 monoubiquitination site (yellow) are indicated by small circles. (B) Schematic of the FANCI protein indicating the Leu (leucine zipper) domain (light blue), ARM (armadillo repeat) domain (pink), and C-terminal EDGE motif (purple) and NLS domain (green). The S/TQ motif (teal) and K523 site of monoubiquitination (yellow) are indicated by small circles. (C) Mouse Fanci-Fancd2 heterodimer crystal structure represented as both surface and ribbons with domains indicated. This structure was solved by the Pavletich group in 2011(PDB ID: 3S4W).Citation35
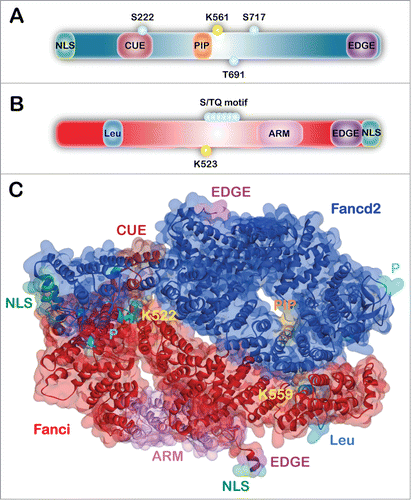
FANCI is a 1,328 amino acid protein with a molecular mass of 149 kDa (). FANCI has an armadillo (ARM) repeat domain, which form superhelical folds involved in mediating protein-protein interactions.Citation10,29 FANCI also possesses an amino-terminal leucine zipper domain, a domain capable of mediating protein-protein or DNA-protein interactions.Citation30,31 The FANCI leucine zipper domain does not bind directly to DNA, but does contribute to FANCI protein stability.Citation31 The conserved carboxy-terminus of FANCI includes a putative EDGE motif and NLS, which have been shown to enhance DNA binding and to promote the nuclear localization of FANCI, respectively.Citation31,32 FANCI also possesses numerous S/TQ motifs, known ATM/ATR phosphorylation consensus sites, and mass spectrometry revealed that at least 5 of these sites are phosphorylated in human and mouse FANCI.Citation15,33,34 The alignment of human and chicken FANCI sequences revealed that 8 S/TQ motifs are highly conserved, with 6 of these motifs clustering proximal to K523, the site of FANCI monoubiquitination.Citation9,10,33
Importantly, the 3.4Å crystal structure of the murine Fancd2-Fanci heterodimer (ID2) was recently solved by the Pavletich laboratory. While this structure continues to be a major resource for FA researchers, it is important to recognize that structural information remains lacking for several peptide stretches of both proteins.Citation35 Fancd2 and Fanci are structurally highly similar and consist largely of antiparallel α-helices that form αα-superhelical structures referred to as solenoids. Fancd2 and Fanci fold into back-to-back, inverted saxophone-like structures with trough-like partially open ends (). Multiple positively charged grooves within these troughs are capable of accommodating single and double-stranded DNA. The sites of monoubiquitination are located in the ID2 interface within solvent accessible tunnels. However, these tunnels are predicted to be too small to accommodate the ubiquitin ligase machinery, raising challenging questions about the temporal, kinetic, and spatial aspects of FANCD2 and FANCI monoubiquitination.Citation35
FANCD2 and FANCI Monoubiquitination
Protein ubiquitination is a multi-step post-translational modification that results in the covalent attachment of a single ubiquitin molecule (monoubiquitination) or chain of linked ubiquitin molecules (polyubiquitination) to a lysine residue on a substrate protein. Mono- and poly-ubiquitination signal a wide range of cellular instructions, the former being most typically associated with the targeting of proteins to specific sub-cellular localizations, while the canonical role of the latter is to target proteins for degradation by the proteasome.Citation36,37 In the general process of ubiquitination, an E1 ubiquitin-activating enzyme activates ubiquitin by forming a thioester bond between a catalytic C of E1 and the C-terminal G76 of ubiquitin. Ubiquitin is then transferred from the E1 to an active site C of an E2 ubiquitin-conjugating enzyme. Ubiquitin can be further transferred to the catalytic site of a HECT (homologous to E6-AP carboxy terminus) domain-containing E3 ubiquitin ligase, and then subsequently transferred to a target K on the substrate. Alternatively, through the action of a RING (really interesting new gene) domain-containing E3 ubiquitin ligase, ubiquitin is transferred directly from the E2 to the substrate. The human genome encodes 2 E1 ubiquitin-activating enzymes, ∼40 E2 ubiquitin-conjugating enzymes, and >600 E3 ubiquitin ligases.Citation38
As mentioned above, when the FANCD2 protein was first characterized by Garcia-Higuera and colleagues in 2001, 2 isoforms of FANCD2 were detected by immunoblotting; a short form approximately 164 kDa and a longer form approximately 173 kDa.Citation3 The latter isoform was determined to represent FANCD2 covalently linked to a single ubiquitin polypeptide on K561.Citation3 Similar to FANCD2, a slower migrating isoform of FANCI was also detected by immunoblotting, following treatment of cells with DNA damaging agents.Citation9 This slower migrating FANCI isoform was subsequently determined to represent FANCI monoubiquitinated on K523.Citation9,10 The monoubiquitination of FANCD2 and FANCI is required for their assembly into discrete nuclear foci, where they co-localize with several proteins including the protein product of one of the major breast and ovarian cancer susceptibility genes BRCA1.Citation3,9,10 This seminal finding provided an important clue as to a potential function for FANCD2, FANCI, and the FA-BRCA pathway, as BRCA1 had previously been determined to play a key role in DNA repair by homologous recombination (HR).Citation39-41 HR is an error-free and conservative DNA repair pathway that utilizes an intact homologous template, such as a sister chromatid or homologous chromosome, to repair DNA damage.Citation42 The ability of FANCD2 and FANCI to promote error-free HR repair was assessed using the DR-GFP HR reporter assay system developed by the Jasin laboratory.Citation43,44 This reporter assay comprises 2 differentially mutated nonfunctional GFP genes oriented in tandem, and separated by a drug selection marker. One GFP gene is interrupted with the rare 18-bp I-Sce I restriction endonuclease recognition sequence, while the other is an 812-bp internal fragment. Following induction of a DSB with the I-Sce I endonuclease, a gene conversion HR event results in the expression of GFP, which can be assayed by flow cytometry or fluorescence microscopy.Citation43 Knockdown of either FANCD2 or FANCI, as well as mutation of FANCD2 K561 or FANCI K523, which precludes the covalent attachment of ubiquitin, leads to decreased HR efficiency.Citation9,10,44 Taken together, this data strongly suggests that monoubiquitinated FANCI and FANCD2 function cooperatively to promote error-free HR repair. Importantly, FANCD2 and FANCI monoubiquitination and nuclear foci formation are interdependent.Citation9,10 For example, siRNA-mediated depletion of FANCI results in decreased FANCD2 monoubiquitination and nuclear foci formation, as well as a reduction in steady state levels of FANCD2.Citation9,10 These findings were suggestive of a physical interaction between FANCD2 and FANCI, and indeed this was verified by co-immunoprecipitation studies as well as by co-crystallization of the murine ID2 heterodimer.Citation10,35 Mutation of FANCD2 K561 renders this protein incompetent for nuclear foci formation, and abrogates its ability to rescue the ICL hypersensitivity of FA-D2 patient cells. Mutation of FANCI K523 also precludes its assembly into nuclear foci.Citation9,10 However, somewhat surprisingly, in contrast to FANCD2 K561, mutation of FANCI K523 does not overtly affect its function in ICL repair.Citation3,9,10,33 In vitro studies have also revealed that mutation of FANCD2 K561 leads to impaired FANCI monoubiquitination, whereas mutation of FANCI K523 has little effect on the monoubiquitination of FANCD2.Citation45
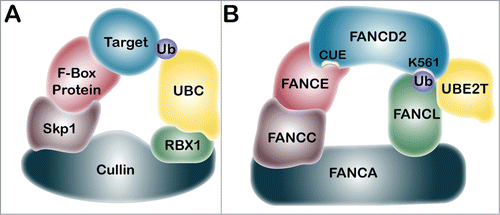
The E2 ubiquitin-conjugating enzyme and E3 ubiquitin ligase for FANCD2 and FANCI monoubiquitination are UBE2T and the RING domain-containing FANCL, respectively.Citation46,47 Biallelic mutations in the FANCL gene underlie the FA-L complementation group, however, FA patient-derived mutations in UBE2T have yet to be uncovered.Citation46,47 In addition to UBE2T and FANCL, the monoubiquitination of FANCD2 and FANCI requires the assembly of the FA core complex, comprising FANCA, FANCB, FANCC, FANCE, FANCF, FANCG, and FANCM, as well as numerous accessory proteins including FAAP20, FAAP24, FAAP100, MHF1, and MHF2.Citation48-53 The precise roles of these ancillary proteins in FANCD2/FANCI monoubiquitination remain to be determined. Indeed, in vitro FANCD2/FANCI monoubiquitination reactions have been reconstituted with FANCL in the absence of these FA core complex components.Citation54-56 Recent studies have identified 3 functional modules within the FA core complex: the FANCB-FANCL-FAAP100 module, which provides the critical catalytic activity, and the FANCA-FANCG-FAAP20 and FANCC-FANCE-FANCF modules, which independently promote the recruitment of the core complex to chromatin.Citation57,58 Accordingly, using isogenic single and double mutants of the HCT116 colorectal carcinoma cell line, Huang et al. established that FANCD2/I monoubiquitination is completely abrogated in the absence of FANCB, FANCL, or FAAP100, and only partially abrogated in the absence of FANCA, FANCG, FANCE, or FAAP20.Citation57 Furthermore, in contrast to FANCD2, the purified FA core complex fails to robustly promote the monoubiquitination of FANCI in the absence or presence of DNA.Citation58 To gain further insight into the relationship between the FA core complex members and FANCD2/I monoubiquitination, it is useful to draw comparisons with other large multi-subunit ubiquitin ligase protein complexes, such as the Skp1/Cullin/F-box protein complex (SCF) ().Citation59 SCF, along with the anaphase-promoting complex or cyclosome (APC/C), plays an intricate and critical role in cell-cycle regulation.Citation60 The F-box protein of the SCF complex functions in substrate recognition and recruitment.Citation59 A good candidate for an FA-pathway protein analogous to the SCF F-box protein is FANCE, a FA core complex member that has been demonstrated to interact with FANCC, FANCF, and FANCD2, thereby bridging the core ubiquitin ligase machinery and the substrate.Citation61 The FANCE-FAND2 interaction, via FANCE F522, is critical for efficient FANCD2 monoubiquitination and ICL repair.Citation62 The Cullin protein functions as a molecular scaffold for the SCF complex, bridging the catalytic RING finger subunit RBX1 to the adaptor and F-box proteins. The FANCA protein, with a molecular mass of 163 kDa, may play a similar structural role in the FA-BRCA pathway. FANCA interacts with FANCC, FANCG, and FAAP20.Citation12,57,58,63,64 The Skp1 protein of the SCF complex functions as an adaptor between the Cullin protein and the F-box protein.Citation65 FANCC interacts with FANCA, FANCE, and FANCF and thus represents a strong candidate Skp1 analog.Citation57,58,61 FANCM is present in very low abundance in the native FA core complex, consistent with its role in replication fork remodeling and chromatin loading of the core complex.Citation58,Citation66-68 The core complex members FANCM, FANCG, and FANCF do not appear to have analogous SCF components. FANCL is analogous to the SCF E3 ubiquitin ligase RBX1. As mentioned above, FANCL, FANCB, and FAAP100 comprise the E3 monoubiquitin ligase catalytic module.Citation57,58 FANCL and RBX1 are both RING E3 ubiquitin ligases, with the RING domain facilitating recruitment of the E2 ubiquitin-conjugating enzyme, UBE2T in the case of the FA-BRCA pathway and UBC3, UBC4, or UBC5 for SCF.Citation46,65,69 FANCL binds to FANCD2 via its DRWD domain, and to UBE2T via its RING domain, to facilitate monoubiquitination.Citation69,70
Figure 2. Comparison of the SCF multi-subunit ubiquitin ligase protein complex and the FA core complex. (A) The Skp1/Cullin/F-box protein (SCF) complex includes the Cullin protein, which acts as a scaffold to bridge the catalytic E3 ubiquitin ligase RBX1 to the adaptor protein Skp1, and the F-box protein.Citation59 The F-box protein recognizes and recruits the target protein for ubiquitination by the E2 ubiquitin-conjugating enzyme, UBC. (B) We propose that the FANCA protein is structurally analogous to Cullin, and may link the E3 ubiquitin ligase FANCL with the putative adaptor protein FANCC. FANCC has been shown to interact with both FANCA and FANCE, indicating that it may function analogously to Skp1.Citation61 FANCE may be analogous to the F-box protein. FANCE is known to interact directly with FANCD2 and may facilitate its monoubiquitination of FANCL and UBE2T.Citation62
Ubiquitination: Mono- vs. Poly-
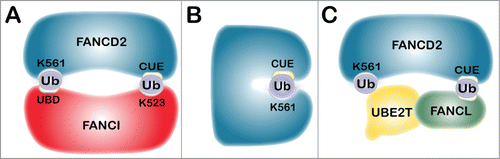
To date, only the monoubiquitination of FANCD2 has been observed, begging the question; how is monoubiquitinated FANCD2 restricted from being polyubiquitinated? Insights into this question can be gained from extensive studies of regulators of endocytosis. Many endocytic proteins are monoubiquitinated in a process referred to as coupled monoubiquitination, whereby substrate recognition and ubiquitination are regulated, at least in part, through noncovalent interactions between ubiquitin and substrate ubiquitin-binding domains (UBDs). One such example is EPS15 (epidermal growth factor receptor pathway substrate 15), an important regulator of the endocytosis of ligand-inducible receptors of the receptor tyrosine kinase (RTK) family, in particular EGFR.Citation71 EPS15 is monoubiquitinated by the NEDD4 HECT E3 ubiquitin ligase and by the Parkin RING E3 ubiquitin ligase. EPS15 harbors 2 ubiquitin-interacting motifs (UIMs) in its carboxy-terminus that are critical for its regulation. One proposed model for why EPS15 is solely monoubiquitinated is that once ubiquitinated, EPS15 undergoes an intramolecular rearrangement enabling covalently attached ubiquitin to engage in a noncovalent interaction with one of its 2 C-terminal UIMs, generating a closed conformation refractory to further ubiquitination.Citation72,73 The crystal structure of the murine ID2 heterodimer has revealed that the mouse Fancd2 K559 and Fanci K522 sites of monoubiquitination are buried within the ID2 interface. A solvent accessible tunnel adjacent to these sites is predicted to be too small to accommodate UBE2T, suggesting that monoubiquitination either precedes heterodimerization or occurs subsequent to ID2 dissociation. Similar to the EPS15 UIM, one possibility is that ID2 heterodimerization occurs following monoubiquitination, and that the ID2 heterodimer is stabilized through a noncovalent interaction between monoubiquitin covalently linked to FANCI K523 and the FANCD2 CUE domain, and quite possibly a reciprocal interaction between monoubiquitinated FANCD2 K561 and an UBD in the carboxy-terminus of FANCI ().Citation27 Thus, FANCD2 and FANCI might be shielded from further ubiquitination via intermolecular association. It is also possible that monoubiquitination promotes an intramolecular association between ubiquitin covalently attached to K561 and the amino-terminal CUE domain,Citation27 potentially blocking the ubiquitin on K561 from further ubiquitination, similar to that for EPS15 (). The latter model would more closely align with recent findings from the Sobeck laboratory indicating that activation of the FA-BRCA pathway coincides with dissociation of FANCD2 and FANCI.Citation74 ID2 dissociation is triggered by ATM/ATR-mediated phosphorylation of a cluster of at least 6 FANCI SQ/TQ motifs, and is followed by the monoubiquitination of FANCD2, see below.Citation33,74
Figure 3. Models for FANCD2 and FANCI monoubiquitination. The schematics depict several potential outcomes upon monoubiquitination of FANCD2 and FANCI, which would preclude further ubiquitination. (A) The ID2 heterodimer inactivation model. Following monoubiquitination, ID2 heterodimerization occurs and is stabilized through a noncovalent interaction between monoubiquitin covalently linked to FANCI K523 and the FANCD2 CUE domain. There is also possibly a reciprocal interaction between monoubiquitinated FANCD2 K561 and an UBD in the carboxy-terminus of FANCI, shielding FANCD2 from further ubiquitination. (B) The FANCD2 self-inactivation model. Monoubiquitination could promote an intramolecular association between ubiquitin covalently attached to K561 and the amino-terminal CUE domain, resulting in a closed conformation. (C) The E3 ubiquitin ligase dissociation model. Once FANCL is autoubiquitinated, the ubiquitin moiety may interact noncovalently with the CUE domain on FANCD2 enabling monoubiquitination of FANCD2 on K561. This interaction is predicted to be weak and short-lived, leading to rapid dissociation of FANCL and FANCD2, precluding further ubiquitination.
The NEDD4 E3 ubiquitin ligase can mono- and polyubiquitinate substrates, and the balance between these 2 posttranslational modifications is determined, at least in part, by the nature of the enzyme-substrate interaction. The tryptophan-tryptophan (WW) motif of NEDD4 can interact with relatively high affinity to a substrate PPxY (where x is any amino acid) motif, leading to substrate polyubiquitination.Citation75,76 EPS15 lacks a PPxY motif and instead interacts with NEDD4 via a weak noncovalent interaction between ubiquitin covalently conjugated to NEDD4 and one of its C-terminal UIMs.Citation77,78 Similarly, EPS15 interacts with the Parkin RING E3 ubiquitin ligase via a weak interaction between an ubiquitin-like domain (UbL) in Parkin and one of its UIMs.Citation79 Previous studies have indicated that the FANCL E3 ubiquitin ligase, as well as the UBE2T E2 ubiquitin-conjugating enzyme, undergoes autoubiquitination.Citation54,80 Therefore, the FANCD2 CUE domain may interact with ubiquitin conjugated to FANCL (or UBE2T) and, because of the weak nature of this interaction, FANCD2 and FANCL may rapidly dissociate following ubiquitination, precluding further ubiquitination ().
Another major factor influencing mono- vs. poly-ubiquitination is the nature of the E2 ubiquitin-conjugating enzyme. There are ∼40 E2 ubiquitin-conjugating enzymes encoded by the human genome, 28 of which are classified as monoubiquitinating E2s. The FANCD2/I E2 ubiquitin-conjugating enzyme UBE2T is classified as a monoubiquitinating E2.Citation46,54,56,80 Several other E2s function as polyubiquitinating E2s and display ubiquitin linkage-specificity. For example, UBE2G1, UBE2G2, UBE2K, UBE2R1, and UBE2R2 promote the assembly of K48-linked polyubiquitin chains, while UBE2S and UBE2N promote the assembly of K11- and K63-linked polyubiquitin chains, respectively.Citation81 In the case of UBE2N, K63-linked polyubiquitin chain assembly is facilitated by the cofactor proteins UBE2V1 and UBE2V2, which structurally align K63 of the acceptor ubiquitin toward the active site C of UBE2N.Citation82,83 Thus, the monoubiquitination fate of FANCD2 and FANCI may simply be determined by the mechanistic nature of UBE2T. Nevertheless, much remains to be learned about the mechanism, structure, and regulation of UBE2T.
Lastly, the concentration of an E3 ubiquitin ligase relative to the concentration of its substrate has also been shown to play an important role in determining if a protein is monoubiquitinated or polyubiquitinated. One example of this mechanism of regulation applies to the ubiquitination of the p53 tumor suppressor protein by its E3 ubiquitin ligase MDM2: When levels of MDM2 are low, p53 is largely monoubiquitinated. Conversely, when levels of MDM2 are high, p53 is predominantly polyubiquitinated.Citation84 Likewise, it is plausible that the cellular concentration of FANCL is sufficient for FANCD2/I monoubiquitination only, precluding further ubiquitination.
FANCD2 and FANCI Deubiquitination
The site-specific monoubiquitination of FANCD2 and FANCI is a reversible process. The deubiquitination of monoubiquitinated FANCD2 and FANCI is mediated by ubiquitin-specific protease 1 (USP1). This important function for USP1 was uncovered in a DUB (deubiquitinating enzyme) gene family RNA interference library screen, whereby knockdown of USP1 resulted in a significant increase in monoubiquitinated FANCD2.Citation85 Deletion of murine Usp1 results in increased perinatal lethality, male infertility, and an FA phenotype.Citation86 Usp1-/- mouse embryonic fibroblasts (MEFs) exhibit elevated levels of Fancd2 monoubiquitination and impaired Fancd2 nuclear foci formation and are defective in HR repair.Citation86 USP1 is activated upon formation of a complex with the UAF1 (USP1 associated factor-1) protein.Citation87 The amino-terminus of UAF1 has a WD40 domain, which binds and activates USP1, while the carboxy-terminus contains 2 SUMO-like domains (SLD1 and SLD2) that mediate substrate binding.Citation87,88 FANCI harbors a highly conserved SUMO-like domain-interacting motif (SLIM) that has been shown to interact with SLD2 of UAF1, targeting USP1/UAF1 to the ID2 complex.Citation88 USP1/UAF1 activity is tightly regulated: upon exposure to UV irradiation, USP1 undergoes inhibitory autocleavage.Citation89 The C-terminal fragment of autocleaved USP1 is subsequently degraded by the R/N-end rule pathway, leading to the effective elimination of deubiquitinating activity.Citation90 The USP1 gene is also transcriptionally repressed upon cellular exposure to DNA damaging agents, a process that requires the function of the p21 cyclin-dependent kinase inhibitor.Citation87,91 Both negative regulatory mechanisms facilitate the timely accumulation of monoubiquitinated FANCD2 and FANCI following the induction of DNA damage.Citation89
FANCD2 Phosphorylation
The FANCD2 protein is also subject to posttranslational modification by phosphorylation. The ATM kinase is a member of the phosphatidylinositol-3-OH-kinase-like family of protein kinases (PIKK). Bilallelic mutations in the ATM gene cause the autosomal recessive disorder Ataxia- telangiectasia (AT).Citation92 AT is clinically characterized by progressive cerebellar ataxia, telangiectasias, immune defects, and increased susceptibility to hematologic cancers of B and T cell origin, as well as central nervous system tumors.Citation93,94 The ATM kinase plays a major coordinating role in the cellular response to DNA DSBs and phosphorylates multiple substrates to halt cell cycle progression and initiate DNA repair. For example, the ATM kinase phosphorylates T68 of the CHK2 kinase following exposure to ionizing radiation (IR) to initiate cell cycle arrest at the G1-S boundary.Citation95 In 2002, Taniguchi and colleagues discovered that ATM phosphorylates FANCD2 on S222, S1401, S1404, and S1418 in response to IR exposure.Citation96 Phosphorylation of FANCD2 S222 is required for the establishment of the IR-inducible S-phase checkpoint, an ability to pause DNA synthesis in the presence of chromosomal DSBs.Citation97 However, ATM-mediated FANCD2 S222 phosphorylation is dispensable for FANCD2 monoubiquitination, nuclear foci formation, and ICL resistance.Citation96
An important role for the ATR kinase in the regulation of FANCD2 has also been established.Citation98,99 While ATM primarily responds to DNA DSBs, ATR is a major regulator of the DNA replication checkpoint, primarily responding to agents that disrupt DNA replication, e.g., hydroxyurea, an inhibitor of deoxyribonucleotide reductase, aphidicolin (APH), a processive DNA polymerase inhibitor, and UV irradiation, which promotes the formation of cyclobutane pyrimidine dimers.Citation100,101 Mutations in ATR underlie Seckel syndrome (SCKL1) a rare autosomal recessive disorder characterized by severe growth retardation, short stature, microcephaly, mental retardation, and increased risk for acute myeloid leukemia (AML), myelodysplasia (MDS), and aplastic anemia, similar to FA.Citation102-104 Following exposure to DNA crosslinking agents, ATR and FANCD2 co-localize in nuclear foci.Citation98,105 ATR also promotes efficient DNA damage-inducible FANCD2 monoubiquitination and nuclear foci formation.Citation98 Accordingly, similar to FA patient cells, ATR-deficient SCKL1 patient cells exhibit increased chromosome aberrations, including radial formations, following exposure to MMC.Citation98,104 The D’Andrea laboratory established that ATR phosphorylates FANCD2 on T691 and S717 in vitro and in vivo.Citation98,99 A FANCD2-T691A/S717A double mutant fails to undergo efficient DNA damage-inducible monoubiquitination and fails to correct the MMC hypersensitivity of FA-D2 patient cells.Citation99 How phosphorylation of these 2 sites promotes FANCD2 monoubiquitination remains unclear. Phosphorylated T691 and S717 may stimulate interaction with the core complex or with FANCL or UBE2T, promoting monoubiquitination. Alternatively, phosphorylation may promote dissociation from FANCI, allowing access to K561, similar to that described for FANCI, see below. Phosphorylation could also promote the interaction of FANCD2 with DNA in chromatin, which has been shown to lead to increased efficiency of monoubiquitination.Citation56,106
FANCI Phosphorylation
FANCI was originally identified as an ATM/ATR substrate in a large-scale SILAC (stable isotope labeling with amino acids in cell culture) proteomics screen.Citation15 As described above, 6 S/TQ motifs are positioned proximal to K523, the sites of monoubiquitination site. These S/TQ sites become phosphorylated in the absence of prior monoubiquitination, indicating that this posttranslational modification acts upstream of monoubiquitination.Citation33 Importantly, mutation of these S/TQ sites results in abrogation of FANCI and FANCD2 monoubiquitination and nuclear foci formation,Citation33 indicating that phosphorylation of the FANCI S/TQ cluster functions as a molecular switch to activate the FA-BRCA pathway. However, the underlying molecular mechanism remains unclear. Recently, it has been suggested that activation of the pathway coincides with disassociation of the ID2 heterodimer, and that ID2 disassociation is triggered by ATM/ATR-mediated phosphorylation of the S/TQ cluster.Citation106 Using cell-free Xenopus laevis egg extracts, Sareen and colleagues demonstrated that a FANCI S/TQ cluster phosphorylation-dead mutant failed to dissociate from FANCD2 and failed to undergo monoubiquitination. Conversely, a phosphorylation-mimetic mutant failed to associate with FANCD2 and exhibited increased monoubiquitination.Citation106 Taken together, these findings indicate that phosphorylation of FANCI at the conserved S/TQ cluster is a key mechanism in the activation of the FA-BRCA pathway. The introduction of a cluster of negatively charged phosphate groups may result in a localized charge repulsion that promotes ID2 disassociation, enabling access of the previously occluded ubiquitin ligase machinery.
FANCD2 and FANCI Function
Replisome surveillance
Early studies indicated that, in addition to being monoubiquitinated following exposure to DNA damaging agents, FANCD2 and FANCI monoubiquitination also occurs during unperturbed cell cycle progression. In addition, monoubiquitinated FANCD2 co-localizes with BRCA1 and RAD51 in nuclear foci during S-phase.Citation97 These findings strongly suggested that monoubiquitinated FANCD2 might play an important role in maintaining genome stability during the process of DNA replication.Citation10,97 Accordingly, the FANCD2 and FANCA proteins are required for the maintenance of common chromosomal fragile site (CFS) stability.Citation107 CFS are chromosomal loci that are prone to breakage when cells are cultured under conditions of DNA replication stress, for example following treatment with low concentrations of the DNA polymerase inhibitor APH.Citation108 CFSs are hot spots for sister chromatid exchanges (SCE), chromosomal translocations, and viral integration, and are frequently rearranged or deleted in cancer.Citation109 The physical localization of FANCD2 and FANCI to FRA3B and FRA16D, the most frequently expressed CFSs, has also been demonstrated.Citation110 Combined immunofluorescence microscopy and FISH analysis of metaphase chromosomes revealed the presence of paired FANCD2/I-stained CFSs on sister chromatids linked by BLM helicase-associated ultra-fine DNA bridges (UFBs).Citation110,111 These paired FANCD2/I sister foci form during S-phase and persist into mitosis, and their presence strongly suggest that FANCD2 and FANCI participate in the resolution of stalled or collapsed DNA replication forks that recurrently arise at particularly unstable genomic loci.Citation107,110,111 In further support of an important role for FANCD2 in the resolution of stalled or collapsed replication forks, Fancd2-/- MEFs are ICL hypersensitive yet do not display increased sensitivity to IR.Citation23 Consistent with a role for the FA-BRCA pathway in the DNA replication stress response, FANCD2 has been shown to interact with several components of the DNA replisome. For example, as mentioned earlier, FANCD2 interacts with the DNA polymerase processivity factor PCNA, via a conserved PIP-box, and this interaction is necessary for efficient FANCD2 monoubiquitination and ICL repair.Citation26 In addition, using a method called iPOND (isolation of proteins on nascent DNA), the Cortez group recently discovered that FANCD2 and FANCI, in addition to ATR, MRE11 and other proteins, are highly enriched at stalled and collapsed replication forks following depletion of deoxyribonucleotide pools.Citation112 Interestingly, FANCI, and not FANCD2, was also shown to accumulate at active replication forks prior to fork staling, suggesting common and independent functions for these proteins. Citation112 In addition, the Vaziri group has recently shown that FANCD2 is necessary for efficient initiation of DNA replication in primary human fibroblasts.Citation113 An unbiased proteomics screen of chromatin-bound FANCD2 immune complexes revealed that FANCD2 physically associates with the minichromosome maintenance 2–7 (MCM2-MCM7) replicative helicases.Citation114,115 FANCD2 interacts with MCM2-MCM7 in response to ATR-mediated DNA replication stress signaling independently of the core FA complex and monoubiquitination.Citation115 In the absence of FANCD2, replication forks stall stochastically and primary cells exhibit increased senescence, indicating that FANCD2 is required for replication fork progression.Citation115 Schlacher and colleagues also showed that FANCD2 is necessary to protect stalled replication forks from MRE11-mediated degradation, and that fork protection in the absence of FANCD2 can be rescued by RAD51.Citation116 Moreover, it was recently discovered that FANCD2, in cooperation with BRCA1 and MRE11, recruits and interacts with CtIP (CtBP-interacting protein) at stalled replication forks to promote replication restart.Citation117,118 CtIP was originally identified on the basis of its ability to bind to CtBP (C-terminal binding protein), a transcriptional co-repressor.Citation118 However, several recent studies have established that CtIP interacts with the MRN (MRE11/RAD50/NBS1) complex and BRCA1 and facilitates the DNA strand resection step of HR repair.Citation118-121 This evidence supports a key role for FANCD2 and FANCI in coordinating the recruitment of HR proteins to sites of stalled replication forks, enabling timely replication fork resolution and progression.
In addition to being directly regulated by the FANCL-UBE2T ubiquitin ligase machinery, several studies have established that FANCD2/I monoubiquitination is indirectly regulated by the RAD18-RAD6 E3/E2-ubiquitin ligase complex.Citation122-126 RAD18-RAD6 catalyzes the monoubiquitination of PCNA on K164, a molecular event necessary for DNA polymerase switching during translesion DNA synthesis (TLS).Citation127,128 TLS polymerases possess larger active sites and exhibit reduced fidelity, and have the ability to catalyze DNA polymerization opposite DNA templates harboring aberrantly modified nucleobases.Citation129 RAD18 and FANCD2, as well as FANCL, function epistatically in the cellular ICL response.Citation122,126 Genetic disruption of RAD18 or RAD18 depletion via siRNA leads to decreased ICL-induced FANCD2/I monoubiquitination and chromatin binding.Citation122-126 Furthermore, RAD18-RAD6 mediated monoubiquitination of PCNA on K164 is necessary for efficient FANCD2 monoubiquitination.Citation122 FANCL was also shown to bind to PCNA directly and PCNA K164 monoubiquitination was shown to be required for FANCL chromatin loading.Citation122 While the molecular mechanism underlying this regulation remains unclear, these findings suggest that the FANCL-UBE2T and RAD18-RAD6 ubiquitin ligases cooperatively regulate FANCD2/I and PCNA monoubiquitination, respectively, to facilitate the coordinated recruitment of HR and TLS factors to stalled or collapsed replication forks.
Independent Functions of FANCD2 and FANCI
BLM complex chromatin assembly
Since the discovery of FANCI, it has generally been accepted that FANCD2 and FANCI function cooperatively in the resolution of ICLs. However, several recent studies have suggested independent functions for these proteins. For example, it has recently been determined that FANCD2, and not FANCI, is an important regulator of the recruitment of the BLM helicase to chromatin.Citation130 Biallelic mutations in the BLM gene underlie Bloom syndrome (BS), a rare recessive disorder characterized by growth retardation, sunlight sensitivity, telangiectasia, hypo- or hyper-pigmentation of the skin, and increased susceptibility to hematologic cancers.Citation131 The BLM helicase is a member of the RecQ family of helicases, which also includes the WRN and RECQ4 proteins, and possesses ATP-dependent 3′-5′ DNA helicase activity.Citation132 Similar to FA patient cells, BS patient cells are hypersensitive to the cytotoxic effects of DNA crosslinking agents as well as agents that inhibit DNA replication.Citation133,134 In addition, FANCD2 and BLM co-localize in nuclear foci following exposure to DNA damaging agents.Citation134 Pichierri and colleagues established that the DNA damage-inducible phosphorylation of BLM and its ability to assemble into nuclear foci are compromised in FA-C and FA-G patient cells.Citation134 In contrast, FANCD2 monoubiquitination and nuclear foci formation, as well as the DNA damage-inducible chromatin localization of FANCA and FANCC, are intact in BS patient cells.Citation134 Similarly, using Xenopus egg extracts, Chaudhury et al recently showed that immunodepletion of both FANCD2 and FANCI leads to a failure of the BLM helicase, and its binding partners TOP3a, RMI1, RM2, and RPA1-3, to associate with chromatin. Notably, FANCD2 alone is sufficient to rescue this defect.Citation130 Furthermore, neither FANCI nor FANCD2 monoubiquitination are required to promote BLM complex chromatin assembly, indicating that this function is monoubiquitination and FANCI-independent.Citation130
Nucleosome Assembly Activity
FANCD2 was recently identified in a proteomics screen of histone H3/H4-interacting proteins.Citation135 Recombinant FANCD2 was subsequently confirmed to bind to H3/H4 in vitro.Citation135 These findings suggested that FANCD2 might play an active role in chromatin dynamics and/or reorganization. Indeed, using a topological assay and a nucleosome assembly assay, FANCD2 was shown to promote nucleosome assembly to an extent comparable to that of the known nucleosome assembly protein Nap1. Similar to the promotion of BLM complex chromatin localization, FANCD2 histone binding and nucleosome assembly activity appear to be FANCI and monoubiquitination-independent.Citation135 However, while FANCI fails to promote nucleosome assembly by itself, it can stimulate FANCD2-mediated nucleosome assembly at low FANCD2 concentrations. These results suggest that the ID2 complex may promote chromatin reorganization during ICL repair. Consistent with this hypothesis, FANCD2 was previously discovered to interact with the 60 kDa Tat-interactive protein (TIP60) chromatin remodeling acetyltransferase and menin, a chromatin remodeling tumor suppressor.Citation136,137 Depletion of TIP60 using siRNA sensitizes cells to ICL cytotoxicity.Citation137 TIP60 is a subunit of the NuA4 chromatin remodeling complex, which comprises multiple enzymatic subunits including TIP60, the p400 motor ATPase, and the RUVBL1 and RUVBL2 helicase-like proteins.Citation138 Upon DSB generation, TIP60 is rapidly recruited to sites of damage where it acetylates multiple substrates including H2A, H4, p53, and ATM.Citation139-142 Menin has also been shown to function in chromatin remodeling, and DNA repair.Citation136,143,144 Cells lacking menin are more sensitive to DNA damage and the interaction between FANCD2 and menin is increased following exposure to IR.Citation136 Collectively, these findings suggest that FANCD2 may function in a chromatin remodeling capacity during the early stages of ICL repair.
FANCD2: Recruitment of Nucleases
Several recent studies have provided important mechanistic insight into the function of monoubiquitinated FANCD2 in ICL repair. The previously uncharacterized KIAA1018 protein was simultaneously identified by several groups as a FANCD2-interacting partner and an important cellular determinant of ICL repair efficiency.Citation145-147 The Jiricny group identified KIAA1018 as a component of the mismatch repair MLH1 and PMS2 interactomes. This protein was predicted to harbor an amino-terminal UBZ (ubiquitin-binding zinc finger) UBD and a C-terminal PD-D/E(X)K-type nuclease domain.Citation146 Similarly, the Huang and Rouse groups identified KIAA1018 on the basis of its UBZ domain and a VRR_nuc (virus-type replication-repair nuclease) domain, which contains a motif frequently found in bacterial and bacteriophage restriction nucleases.Citation145,148 Importantly, UBZ domains have been uncovered in several DNA damage response proteins, including RAD18 and DNA polymerase κ (POL κ).Citation149,150 In another approach, Smogorzewska and colleagues discovered KIAA1018 in an unbiased genome-wide shRNA screen of proteins necessary for efficient ICL repair.Citation147 KIAA1018 was found to co-localize with FANCD2 in nuclear foci following ICL exposure and to interact with FANCD2 via co-immunoprecipitation analysis, and was hence renamed FAN1 (FANCD2-associated nuclease 1).Citation145-148 FAN1 was shown to display 5′-3′ exonuclease activity and structure-specific endonuclease activity, preferentially toward 5′ flap structures.Citation145-148 Furthermore, the FAN1 UBZ domain was shown to interact noncovalently with monoubiquitin conjugated to FANCD2 K561. The FAN1 UBZ domain and the monoubiquitination of FANCD2 were shown to be required for the recruitment of FAN1 to sites of DNA damage, and for efficient ICL repair.Citation145 While FAN1 mutations have yet to be uncovered in FA patients, mutations in FAN1 have been identified as a cause of karyomegalic interstitial nephritis, a chronic kidney disease.Citation151
Mutations in the SLX4 gene were recently uncovered in FA complementation group P.Citation152-154 FANCP/SLX4 is a ∼200 kDa multidomain protein that, similar to FAN1, harbors an amino-terminal UBZ domain.Citation155 FANCP/SLX4 functions as a molecular platform to coordinate the activities of the structure-specific endonucleases XPF-ERCC1, MUS81-EME1, and SLX1, with XPF-ERCC1 playing a major role in ICL repair, and MUS81-EME1 and SLX1 playing less critical roles.Citation156-160 Similar to FAN1 and FANCD2, FANCP/SLX4 and FANCD2 co-localize in nuclear foci and co-immunoprecipitate.Citation154 While Yamamoto and colleagues established that the FANCP/SLX4 UBZ domain and the monoubiquitination of FANCD2 are required for FANCP/SLX4 targeting to nuclear foci, a recent report has contradicted these findings and shown that SLX4 recruitment occurs independent of FANCD2 monoubiquitination.Citation154,161 Taken together, these findings suggest that one major function for chromatin-bound monoubiquitinated FANCD2 may be the recruitment of structure-specific endonucleases that mediate key incision steps during the process of ICL repair.Citation162
FANCD2 and FANCI: DNA Binding
Several studies have established that FANCD2 and FANCI physically bind to DNA. Using FANCD2 purified from baculovirus-infected SF9 insect cells, the Parvin group originally determined that full-length FANCD2 binds to dsDNA and Holliday junctions (HJs), quadruplex heteroduplex DNA intermediates that arise during HR.Citation163 Similarly, the Zhang group purified full-length FANCI and demonstrated that it can bind to multiple DNA substrates, including HJs, dsDNA, ssDNA, and flap structures, without any apparent structural preference.Citation31 Longerich et al also demonstrated that purified FANCI is capable of binding to dsDNA, ssDNA, and HJs.Citation80 Importantly, however, co-purified ID2 displayed a much lower affinity for dsDNA and ssDNA, yet maintained a robust binding activity toward HJs and various branched DNA structures.Citation31 Consistent with these findings, the Pavletich group described the structure of co-crystals of FANCI bound to splayed Y DNA at a resolution of 7.8 Å.Citation35 Based on this structure and the structural homology between FANCD2 and FANCI, the authors proposed that the ID2 heterodimer could have 2 sets of pseudo-symmetrical dsDNA/ssDNA binding sites, structures that could arise upon stalling of one or 2 replication forks at an ICL.Citation35 Notably, the Patel group discovered that, in addition to binding to dsDNA and ssDNA, FANCD2 exhibits 3′ to 5′ exonucleolytic activity toward ssDNA in the presence of MgCitation2+ or MnCitation2+.Citation164 However, the functional significance for this activity remains to be established.
In addition, recent studies have established that FANCD2 monoubiquitination is strongly stimulated through DNA binding. For example, the Kurumizaka and Sung laboratories demonstrated that several forms of DNA, including dsDNA, ssDNA, and Holliday junctions, could robustly stimulate the in vitro monoubiquitination of FANCD2.Citation45,56 Of note, this DNA-dependent stimulation of FANCD2 monoubiquitination required both the presence of FANCI and its ability to bind to DNA.Citation56 Using Xenopus egg extracts, Sobeck and colleagues also demonstrated that the monoubiquitination of FANCD2 could be stimulated with linear and branched dsDNA structures but not with ssDNA or Y-shaped DNA.Citation165 Expanding on these studies, the Sobeck group recently demonstrated that while FANCD2 monoubiquitination is strongly stimulated by circular and linear dsDNA, the monoubiquitination of FANCI is strongly stimulated by ssDNA and only weakly induced by circular and linear dsDNA, indicating a dichotomy in substrate specificity for both proteins.Citation74 In addition, through a time course study, the same group determined that the monoubiquitination of FANCD2 in the presence of circular dsDNA and ssDNA occurs significantly earlier than the monoubiquitination of FANCI.Citation74 FANCD2 was also found to localize to chromatin earlier than, and independent of, FANCI. In contrast, the localization of FANCI to chromatin is FANCD2-dependent.Citation74
Finally, recent studies from the D’Andrea laboratory have established that FANCD2 promotes RAS oncogene-induced senescence in part by promoting the transcriptional activation of the TAp63 tumor suppressor gene.Citation166 Monoubiquitinated FANCD2 and FANCP/SLX4 bind to the TAp63 promoter and cooperate in the transcriptional activation of TAp63.Citation166 Genome-wide ChIP sequencing (ChIP-seq) analysis revealed that following exposure to DNA damaging agents, FANCD2-Ub binds to cis-regulatory sequences of numerous genes essential for epithelial development and morphogenesis.Citation166 This newly discovered function for FANCD2 in transcriptional regulation is supported by other studies describing the binding of FANCD2 to the TNFα gene as well as the antioxidant genes GPX1 and TXNRD1, and constitutes a novel dimension to the portfolio of FANCD2 functions.Citation167,168
Conclusions
The FANCD2 and FANCI proteins function at a pivotal point in the FA-BRCA pathway and play an essential role in maintaining chromosome stability. Both proteins are critical for haematopoietic stem cell homeostasis and the prevention of cellular transformation. These key functions are achieved through what could be considered disparate means: the promotion of error-free HR repair through the recruitment of structure-specific endonucleases, replisome surveillance and resolution, nucleosome remodeling, as well as transcriptional regulation. However, these functions are not necessarily mutually exclusive and it is conceivable that FANCD2 and FANCI facilitate these processes through the recruitment of multiple binding partners and/or via a fundamental chromatin remodeling capacity. Many outstanding questions remain to be addressed, including 1) the identification and characterization of structural and functional domains of FANCD2 and FANCI, 2) cataloging the full spectrum and functional significance of additional FANCD2 and FANCI posttranslational modifications, 3) identification of the FANCI catalytic ubiquitin ligase machinery, 4) determination of the function of monoubiquitinated FANCI (and FANCD2), and 5) elucidation of the roles of FANCB and FAAP100 in the regulation of FANCL activity. Much work remains to be done to clearly elucidate the structural and regulatory mechanisms of these functions. The achievement of these goals will no doubt greatly inform our understanding of the molecular etiology of FA and offer much needed direction in the urgent quest for potential therapeutics.
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
References
- Timmers C, Taniguchi T, Hejna J, Reifsteck C, Lucas L, Bruun D, Thayer M, Cox B, Olson S, D'Andrea AD, et al. Positional cloning of a novel Fanconi anemia gene, FANCD2. Mol Cell 2001; 7:241-8; PMID:11239453; http://dx.doi.org/10.1016/S1097-2765(01)00172-1
- Hejna JA, Timmers CD, Reifsteck C, Bruun DA, Lucas LW, Jakobs PM, Toth-Fejel S, Unsworth N, Clemens SL, Garcia DK, et al. Localization of the Fanconi anemia complementation group D gene to a 200-kb region on chromosome 3p25.3. Am J Hum Genet 2000; 66:1540-51; PMID:10762542; http://dx.doi.org/10.1086/302896
- Garcia-Higuera I, Taniguchi T, Ganesan S, Meyn MS, Timmers C, Hejna J, Grompe M, D'Andrea AD. Interaction of the Fanconi anemia proteins and BRCA1 in a common pathway. Mol Cell 2001; 7:249-62; PMID:11239454; http://dx.doi.org/10.1016/S1097-2765(01)00173-3
- Whitney M, Thayer M, Reifsteck C, Olson S, Smith L, Jakobs PM, Leach R, Naylor S, Joenje H, Grompe M. Microcell mediated chromosome transfer maps the Fanconi anaemia group D gene to chromosome 3p. Nat Genet 1995; 11:341-3; PMID:7581463; http://dx.doi.org/10.1038/ng1195-341
- Jakobs PM, Fiddler-Odell E, Reifsteck C, Olson S, Moses RE, Grompe M. Complementation group assignments in Fanconi anemia fibroblast cell lines from North America. Somat Cell Mol Genet 1997; 23:1-7; PMID:9217996; http://dx.doi.org/10.1007/BF02679950
- Jakobs PM, Sahaayaruban P, Saito H, Reifsteck C, Olson S, Joenje H, Moses RE, Grompe M. Immortalization of four new Fanconi anemia fibroblast cell lines by an improved procedure. Somat Cell Mol Genet 1996; 22:151-7; PMID:8782494; http://dx.doi.org/10.1007/BF02369905
- Levitus M, Rooimans MA, Steltenpool J, Cool NF, Oostra AB, Mathew CG, Hoatlin ME, Waisfisz Q, Arwert F, de Winter JP, et al. Heterogeneity in Fanconi anemia: evidence for 2 new genetic subtypes. Blood 2004; 103:2498-503; PMID:14630800; http://dx.doi.org/10.1182/blood-2003-08-2915
- Dorsman JC, Levitus M, Rockx D, Rooimans MA, Oostra AB, Haitjema A, Bakker ST, Steltenpool J, Schuler D, Mohan S, et al. Identification of the Fanconi anemia complementation group I gene, FANCI. Cell Oncol 2007; 29:211-8; PMID:17452773
- Sims AE, Spiteri E, Sims RJ, 3rd, Arita AG, Lach FP, Landers T, Wurm M, Freund M, Neveling K, Hanenberg H, et al. FANCI is a second monoubiquitinated member of the Fanconi anemia pathway. Nat Struct Mol Biol 2007; 14:564-7; PMID:17460694; http://dx.doi.org/10.1038/nsmb1252
- Smogorzewska A, Matsuoka S, Vinciguerra P, McDonald ER, 3rd, Hurov KE, Luo J, Ballif BA, Gygi SP, Hofmann K, D'Andrea AD, et al. Identification of the FANCI protein, a monoubiquitinated FANCD2 paralog required for DNA repair. Cell 2007; 129:289-301; PMID:17412408; http://dx.doi.org/10.1016/j.cell.2007.03.009
- Kupfer GM, Naf D, Suliman A, Pulsipher M, D'Andrea AD. The Fanconi anaemia proteins, FAA and FAC, interact to form a nuclear complex. Nat Genet 1997; 17:487-90; PMID:9398857; http://dx.doi.org/10.1038/ng1297-487
- Garcia-Higuera I, Kuang Y, Naf D, Wasik J, D'Andrea AD. Fanconi anemia proteins FANCA, FANCC, and FANCG/XRCC9 interact in a functional nuclear complex. Mol Cell Biol 1999; 19:4866-73; PMID:10373536
- de Winter JP, van der Weel L, de Groot J, Stone S, Waisfisz Q, Arwert F, Scheper RJ, Kruyt FA, Hoatlin ME, Joenje H. The Fanconi anemia protein FANCF forms a nuclear complex with FANCA, FANCC and FANCG. Hum Mol Genet 2000; 9:2665-74; PMID:11063725; http://dx.doi.org/10.1093/hmg/9.18.2665
- Waisfisz Q, de Winter JP, Kruyt FA, de Groot J, van der Weel L, Dijkmans LM, Zhi Y, Arwert F, Scheper RJ, Youssoufian H, et al. A physical complex of the Fanconi anemia proteins FANCG/XRCC9 and FANCA. Proc Nat Acad Sci U S A 1999; 96:10320-5; PMID:10468606; http://dx.doi.org/10.1073/pnas.96.18.10320
- Matsuoka S, Ballif BA, Smogorzewska A, McDonald ER, 3rd, Hurov KE, Luo J, Bakalarski CE, Zhao Z, Solimini N, Lerenthal Y, et al. ATM and ATR substrate analysis reveals extensive protein networks responsive to DNA damage. Science 2007; 316:1160-6; PMID:17525332; http://dx.doi.org/10.1126/science.1140321
- Swift M. Fanconi's anaemia in the genetics of neoplasia. Nature 1971; 230:370-3; PMID:4927726; http://dx.doi.org/10.1038/230370a0
- Rosenberg PS, Tamary H, Alter BP. How high are carrier frequencies of rare recessive syndromes? Contemporary estimates for Fanconi Anemia in the United States and Israel. Am J Med Genet A 2011; 155A:1877-83; PMID:21739583; http://dx.doi.org/10.1002/ajmg.a.34087
- Tischkowitz M, Dokal I. Fanconi anaemia and leukaemia - clinical and molecular aspects. Br J Haematol 2004; 126:176-91; PMID:15238138; http://dx.doi.org/10.1111/j.1365-2141.2004.05023.x
- Taniguchi T, D'Andrea AD. Molecular pathogenesis of Fanconi anemia: recent progress. Blood 2006; 107:4223-33; PMID:16493006; http://dx.doi.org/10.1182/blood-2005-10-4240
- Kalb R, Neveling K, Hoehn H, Schneider H, Linka Y, Batish SD, Hunt C, Berwick M, Callen E, Surralles J, et al. Hypomorphic mutations in the gene encoding a key Fanconi anemia protein, FANCD2, sustain a significant group of FA-D2 patients with severe phenotype. Am J Hum Genet 2007; 80:895-910; PMID:17436244; http://dx.doi.org/10.1086/517616
- De Kerviler E, Guermazi A, Zagdanski AM, Gluckman E, Frija J. The clinical and radiological features of Fanconi's anaemia. Clin Radiol 2000; 55:340-5; PMID:10816398; http://dx.doi.org/10.1053/crad.2000.0445
- Trivin C, Gluckman E, Leblanc T, Cousin MN, Soulier J, Brauner R. Factors and markers of growth hormone secretion and gonadal function in Fanconi anemia. Growth Horm IGF Res: Off J Growth Horm Res Soc Int IGF Res Soc 2007; 17:122-9; PMID:17336561; http://dx.doi.org/10.1016/j.ghir.2006.12.007
- Houghtaling S, Timmers C, Noll M, Finegold MJ, Jones SN, Meyn MS, Grompe M. Epithelial cancer in Fanconi anemia complementation group D2 (Fancd2) knockout mice. Genes Dev 2003; 17:2021-35; PMID:12893777; http://dx.doi.org/10.1101/gad.1103403
- Scheckenbach K, Morgan M, Filger-Brillinger J, Sandmann M, Strimling B, Scheurlen W, Schindler D, Gobel U, Hanenberg H. Treatment of the bone marrow failure in Fanconi anemia patients with danazol. Blood Cells Mol Dis 2012; 48:128-31; PMID:22178060; http://dx.doi.org/10.1016/j.bcmd.2011.11.006
- Montes de Oca R, Andreassen PR, Margossian SP, Gregory RC, Taniguchi T, Wang X, Houghtaling S, Grompe M, D'Andrea AD. Regulated interaction of the Fanconi anemia protein, FANCD2, with chromatin. Blood 2005; 105:1003-9; PMID:15454491; http://dx.doi.org/10.1182/blood-2003-11-3997
- Howlett NG, Harney JA, Rego MA, Kolling FWt, Glover TW. Functional interaction between the Fanconi Anemia D2 protein and proliferating cell nuclear antigen (PCNA) via a conserved putative PCNA interaction motif. J Biol Chem 2009; 284:28935-42; PMID:19704162; http://dx.doi.org/10.1074/jbc.M109.016352
- Rego MA, Kolling FWt, Vuono EA, Mauro M, Howlett NG. Regulation of the Fanconi anemia pathway by a CUE ubiquitin-binding domain in the FANCD2 protein. Blood 2012; 120:2109-17; PMID:22855611; http://dx.doi.org/10.1182/blood-2012-02-410472
- Boisvert RA, Rego MA, Azzinaro PA, Mauro M, Howlett NG. Coordinate Nuclear Targeting of the FANCD2 and FANCI Proteins via a FANCD2 Nuclear Localization Signal. PLoS One 2013; 8:e81387; http://dx.doi.org/10.1371/journal.pone.0081387
- Xu W, Kimelman D. Mechanistic insights from structural studies of beta-catenin and its binding partners. J Cell Sci 2007; 120:3337-44; PMID:17881495; http://dx.doi.org/10.1242/jcs.013771
- Landschulz WH, Johnson PF, McKnight SL. The leucine zipper: a hypothetical structure common to a new class of DNA binding proteins. Science 1988; 240:1759-64; PMID:3289117; http://dx.doi.org/10.1126/science.3289117
- Yuan F, El Hokayem J, Zhou W, Zhang Y. FANCI protein binds to DNA and interacts with FANCD2 to recognize branched structures. The J Biol Chem 2009; 284:24443-52; PMID:19561358; http://dx.doi.org/10.1074/jbc.M109.016006
- Colnaghi L, Jones MJ, Cotto-Rios XM, Schindler D, Hanenberg H, Huang TT. Patient-derived C-terminal mutation of FANCI causes protein mislocalization and reveals putative EDGE motif function in DNA repair. Blood 2011; 117:2247-56; PMID:20971953; http://dx.doi.org/10.1182/blood-2010-07-295758
- Ishiai M, Kitao H, Smogorzewska A, Tomida J, Kinomura A, Uchida E, Saberi A, Kinoshita E, Kinoshita-Kikuta E, Koike T, et al. FANCI phosphorylation functions as a molecular switch to turn on the Fanconi anemia pathway. Nat Struct Mol Biol 2008; 15:1138-46; PMID:18931676; http://dx.doi.org/10.1038/nsmb.1504
- Kim ST, Lim DS, Canman CE, Kastan MB. Substrate specificities and identification of putative substrates of ATM kinase family members. J Biol Chem 1999; 274:37538-43; PMID:10608806; http://dx.doi.org/10.1074/jbc.274.53.37538
- Joo W, Xu G, Persky NS, Smogorzewska A, Rudge DG, Buzovetsky O, Elledge SJ, Pavletich NP. Structure of the FANCI-FANCD2 complex: insights into the Fanconi anemia DNA repair pathway. Science 2011; 333:312-6; PMID:21764741; http://dx.doi.org/10.1126/science.1205805
- Chau V, Tobias JW, Bachmair A, Marriott D, Ecker DJ, Gonda DK, Varshavsky A. A multiubiquitin chain is confined to specific lysine in a targeted short-lived protein. Science 1989; 243:1576-83; PMID:2538923; http://dx.doi.org/10.1126/science.2538923
- Sigismund S, Polo S, Di Fiore PP. Signaling through monoubiquitination. Curr Top Microbiol Immunol 2004; 286:149-85; PMID:15645713
- Deshaies RJ, Joazeiro CA. RING domain E3 ubiquitin ligases. Annu Rev Biochem 2009; 78:399-434; PMID:19489725; http://dx.doi.org/10.1146/annurev.biochem.78.101807.093809
- Moynahan ME, Chiu JW, Koller BH, Jasin M. Brca1 controls homology-directed DNA repair. Mol Cell 1999; 4:511-8; PMID:10549283; http://dx.doi.org/10.1016/S1097-2765(00)80202-6
- Moynahan ME, Cui TY, Jasin M. Homology-directed dna repair, mitomycin-c resistance, and chromosome stability is restored with correction of a Brca1 mutation. Cancer Res 2001; 61:4842-50; PMID:11406561
- Scully R, Chen J, Plug A, Xiao Y, Weaver D, Feunteun J, Ashley T, Livingston DM. Association of BRCA1 with Rad51 in mitotic and meiotic cells. Cell 1997; 88:265-75; PMID:9008167; http://dx.doi.org/10.1016/S0092-8674(00)81847-4
- Mazon G, Mimitou EP, Symington LS. SnapShot: homologous recombination in DNA double-strand break repair. Cell 2010; 142:646, e1; PMID:20723763; http://dx.doi.org/10.1016/j.cell.2010.08.006
- Pierce AJ, Johnson RD, Thompson LH, Jasin M. XRCC3 promotes homology-directed repair of DNA damage in mammalian cells. Genes Dev 1999; 13:2633-8; PMID:10541549; http://dx.doi.org/10.1101/gad.13.20.2633
- Nakanishi K, Yang YG, Pierce AJ, Taniguchi T, Digweed M, D'Andrea AD, Wang ZQ, Jasin M. Human Fanconi anemia monoubiquitination pathway promotes homologous DNA repair. Proc Nat Acad Sci U S A 2005; 102:1110-5; PMID:15650050; http://dx.doi.org/10.1073/pnas.0407796102
- Longerich S, Kwon Y, Tsai MS, Hlaing AS, Kupfer GM, Sung P. Regulation of FANCD2 and FANCI monoubiquitination by their interaction and by DNA. Nucleic Acids Res 2014; 42:5657-70; PMID:24623813; http://dx.doi.org/10.1093/nar/gku198
- Machida YJ, Machida Y, Chen Y, Gurtan AM, Kupfer GM, D'Andrea AD, Dutta A. UBE2T is the E2 in the Fanconi anemia pathway and undergoes negative autoregulation. Mol Cell 2006; 23:589-96; PMID:16916645; http://dx.doi.org/10.1016/j.molcel.2006.06.024
- Meetei AR, Sechi S, Wallisch M, Yang D, Young MK, Joenje H, Hoatlin ME, Wang W. A multiprotein nuclear complex connects Fanconi anemia and Bloom syndrome. Mol Cell Biol 2003; 23:3417-26; PMID:12724401; http://dx.doi.org/10.1128/MCB.23.10.3417-3426.2003
- Ali AM, Pradhan A, Singh TR, Du C, Li J, Wahengbam K, Grassman E, Auerbach AD, Pang Q, Meetei AR. FAAP20: a novel ubiquitin-binding FA nuclear core-complex protein required for functional integrity of the FA-BRCA DNA repair pathway. Blood 2012; 119:3285-94; PMID:22343915; http://dx.doi.org/10.1182/blood-2011-10-385963
- Ciccia A, Ling C, Coulthard R, Yan Z, Xue Y, Meetei AR, Laghmani el H, Joenje H, McDonald N, de Winter JP, et al. Identification of FAAP24, a Fanconi anemia core complex protein that interacts with FANCM. Mol Cell 2007; 25:331-43; PMID:17289582; http://dx.doi.org/10.1016/j.molcel.2007.01.003
- Kim H, D'Andrea AD. Regulation of DNA cross-link repair by the Fanconi anemia/BRCA pathway. Genes Dev 2012; 26:1393-408; PMID:22751496; http://dx.doi.org/10.1101/gad.195248.112
- Kim Y, Spitz GS, Veturi U, Lach FP, Auerbach AD, Smogorzewska A. Regulation of multiple DNA repair pathways by the Fanconi anemia protein SLX4. Blood 2013; 121:54-63.
- Ling C, Ishiai M, Ali AM, Medhurst AL, Neveling K, Kalb R, Yan Z, Xue Y, Oostra AB, Auerbach AD, et al. FAAP100 is essential for activation of the Fanconi anemia-associated DNA damage response pathway. EMBO J 2007; 26:2104-14; PMID:17396147; http://dx.doi.org/10.1038/sj.emboj.7601666
- Singh TR, Saro D, Ali AM, Zheng XF, Du CH, Killen MW, Sachpatzidis A, Wahengbam K, Pierce AJ, Xiong Y, et al. MHF1-MHF2, a histone-fold-containing protein complex, participates in the Fanconi anemia pathway via FANCM. Mol Cell 2010; 37:879-86; PMID:20347429; http://dx.doi.org/10.1016/j.molcel.2010.01.036
- Alpi AF, Pace PE, Babu MM, Patel KJ. Mechanistic insight into site-restricted monoubiquitination of FANCD2 by Ube2t, FANCL, and FANCI. Mol Cell 2008; 32:767-77; PMID:19111657; http://dx.doi.org/10.1016/j.molcel.2008.12.003
- Longerich S, San Filippo J, Liu D, Sung P. FANCI binds branched DNA and is monoubiquitinated by UBE2T-FANCL. J Biol Chem 2009; 284:23182-6; PMID:19589784; http://dx.doi.org/10.1074/jbc.C109.038075
- Sato K, Toda K, Ishiai M, Takata M, Kurumizaka H. DNA robustly stimulates FANCD2 monoubiquitylation in the complex with FANCI. Nucleic Acids Res 2012; 40:4553-61; PMID:22287633; http://dx.doi.org/10.1093/nar/gks053
- Huang Y, Leung JW, Lowery M, Matsushita N, Wang Y, Shen X, Huong D, Takata M, Chen J, Li L. Modularized functions of the fanconi anemia core complex. Cell Rep 2014; 7:1849-57; PMID:24910428; http://dx.doi.org/10.1016/j.celrep.2014.04.029
- Rajendra E, Oestergaard VH, Langevin F, Wang M, Dornan GL, Patel KJ, Passmore LA. The genetic and biochemical basis of FANCD2 monoubiquitination. Mol Cell 2014; 54:858-69; PMID:24905007; http://dx.doi.org/10.1016/j.molcel.2014.05.001
- Skowyra D, Craig KL, Tyers M, Elledge SJ, Harper JW. F-box proteins are receptors that recruit phosphorylated substrates to the SCF ubiquitin-ligase complex. Cell 1997; 91:209-19; PMID:9346238; http://dx.doi.org/10.1016/S0092-8674(00)80403-1
- Vodermaier HC. APC/C and SCF: controlling each other and the cell cycle. Curr Biol 2004; 14:R787-96; PMID:15380093; http://dx.doi.org/10.1016/j.cub.2004.09.020
- Pace P, Johnson M, Tan WM, Mosedale G, Sng C, Hoatlin M, de Winter J, Joenje H, Gergely F, Patel KJ. FANCE: the link between Fanconi anaemia complex assembly and activity. EMBO J 2002; 21:3414-23; PMID:12093742; http://dx.doi.org/10.1093/emboj/cdf355
- Polito D, Cukras S, Wang X, Spence P, Moreau L, D'Andrea AD, Kee Y. The carboxy terminus of FANCE recruits FANCD2 to the FA E3 ligase complex to promote the Fanconi Anemia DNA repair pathway. J Biol Chem 2014; PMID:24451376
- Garcia-Higuera I, Kuang Y, Denham J, D'Andrea AD. The fanconi anemia proteins FANCA and FANCG stabilize each other and promote the nuclear accumulation of the Fanconi anemia complex. Blood 2000; 96:3224-30; PMID:11050007
- Garcia-Higuera I, D'Andrea AD. Regulated binding of the Fanconi anemia proteins, FANCA and FANCC. Blood 1999; 93:1430-2; PMID:10075454
- Cardozo T, Pagano M. The SCF ubiquitin ligase: insights into a molecular machine. Nat Rev Mol Cell Biol 2004; 5:739-51; PMID:15340381; http://dx.doi.org/10.1038/nrm1471
- Gari K, Decaillet C, Stasiak AZ, Stasiak A, Constantinou A. The Fanconi anemia protein FANCM can promote branch migration of Holliday junctions and replication forks. Mol Cell 2008; 29:141-8; PMID:18206976; http://dx.doi.org/10.1016/j.molcel.2007.11.032
- Kim JM, Kee Y, Gurtan A, D'Andrea AD. Cell cycle-dependent chromatin loading of the Fanconi anemia core complex by FANCM/FAAP24. Blood 2008; 111:5215-22; PMID:18174376; http://dx.doi.org/10.1182/blood-2007-09-113092
- Gari K, Decaillet C, Delannoy M, Wu L, Constantinou A. Remodeling of DNA replication structures by the branch point translocase FANCM. Proc Nat Acad Sci U S A 2008; 105:16107-12; PMID:18843105; http://dx.doi.org/10.1073/pnas.0804777105
- Hodson C, Cole AR, Lewis LP, Miles JA, Purkiss A, Walden H. Structural analysis of human FANCL, the E3 ligase in the Fanconi anemia pathway. J Biol Chem 2011; 286:32628-37; PMID:21775430; http://dx.doi.org/10.1074/jbc.M111.244632
- Cole AR, Lewis LP, Walden H. The structure of the catalytic subunit FANCL of the Fanconi anemia core complex. Nat Struct Mol Biol 2010; 17:294-8; PMID:20154706; http://dx.doi.org/10.1038/nsmb.1759
- Carbone R, Fre S, Iannolo G, Belleudi F, Mancini P, Pelicci PG, Torrisi MR, Di Fiore PP. eps15 and eps15R are essential components of the endocytic pathway. Cancer Res 1997; 57:5498-504; PMID:9407958
- Ramanathan HN, Ye Y. Cellular strategies for making monoubiquitin signals. Crit Rev Biochem Mol Biol 2012; 47:17-28; PMID:21981143; http://dx.doi.org/10.3109/10409238.2011.620943
- Hoeller D, Crosetto N, Blagoev B, Raiborg C, Tikkanen R, Wagner S, Kowanetz K, Breitling R, Mann M, Stenmark H, et al. Regulation of ubiquitin-binding proteins by monoubiquitination. Nat Cell Biol 2006; 8:163-9; PMID:16429130; http://dx.doi.org/10.1038/ncb1354
- Sareen A, Chaudhury I, Adams N, Sobeck A. Fanconi anemia proteins FANCD2 and FANCI exhibit different DNA damage responses during S-phase. Nucleic Acids Res 2012; 40:8425-39; PMID:22753026; http://dx.doi.org/10.1093/nar/gks638
- Staub O, Abriel H, Plant P, Ishikawa T, Kanelis V, Saleki R, Horisberger JD, Schild L, Rotin D. Regulation of the epithelial Na+ channel by Nedd4 and ubiquitination. Kidney Int 2000; 57:809-15; PMID:10720933; http://dx.doi.org/10.1046/j.1523-1755.2000.00919.x
- Lott JS, Coddington-Lawson SJ, Teesdale-Spittle PH, McDonald FJ. A single WW domain is the predominant mediator of the interaction between the human ubiquitin-protein ligase Nedd4 and the human epithelial sodium channel. Biochem J 2002; 361:481-8; PMID:11802777; http://dx.doi.org/10.1042/0264-6021:3610481
- Polo S, Sigismund S, Faretta M, Guidi M, Capua MR, Bossi G, Chen H, De Camilli P, Di Fiore PP. A single motif responsible for ubiquitin recognition and monoubiquitination in endocytic proteins. Nature 2002; 416:451-5; PMID:11919637; http://dx.doi.org/10.1038/416451a
- Di Fiore PP, Polo S, Hofmann K. When ubiquitin meets ubiquitin receptors: a signalling connection. Nat Rev Mol Cell Biol 2003; 4:491-7; PMID:12778128; http://dx.doi.org/10.1038/nrm1124
- Fallon L, Belanger CM, Corera AT, Kontogiannea M, Regan-Klapisz E, Moreau F, Voortman J, Haber M, Rouleau G, Thorarinsdottir T, et al. A regulated interaction with the UIM protein Eps15 implicates parkin in EGF receptor trafficking and PI(3)K-Akt signalling. Nat Cell Biol 2006; 8:834-42; PMID:16862145; http://dx.doi.org/10.1038/ncb1441
- Longerich S, San Filippo J, Liu D, Sung P. FANCI binds branched DNA and is mono-ubiquitinated by UBE2T-FANCL. J Biol Chem 2009; PMID:19589784
- David Y, Ziv T, Admon A, Navon A. The E2 ubiquitin-conjugating enzymes direct polyubiquitination to preferred lysines. J Biol Chem 2010; 285:8595-604; PMID:20061386; http://dx.doi.org/10.1074/jbc.M109.089003
- Kulathu Y, Komander D. Atypical ubiquitylation - the unexplored world of polyubiquitin beyond Lys48 and Lys63 linkages. Nat Rev Mol Cell Biol 2012; 13:508-23; PMID:22820888; http://dx.doi.org/10.1038/nrm3394
- Moraes TF, Edwards RA, McKenna S, Pastushok L, Xiao W, Glover JN, Ellison MJ. Crystal structure of the human ubiquitin conjugating enzyme complex, hMms2-hUbc13. Nat Struct Biol 2001; 8:669-73; PMID:11473255; http://dx.doi.org/10.1038/90373
- Li M, Brooks CL, Wu-Baer F, Chen D, Baer R, Gu W. Mono- versus polyubiquitination: differential control of p53 fate by Mdm2. Science 2003; 302:1972-5; PMID:14671306; http://dx.doi.org/10.1126/science.1091362
- Nijman SM, Huang TT, Dirac AM, Brummelkamp TR, Kerkhoven RM, D'Andrea AD, Bernards R. The deubiquitinating enzyme USP1 regulates the Fanconi anemia pathway. Mol Cell 2005; 17:331-9; PMID:15694335; http://dx.doi.org/10.1016/j.molcel.2005.01.008
- Kim JM, Parmar K, Huang M, Weinstock DM, Ruit CA, Kutok JL, D'Andrea AD. Inactivation of murine Usp1 results in genomic instability and a Fanconi anemia phenotype. Dev Cell 2009; 16:314-20; PMID:19217432; http://dx.doi.org/10.1016/j.devcel.2009.01.001
- Cohn MA, Kowal P, Yang K, Haas W, Huang TT, Gygi SP, D'Andrea AD. A UAF1-containing multisubunit protein complex regulates the Fanconi anemia pathway. Mol Cell 2007; 28:786-97; PMID:18082604; http://dx.doi.org/10.1016/j.molcel.2007.09.031
- Yang K, Moldovan GL, Vinciguerra P, Murai J, Takeda S, D'Andrea AD. Regulation of the Fanconi anemia pathway by a SUMO-like delivery network. Genes Dev 2011; 25:1847-58; PMID:21896657; http://dx.doi.org/10.1101/gad.17020911
- Huang TT, Nijman SM, Mirchandani KD, Galardy PJ, Cohn MA, Haas W, Gygi SP, Ploegh HL, Bernards R, D'Andrea AD. Regulation of monoubiquitinated PCNA by DUB autocleavage. Nat Cell Biol 2006; 8:339-47; PMID:16531995
- Piatkov KI, Colnaghi L, Bekes M, Varshavsky A, Huang TT. The auto-generated fragment of the usp1 deubiquitylase is a physiological substrate of the N-end rule pathway. Mol Cell 2012; 48:926-33; PMID:23159736; http://dx.doi.org/10.1016/j.molcel.2012.10.012
- Rego MA, Harney JA, Mauro M, Shen M, Howlett NG. Regulation of the activation of the Fanconi anemia pathway by the p21 cyclin-dependent kinase inhibitor. Oncogene 2012; 31:366-75; PMID:21685936; http://dx.doi.org/10.1038/onc.2011.237
- Savitsky K, Bar-Shira A, Gilad S, Rotman G, Ziv Y, Vanagaite L, Tagle DA, Smith S, Uziel T, Sfez S, et al. A single ataxia telangiectasia gene with a product similar to PI-3 kinase. Science 1995; 268:1749-53; PMID:7792600; http://dx.doi.org/10.1126/science.7792600
- Chun HH, Gatti RA. Ataxia-telangiectasia, an evolving phenotype. DNA Repair (Amst) 2004; 3:1187-96; PMID:15279807; http://dx.doi.org/10.1016/j.dnarep.2004.04.010
- Taylor AM, Metcalfe JA, Thick J, Mak YF. Leukemia and lymphoma in ataxia telangiectasia. Blood 1996; 87:423-38; PMID:8555463
- Matsuoka S, Rotman G, Ogawa A, Shiloh Y, Tamai K, Elledge SJ. Ataxia telangiectasia-mutated phosphorylates Chk2 in vivo and in vitro. Proc Natl Acad Sci U S A 2000; 97:10389-94; PMID:10973490; http://dx.doi.org/10.1073/pnas.190030497
- Taniguchi T, Garcia-Higuera I, Xu B, Andreassen PR, Gregory RC, Kim ST, Lane WS, Kastan MB, D'Andrea AD. Convergence of the fanconi anemia and ataxia telangiectasia signaling pathways. Cell 2002; 109:459-72; PMID:12086603; http://dx.doi.org/10.1016/S0092-8674(02)00747-X
- Taniguchi T, Garcia-Higuera I, Andreassen PR, Gregory RC, Grompe M, D'Andrea AD. S-phase-specific interaction of the Fanconi anemia protein, FANCD2, with BRCA1 and RAD51. Blood 2002; 100:2414-20; PMID:12239151; http://dx.doi.org/10.1182/blood-2002-01-0278
- Andreassen PR, D'Andrea AD, Taniguchi T. ATR couples FANCD2 monoubiquitination to the DNA-damage response. Genes Dev 2004; 18:1958-63; PMID:15314022; http://dx.doi.org/10.1101/gad.1196104
- Ho GP, Margossian S, Taniguchi T, D'Andrea AD. Phosphorylation of FANCD2 on two novel sites is required for mitomycin C resistance. Mol Cell Biol 2006; 26:7005-15; PMID:16943440; http://dx.doi.org/10.1128/MCB.02018-05
- Cimprich KA, Cortez D. ATR: an essential regulator of genome integrity. Nat Rev Mol Cell Biol 2008; 9:616-27; PMID:18594563; http://dx.doi.org/10.1038/nrm2450
- Paulsen RD, Cimprich KA. The ATR pathway: fine-tuning the fork. DNA Repair (Amst) 2007; 6:953-66; PMID:17531546; http://dx.doi.org/10.1016/j.dnarep.2007.02.015
- Goodship J, Gill H, Carter J, Jackson A, Splitt M, Wright M. Autozygosity mapping of a seckel syndrome locus to chromosome 3q22. 1-q24. Am J Hum Genet 2000; 67:498-503; PMID:10889046; http://dx.doi.org/10.1086/303023
- Hayani A, Suarez CR, Molnar Z, LeBeau M, Godwin J. Acute myeloid leukaemia in a patient with Seckel syndrome. J Med Genet 1994; 31:148-9; PMID:8182723; http://dx.doi.org/10.1136/jmg.31.2.148
- O'Driscoll M, Ruiz-Perez VL, Woods CG, Jeggo PA, Goodship JA. A splicing mutation affecting expression of ataxia-telangiectasia and Rad3-related protein (ATR) results in Seckel syndrome. Nat Genet 2003; 33:497-501; PMID:12640452; http://dx.doi.org/10.1038/ng1129
- Pichierri P, Rosselli F. The DNA crosslink-induced S-phase checkpoint depends on ATR-CHK1 and ATR-NBS1-FANCD2 pathways. EMBO J 2004; 23:1178-87; PMID:14988723; http://dx.doi.org/10.1038/sj.emboj.7600113
- Sareen A, Chaudhury I, Adams N, Sobeck A. Fanconi anemia proteins FANCD2 and FANCI exhibit different DNA damage responses during S-phase. Nucleic Acids Res 2012; 40:8425-39; PMID:22753026
- Howlett NG, Taniguchi T, Durkin SG, D'Andrea AD, Glover TW. The Fanconi anemia pathway is required for the DNA replication stress response and for the regulation of common fragile site stability. Hum Mol Genet 2005; 14:693-701; PMID:15661754; http://dx.doi.org/10.1093/hmg/ddi065
- Glover TW, Berger C, Coyle J, Echo B. DNA polymerase alpha inhibition by aphidicolin induces gaps and breaks at common fragile sites in human chromosomes. Human genetics 1984; 67:136-42; PMID:6430783; http://dx.doi.org/10.1007/BF00272988
- Arlt MF, Durkin SG, Ragland RL, Glover TW. Common fragile sites as targets for chromosome rearrangements. DNA Repair (Amst) 2006; 5:1126-35; PMID:16807141; http://dx.doi.org/10.1016/j.dnarep.2006.05.010
- Chan KL, Palmai-Pallag T, Ying S, Hickson ID. Replication stress induces sister-chromatid bridging at fragile site loci in mitosis. Nat Cell Biol 2009; 11:753-60; PMID:19465922; http://dx.doi.org/10.1038/ncb1882
- Naim V, Rosselli F. The FANC pathway and BLM collaborate during mitosis to prevent micro-nucleation and chromosome abnormalities. Nat Cell Biol 2009; 11:761-8; PMID:19465921; http://dx.doi.org/10.1038/ncb1883
- Sirbu BM, McDonald WH, Dungrawala H, Badu-Nkansah A, Kavanaugh GM, Chen Y, Tabb DL, Cortez D. Identification of proteins at active, stalled, and collapsed replication forks using isolation of proteins on nascent DNA (iPOND) coupled with mass spectrometry. J Biol Chem 2013; 288:31458-67; PMID:24047897; http://dx.doi.org/10.1074/jbc.M113.511337
- Song IY, Barkley LR, Day TA, Weiss RS, Vaziri C. A novel role for Fanconi anemia (FA) pathway effector protein FANCD2 in cell cycle progression of untransformed primary human cells. Cell Cycle 2010; 9:2375-88; PMID:20519958; http://dx.doi.org/10.4161/cc.9.12.11900
- Cortez D, Glick G, Elledge SJ. Minichromosome maintenance proteins are direct targets of the ATM and ATR checkpoint kinases. Proc Nat Acad Sci U S A 2004; 101:10078-83; PMID:15210935; http://dx.doi.org/10.1073/pnas.0403410101
- Lossaint G, Larroque M, Ribeyre C, Bec N, Larroque C, Decaillet C, Gari K, Constantinou A. FANCD2 binds MCM proteins and controls replisome function upon activation of s phase checkpoint signaling. Mol Cell 2013; 51:678-90; PMID:23993743; http://dx.doi.org/10.1016/j.molcel.2013.07.023
- Schlacher K, Wu H, Jasin M. A distinct replication fork protection pathway connects Fanconi anemia tumor suppressors to RAD51-BRCA1/2. Cancer Cell 2012; 22:106-16; PMID:22789542; http://dx.doi.org/10.1016/j.ccr.2012.05.015
- Yeo JE, Lee EH, Hendrickson E, Sobeck A. CtIP mediates replication fork recovery in a FANCD2-regulated manner. Hum Mol Genet 2014; 23:3695-705; PMID:24556218; http://dx.doi.org/10.1093/hmg/ddu078
- Schaeper U, Subramanian T, Lim L, Boyd JM, Chinnadurai G. Interaction between a cellular protein that binds to the C-terminal region of adenovirus E1A (CtBP) and a novel cellular protein is disrupted by E1A through a conserved PLDLS motif. J Biol Chem 1998; 273:8549-52; PMID:9535825; http://dx.doi.org/10.1074/jbc.273.15.8549
- Wong AK, Ormonde PA, Pero R, Chen Y, Lian L, Salada G, Berry S, Lawrence Q, Dayananth P, Ha P, et al. Characterization of a carboxy-terminal BRCA1 interacting protein. Oncogene 1998; 17:2279-85; PMID:9811458; http://dx.doi.org/10.1038/sj.onc.1202150
- You Z, Bailis JM. DNA damage and decisions: CtIP coordinates DNA repair and cell cycle checkpoints. Trends Cell Biol 2010; 20:402-9; PMID:20444606; http://dx.doi.org/10.1016/j.tcb.2010.04.002
- Escribano-Diaz C, Orthwein A, Fradet-Turcotte A, Xing M, Young JT, Tkac J, Cook MA, Rosebrock AP, Munro M, Canny MD, et al. A cell cycle-dependent regulatory circuit composed of 53BP1-RIF1 and BRCA1-CtIP controls DNA repair pathway choice. Mol Cell 2013; 49:872-83; PMID:23333306; http://dx.doi.org/10.1016/j.molcel.2013.01.001
- Geng L, Huntoon CJ, Karnitz LM. RAD18-mediated ubiquitination of PCNA activates the Fanconi anemia DNA repair network. J Cell Biol 2010; 191:249-57; PMID:20937699; http://dx.doi.org/10.1083/jcb.201005101
- Palle K, Vaziri C. Rad18 E3 ubiquitin ligase activity mediates Fanconi anemia pathway activation and cell survival following DNA Topoisomerase 1 inhibition. Cell Cycle 2011; 10:1625-38; PMID:21478670; http://dx.doi.org/10.4161/cc.10.10.15617
- Park HK, Wang H, Zhang J, Datta S, Fei P. Convergence of Rad6/Rad18 and Fanconi anemia tumor suppressor pathways upon DNA damage. PloS One 2010; 5:e13313; PMID:20967207; http://dx.doi.org/10.1371/journal.pone.0013313
- Song IY, Palle K, Gurkar A, Tateishi S, Kupfer GM, Vaziri C. Rad18-mediated translesion synthesis of bulky DNA adducts is coupled to activation of the Fanconi anemia DNA repair pathway. J Biol Chem 2010; 285:31525-36; PMID:20675655; http://dx.doi.org/10.1074/jbc.M110.138206
- Williams SA, Longerich S, Sung P, Vaziri C, Kupfer GM. The E3 ubiquitin ligase RAD18 regulates ubiquitylation and chromatin loading of FANCD2 and FANCI. Blood 2011; 117:5078-87; PMID:21355096; http://dx.doi.org/10.1182/blood-2010-10-311761
- Hoege C, Pfander B, Moldovan GL, Pyrowolakis G, Jentsch S. RAD6-dependent DNA repair is linked to modification of PCNA by ubiquitin and SUMO. Nature 2002; 419:135-41; PMID:12226657; http://dx.doi.org/10.1038/nature00991
- Kannouche PL, Wing J, Lehmann AR. Interaction of human DNA polymerase eta with monoubiquitinated PCNA: a possible mechanism for the polymerase switch in response to DNA damage. Mol Cell 2004; 14:491-500; PMID:15149598; http://dx.doi.org/10.1016/S1097-2765(04)00259-X
- Sale JE. Translesion DNA synthesis and mutagenesis in eukaryotes. Cold Spring Harb Perspect Biol 2013; 5:a012708; PMID:23457261; http://dx.doi.org/10.1101/cshperspect.a012708
- Chaudhury I, Sareen A, Raghunandan M, Sobeck A. FANCD2 regulates BLM complex functions independently of FANCI to promote replication fork recovery. Nucleic Acids Res 2013; 41:6444-59; PMID:23658231; http://dx.doi.org/10.1093/nar/gkt348
- Amor-Gueret M. Bloom syndrome, genomic instability and cancer: the SOS-like hypothesis. Cancer Lett 2006; 236:1-12; PMID:15950375; http://dx.doi.org/10.1016/j.canlet.2005.04.023
- Cheok CF, Bachrati CZ, Chan KL, Ralf C, Wu L, Hickson ID. Roles of the Bloom's syndrome helicase in the maintenance of genome stability. Biochem Soc Trans 2005; 33:1456-9; PMID:16246145; http://dx.doi.org/10.1042/BST20051456
- Hemphill AW, Akkari Y, Newell AH, Schultz RA, Grompe M, North PS, Hickson ID, Jakobs PM, Rennie S, Pauw D, et al. Topo IIIalpha and BLM act within the Fanconi anemia pathway in response to DNA-crosslinking agents. Cytogenet Genome Res 2009; 125:165-75; PMID:19738377; http://dx.doi.org/10.1159/000230001
- Pichierri P, Franchitto A, Rosselli F. BLM and the FANC proteins collaborate in a common pathway in response to stalled replication forks. EMBO J 2004; 23:3154-63; PMID:15257300; http://dx.doi.org/10.1038/sj.emboj.7600277
- Sato K, Ishiai M, Toda K, Furukoshi S, Osakabe A, Tachiwana H, Takizawa Y, Kagawa W, Kitao H, Dohmae N, et al. Histone chaperone activity of Fanconi anemia proteins, FANCD2 and FANCI, is required for DNA crosslink repair. EMBO J 2012; 31:3524-36; PMID:22828868; http://dx.doi.org/10.1038/emboj.2012.197
- Jin S, Mao H, Schnepp RW, Sykes SM, Silva AC, D'Andrea AD, Hua X. Menin associates with FANCD2, a protein involved in repair of DNA damage. Cancer Res 2003; 63:4204-10; PMID:12874027
- Hejna J, Holtorf M, Hines J, Mathewson L, Hemphill A, Al-Dhalimy M, Olson SB, Moses RE. Tip60 is required for DNA interstrand cross-link repair in the Fanconi anemia pathway. J Biol Chem 2008; 283:9844-51; PMID:18263878; http://dx.doi.org/10.1074/jbc.M709076200
- Doyon Y, Cote J. The highly conserved and multifunctional NuA4 HAT complex. Curr Opin Genet Dev 2004; 14:147-54; PMID:15196461; http://dx.doi.org/10.1016/j.gde.2004.02.009
- Sykes SM, Mellert HS, Holbert MA, Li K, Marmorstein R, Lane WS, McMahon SB. Acetylation of the p53 DNA-binding domain regulates apoptosis induction. Mol Cell 2006; 24:841-51; PMID:17189187; http://dx.doi.org/10.1016/j.molcel.2006.11.026
- Ikura T, Tashiro S, Kakino A, Shima H, Jacob N, Amunugama R, Yoder K, Izumi S, Kuraoka I, Tanaka K, et al. DNA damage-dependent acetylation and ubiquitination of H2AX enhances chromatin dynamics. Mol Cell Biol 2007; 27:7028-40; PMID:17709392; http://dx.doi.org/10.1128/MCB.00579-07
- Sun Y, Jiang X, Chen S, Fernandes N, Price BD. A role for the Tip60 histone acetyltransferase in the acetylation and activation of ATM. Proc Nat Acad Sci U S A 2005; 102:13182-7; PMID:16141325; http://dx.doi.org/10.1073/pnas.0504211102
- Bird AW, Yu DY, Pray-Grant MG, Qiu Q, Harmon KE, Megee PC, Grant PA, Smith MM, Christman MF. Acetylation of histone H4 by Esa1 is required for DNA double-strand break repair. Nature 2002; 419:411-5; PMID:12353039; http://dx.doi.org/10.1038/nature01035
- Hughes CM, Rozenblatt-Rosen O, Milne TA, Copeland TD, Levine SS, Lee JC, Hayes DN, Shanmugam KS, Bhattacharjee A, Biondi CA, et al. Menin associates with a trithorax family histone methyltransferase complex and with the hoxc8 locus. Mol Cell 2004; 13:587-97; PMID:14992727; http://dx.doi.org/10.1016/S1097-2765(04)00081-4
- Kaji H, Canaff L, Goltzman D, Hendy GN. Cell cycle regulation of menin expression. Cancer Res 1999; 59:5097-101; PMID:10537281
- MacKay C, Declais AC, Lundin C, Agostinho A, Deans AJ, MacArtney TJ, Hofmann K, Gartner A, West SC, Helleday T, et al. Identification of KIAA1018/FAN1, a DNA repair nuclease recruited to DNA damage by monoubiquitinated FANCD2. Cell 2010; 142:65-76; PMID:20603015; http://dx.doi.org/10.1016/j.cell.2010.06.021
- Kratz K, Schopf B, Kaden S, Sendoel A, Eberhard R, Lademann C, Cannavo E, Sartori AA, Hengartner MO, Jiricny J. Deficiency of FANCD2-associated nuclease KIAA1018/FAN1 sensitizes cells to interstrand crosslinking agents. Cell 2010; 142:77-88; PMID:20603016; http://dx.doi.org/10.1016/j.cell.2010.06.022
- Smogorzewska A, Desetty R, Saito TT, Schlabach M, Lach FP, Sowa ME, Clark AB, Kunkel TA, Harper JW, Colaiacovo MP, et al. A genetic screen identifies FAN1, a Fanconi anemia-associated nuclease necessary for DNA interstrand crosslink repair. Mol Cell 2010; 39:36-47; PMID:20603073; http://dx.doi.org/10.1016/j.molcel.2010.06.023
- Liu T, Ghosal G, Yuan J, Chen J, Huang J. FAN1 acts with FANCI-FANCD2 to promote DNA interstrand cross-link repair. Science 2010; 329:693-6; PMID:20671156; http://dx.doi.org/10.1126/science.1192656
- Notenboom V, Hibbert RG, van Rossum-Fikkert SE, Olsen JV, Mann M, Sixma TK. Functional characterization of Rad18 domains for Rad6, ubiquitin, DNA binding and PCNA modification. Nucleic Acids Res 2007; 35:5819-30; PMID:17720710; http://dx.doi.org/10.1093/nar/gkm615
- Guo C, Tang TS, Bienko M, Dikic I, Friedberg EC. Requirements for the interaction of mouse Polkappa with ubiquitin and its biological significance. J Biol Chem 2008; 283:4658-64; PMID:18162470; http://dx.doi.org/10.1074/jbc.M709275200
- Zhou W, Otto EA, Cluckey A, Airik R, Hurd TW, Chaki M, Diaz K, Lach FP, Bennett GR, Gee HY, et al. FAN1 mutations cause karyomegalic interstitial nephritis, linking chronic kidney failure to defective DNA damage repair. Nat Genet 2012; 44:910-5; PMID:22772369; http://dx.doi.org/10.1038/ng.2347
- Kim Y, Lach FP, Desetty R, Hanenberg H, Auerbach AD, Smogorzewska A. Mutations of the SLX4 gene in Fanconi anemia. Nat Genet 2011; 43:142-6; PMID:21240275; http://dx.doi.org/10.1038/ng.750
- Stoepker C, Hain K, Schuster B, Hilhorst-Hofstee Y, Rooimans MA, Steltenpool J, Oostra AB, Eirich K, Korthof ET, Nieuwint AW, et al. SLX4, a coordinator of structure-specific endonucleases, is mutated in a new Fanconi anemia subtype. Nat Genet 2011; 43:138-41; PMID:21240277; http://dx.doi.org/10.1038/ng.751
- Yamamoto KN, Kobayashi S, Tsuda M, Kurumizaka H, Takata M, Kono K, Jiricny J, Takeda S, Hirota K. Involvement of SLX4 in interstrand cross-link repair is regulated by the Fanconi anemia pathway. Proc Natl Acad Sci U S A 2011; 108:6492-6; PMID:21464321; http://dx.doi.org/10.1073/pnas.1018487108
- Hofmann K. Ubiquitin-binding domains and their role in the DNA damage response. DNA Repair (Amst) 2009; 8:544-56; PMID:19213613; http://dx.doi.org/10.1016/j.dnarep.2009.01.003
- Munoz IM, Hain K, Declais AC, Gardiner M, Toh GW, Sanchez-Pulido L, Heuckmann JM, Toth R, Macartney T, Eppink B, et al. Coordination of structure-specific nucleases by human SLX4/BTBD12 is required for DNA repair. Mol Cell 2009; 35:116-27; PMID:19595721; http://dx.doi.org/10.1016/j.molcel.2009.06.020
- Svendsen JM, Smogorzewska A, Sowa ME, O'Connell BC, Gygi SP, Elledge SJ, Harper JW. Mammalian BTBD12/SLX4 assembles a Holliday junction resolvase and is required for DNA repair. Cell 2009; 138:63-77; PMID:19596235; http://dx.doi.org/10.1016/j.cell.2009.06.030
- Hodskinson MR, Silhan J, Crossan GP, Garaycoechea JI, Mukherjee S, Johnson CM, Scharer OD, Patel KJ. Mouse SLX4 Is a Tumor Suppressor that Stimulates the Activity of the Nuclease XPF-ERCC1 in DNA Crosslink Repair. Mol Cell 2014; PMID:24726326
- Kim Y, Spitz GS, Veturi U, Lach FP, Auerbach AD, Smogorzewska A. Regulation of multiple DNA repair pathways by the Fanconi anemia protein SLX4. Blood 2013; 121:54-63; PMID:23093618; http://dx.doi.org/10.1182/blood-2012-07-441212
- Klein Douwel D, Boonen RA, Long DT, Szypowska AA, Raschle M, Walter JC, Knipscheer P. XPF-ERCC1 Acts in Unhooking DNA Interstrand Crosslinks in Cooperation with FANCD2 and FANCP/SLX4. Mol Cell 2014; 54:460-71; PMID:24726325; http://dx.doi.org/10.1016/j.molcel.2014.03.015
- Lachaud C, Castor D, Hain K, Munoz I, Wilson J, MacArtney TJ, Schindler D, Rouse J. Distinct functional roles for the two SLX4 ubiquitin-binding UBZ domains mutated in Fanconi anemia. J Cell Sci 2014; 127:2811-7; PMID:24794496; http://dx.doi.org/10.1242/jcs.146167
- Cybulski KE, Howlett NG. FANCP/SLX4: a Swiss army knife of DNA interstrand crosslink repair. Cell Cycle 2011; 10:1757-63; PMID:21527828; http://dx.doi.org/10.4161/cc.10.11.15818
- Park WH, Margossian S, Horwitz AA, Simons AM, D'Andrea AD, Parvin JD. Direct DNA binding activity of the fanconi anemia D2 protein. J Biol Chem 2005.
- Pace P, Mosedale G, Hodskinson MR, Rosado IV, Sivasubramaniam M, Patel KJ. Ku70 corrupts DNA repair in the absence of the Fanconi anemia pathway. Science 2010; 329:219-23; PMID:20538911; http://dx.doi.org/10.1126/science.1192277
- Sobeck A, Stone S, Hoatlin ME. DNA structure-induced recruitment and activation of the Fanconi anemia pathway protein FANCD2. Mol Cell Biol 2007; 27:4283-92; PMID:17420278; http://dx.doi.org/10.1128/MCB.02196-06
- Park E, Kim H, Kim JM, Primack B, Vidal-Cardenas S, Xu Y, Price BD, Mills AA, D'Andrea AD. FANCD2 activates transcription of TAp63 and suppresses tumorigenesis. Mol Cell 2013; 50:908-18; PMID:23806336; http://dx.doi.org/10.1016/j.molcel.2013.05.017
- Matsushita N, Endo Y, Sato K, Kurumizaka H, Yamashita T, Takata M, Yanagi S. Direct inhibition of TNF-alpha promoter activity by Fanconi anemia protein FANCD2. PloS one 2011; 6:e23324; PMID:21912593; http://dx.doi.org/10.1371/journal.pone.0023324
- Du W, Rani R, Sipple J, Schick J, Myers KC, Mehta P, Andreassen PR, Davies SM, Pang Q. The FA pathway counteracts oxidative stress through selective protection of antioxidant defense gene promoters. Blood 2012; 119:4142-51; PMID:22408259; http://dx.doi.org/10.1182/blood-2011-09-381970
