Abstract
The bZIP transcription factor, C/EBPα is highly inducible by UVB and other DNA damaging agents in keratinocytes. C/EBPα-deficient keratinocytes fail to undergo cell cycle arrest in G1 in response to UVB-induced DNA damage and mice lacking epidermal C/EBPα are highly susceptible to UVB-induced skin cancer. The mechanism through which C/EBPα regulates the cell cycle checkpoint in response to DNA damage is unknown. Here we report untreated C/EBPα-deficient keratinocytes have normal levels of the cyclin-dependent kinase inhibitor, p21, however, UVB-treated C/EBPα-deficient keratinocytes fail to up-regulate nuclear p21 protein levels despite normal up-regulation of Cdkn1a mRNA levels. UVB-treated C/EBPα-deficient keratinocytes displayed a 4-fold decrease in nuclear p21 protein half-life due to the increased proteasomal degradation of p21 via the E3 ubiquitin ligase CRL4Cdt2. Cdt2 is the substrate recognition subunit of CRL4Cdt2 and Cdt2 mRNA and protein levels were up-regulated in UVB-treated C/EBPα-deficient keratinocytes. Knockdown of Cdt2 restored p21 protein levels in UVB-treated C/EBPα-deficient keratinocytes. Lastly, the failure to accumulate p21 in response to UVB in C/EBPα-deficient keratinocytes resulted in decreased p21 interactions with critical cell cycle regulatory proteins, increased CDK2 activity, and inappropriate entry into S-phase. These findings reveal C/EBPα regulates G1/S cell cycle arrest in response to DNA damage via the control of CRL4Cdt2 mediated degradation of p21.
Abbreviations
| C/EBPα | = | CCAAT/enhancer binding protein α |
| UVB | = | ultraviolet B |
| SCF | = | Skp, Cullin, F-box containing complex |
| CRL | = | Cullin-RING ubiquitin ligases |
| DCAF | = | DDB1/CUL4-associated factor; Cdt2, Cdc10 dependent transcript 2 |
Introduction
Nonmelanoma skin cancer (NMSC) is the most common cancer in the United States.Citation1 The majority of NMSC is caused by UVB radiation from sunlight. Exposure to UVB results in DNA damage in the form of; cyclobutane pyrimidine dimers, 6–4 photoproducts, DNA strand breaks, and DNA crosslinks.Citation2,3 Keratinocytes must respond to UVB-induced DNA damage by engaging the DNA damage response which involves cell cycle arrest, DNA repair and apoptosis. If the UVB-induced DNA damage is not repaired or is misrepaired, mutations can result and contribute to the development of skin cancer.Citation4-6 Therefore, the ability of cells to correctly and accurately respond to UVB-induced DNA damage is essential to ensure the integrity of the genome.
The G1/S DNA damage checkpoint is essential to prevent cells with damaged DNA from entering S-phase.Citation7,8 A critical component of the G1/S checkpoint response is the DNA damage-induced accumulation of the cyclin dependent kinase (CDK) inhibitor p21WAF1/CIP1. p21 is highly induced by UVB DNA damage through transcriptional upregulation of Cdkn1a (gene encoding p21) primarily due to the tumor suppressor protein p53.Citation9-12 p21 functions as a negative regulator of G1/S progression and as a promoter of cell cycle arrest induced by DNA damage through two distinct mechanisms; (1) p21 regulates cell cycle progression by binding to and inhibiting the kinase activity of CDKs, kinases required for cell cycle progressionCitation13-16 and (2) p21 binds to proliferating cell nuclear antigen (PCNA) and blocks DNA polymerase progression and DNA replication.Citation17-19 Cells deficient in p21 have a defective G1/S checkpoint and fail to arrest in G1 following UVB exposure.Citation20-22
The basic leucine zipper transcription factor, CCAAT/enhancer binding protein α (C/EBPα) is abundantly expressed in keratinocytes of the skin.Citation23,24 Previously, we reported that UVB radiation is a potent inducer of C/EBPα in human and mouse keratinocytes, as well as in mouse skin in vivo. Moreover, C/EBPα-deficient keratinocytes in culture or in vivo in mouse skin fail to undergo cell cycle arrest in G1 in response to UVB-induced DNA damage, thus allowing cells with damaged DNA to inappropriately enter S-phase.Citation25,26 Consistent with these observations, mice containing a skin specific ablation of C/EBPα are highly susceptible to skin cancer development following chronic low doses of UVB radiation.Citation25
Despite these critical and novel roles for C/EBPα in prevention of UVB-induced skin cancer and the cellular response to DNA damage involving cell cycle arrest, the molecular mechanisms and key players involved downstream of C/EBPα in the DNA damage G1/S checkpoint response remain uncharacterized. Our novel findings reveal C/EBPα regulates G1/S cell cycle arrest in response to DNA damage via the control of CRL4Cdt2 E3 ubiquitin ligase mediated degradation of p21.
Results
C/EBPα is required for UVB-induced up-regulation of p21 protein levels
Initially we examined the protein levels of the cyclin-dependent kinase inhibitor, p21, in untreated C/EBPα-deficient keratinocytes in culture (siRNA knockdown) and in vivo in the epidermis of K5Cretg/+;C/EBPαflox/flox mice (epidermal specific knockout). As shown in , p21 protein levels were normal and not altered by C/EBPα deficiency in cultured keratinocytes or in the epidermis of K5Cretg/+;C/EBPαflox/flox mice. However, UVB-treated C/EBPα-deficient keratinocytes in culture () and in the epidermis of K5Cretg/+;C/EBPαflox/flox mice () failed to up-regulate p21 protein levels despite normal upregulation of Cdkn1a mRNA levels (). The defect in p21 protein accumulation is conserved in UVB-treated human keratinocytes (), also occurs in response to other types of DNA damaging agents such as the DNA alkylating agent MNNG (), and occurs despite normal p53 protein levels and activity (). These data demonstrate that despite the normal transcriptional up-regulation of Cdkn1a in response to DNA damage, C/EBPα-deficient keratinocytes fail to accumulate p21 protein levels indicating that C/EBPα is required for the post-transcriptional regulation of p21 in response to DNA damage.
Figure 1. C/EBPα is required for UVB-induced upregulation of p21 protein levels. (A) Immunoblot analysis of untreated control and C/EBPα siRNA treated Balb/MK2 keratinocytes and K5Cretg/+ and K5Cretg/+;C/EBPαflox/flox epidermal lysates. (B) Immunoblot analysis of lysates from Balb/MK2 keratinocytes treated with C/EBPα siRNA or control siRNA and collected at the indicated times after exposure to 5 mJ/cm2 UVB. (C) IHC staining for p21 in formalin-fixed paraffin-embedded sections of mouse skin from K5Cretg/+ and K5Cretg/+;C/EBPαflox/flox mice 4 h after treatment with 50 mJ/cm2 UVB. (D) Immunoblot analysis of p21, and C/EBPα from lysates isolated from K5Cretg/+ and K5Cretg/+;C/EBPαflox/flox mouse epidermis collected at the indicated times following exposure to 100 mJ/cm2 UVB. (E and F) Relative Cdkn1a (p21) mRNA levels in (E) Balb/MK2 cells treated with C/EBPα siRNA or control siRNA and (F) K5Cre and K5Cre;C/EBPαflox/flox in mouse epidermis collected at the indicated times following UVB. Data are expressed as the mean normalized to Gapdh ±S .D. (N ≥ 3). There were no statistically significant differences between control and C/EBPα-deficient keratinocytes in culture or in vivo as measured and calculated by Student's T test. (G) Immunoblot analysis of lysates of NHEK cells treated with C/EBPα siRNA or control siRNA and collected 10 h post 10 mJ/cm2 UVB. (H) Immunoblot analysis of Balb/MK2 treated with C/EBPα siRNA or control siRNA and collected 6 or 12 h after 25 μM MNNG treatment. (I) Immunoblot analysis of Balb/MK2 treated with C/EBPα siRNA or control siRNA and collected 8 h after exposure to 5 mJ/cm2 UVB.
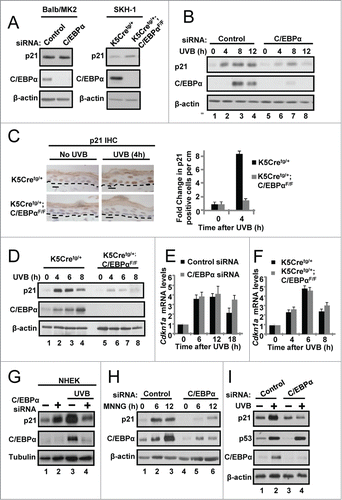
C/EBPα regulates nuclear p21 protein stability following UVB-induced DNA damage
The anti-proliferative function of p21 in the DNA damage-induced G1/S checkpoint is linked to its nuclear localization.Citation34 Therefore, we prepared cytosolic and nuclear protein extracts from UVB-treated control and C/EBPα knockdown keratinocytes to determine whether C/EBPα deficiency preferentially affects nuclear p21 protein levels. The knockdown of C/EBPα had only a modest effect on UVB-induced cytosolic p21 levels, however, nuclear p21 protein levels failed to increase after UVB exposure in C/EBPα-deficient keratinocytes () and this profound effect was sustained up to 20 h post UVB (). To examine the half-life or nuclear p21, we treated control and C/EBPα-deficient keratinocytes with cycloheximide (CHX) to block new protein synthesis and then examined nuclear p21 protein post-CHX and UVB treatment. UVB- and CHX-treated C/EBPα-deficient keratinocytes displayed a ∼4-fold decrease in nuclear p21 stability compared to similarly treated control keratinocytes (). This effect of C/EBPα deficiency and DNA damage on nuclear p21 stability (4-fold decrease in half-life) was confirmed by pulse-chase experiments (Fig. S1A). Importantly, the observed effect of C/EBPα deficiency on decreased nuclear p21 protein stability was completely dependent upon UVB-induced DNA damage as C/EBPα deficiency had no effect on nuclear p21 protein stability in the absence of UVB ().
Figure 2. C/EBPα regulates nuclear p21 protein stability following UVB-induced DNA damage. (A) Immunoblot analysis of cytosolic and nuclear fractions prepared from siRNA treated Balb/MK2 keratinocytes collected 8 h post 5 mJ/cm2 UVB. (B) Immunoblot analysis of nuclear fractions prepared from siRNA treated Balb/MK2 collected at the indicated times post UVB. (C) Immunoblot analysis of p21 and C/EBPα in UVB and CHX treated nuclear extracts. Five h post 5 mJ/cm2 UVB cells were treated with 40 μg/mL CHX (t = 0) and collected at the indicated times. Values below the p21 image represent the fraction of p21 remaining at that time point as measure by densitometry normalized to β-actin. Values are also plotted to the right. (D) Immunoblot analysis of p21 and C/EBPα in CHX treated nuclear extracts. C/EBPα or control siRNA cells were treated with CHX. Values below the p21 image represent the fraction of p21 remaining at that time point as measure by densitometry normalized to β-actin. (E) Immunoblot analysis of p21 co-IP from nuclear extracts 6 h post 5 mJ/cm2 UVB.
A previous study reported that C/EBPα could bind to p21 and stabilize p21.Citation35 To test whether C/EBPα and p21 form a complex in keratinocytes, we immunoprecipitated p21 from nuclear lysates of UVB-treated and untreated keratinocytes. Despite highly efficient immunoprecipitation of p21, we failed to detect any interactions between p21 and C/EBPα () but readily detected p21-CDK2 protein complexes. To further investigate possible p21 and C/EBPα interactions, we immunoprecipitated p21 from nuclear lysates of UVB-treated keratinocytes and conducted LC MS/MS analysis of proteins that co-immunoprecipitated with p21. LC/MS/MS analysis revealed numerous known p21 binding of proteins, however, C/EBPα was not detected (Fig. S1B). Collectively, these data demonstrate that C/EBPα regulates nuclear p21 protein stability following UVB-induced DNA damage independent of its binding to p21.
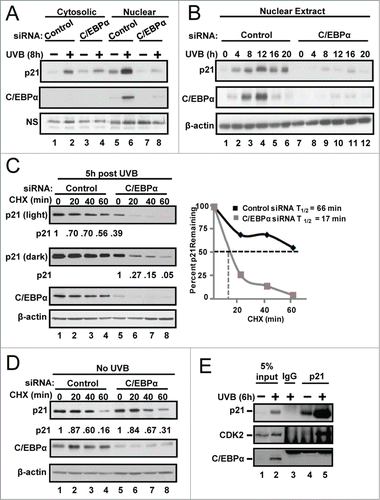
C/EBPα regulates CRL4Cdt2-mediated degradation of p21 in response to UVB
To determine whether the decreased p21 protein stability in UVB-treated C/EBPα-deficient keratinocytes is due to increased proteasomal-mediated degradation of p21, we conducted experiments with the proteasome inhibitor MG132. The addition of MG132 resulted in a dramatic recovery of p21 protein in the UVB-treated C/EBPα-deficient cells () indicating that decreased p21 protein stability in UVB-treated C/EBPα-deficient keratinocytes is due to increased proteasome-mediated degradation of p21. MLN4924, is a potent small molecule inhibitor of the Nedd8 activating enzyme (NAE) which blocks the neddylation and the activity of the SCF (Skp, Cullin, F-box containing complex) and CRL (Cullin-RING ubiquitin ligases) E3 ligases.Citation36,37 The addition of MLN4924 resulted in a dramatic recovery of p21 protein in the UVB-treated C/EBPα-deficient cells () indicating that the decreased p21 protein stability in UVB-treated C/EBPα-deficient keratinocytes is dependent on SCF or CRL E3 ubiquitin ligase activity. The knockdown of Skp2, a critical component of the SCFSkp2 E3 ligase, did not rescue p21 protein levels in C/EBPα-depleted keratinocytes (Fig. S2). In contrast, the knockdown of Cdt2, the substrate binding subunit of the Cul4-RING protein ligase CRL4, rescued p21 protein levels in UVB-exposed C/EBPα-deficient keratinocytes (). We observed that following UVB exposure, Cdt2 is down-regulated at both the protein and mRNA level in control keratinocytes (), however, C/EBPα-deficient keratinocytes failed to down-regulate Cdt2 mRNA and protein levels and thus displayed elevated Cdt2 levels after UVB exposure (). These results indicate that C/EBPα regulates Cdt2 levels and CRL4Cdt2-mediated proteasomal degradation of p21 in response to UVB.
Figure 3. C/EBPα regulates CRL4Cdt2-mediated degradation of p21 in response to UVB. (A) Immunoblot analysis of control and C/EBPα siRNA nuclear extracts 8 h after 5 mJ/cm2 UVB. Ten μM MG132 was added 2 h before collection. (B) Immunoblot analysis of control and C/EBPα siRNA nuclear extracts 6 h after exposure to 5 mJ/cm2 UVB. Cells were treated with 800 nM MLN4924 30 min before UVB exposure. (C) Immunoblot analysis of control, C/EBPα, and Cdt2 siRNA nuclear extracts 6 h after 5 mJ/cm2 UVB. (D) Relative Cdt2 mRNA levels in control and C/EBPα siRNA 6 h following 5 mJ/cm2 UVB. Data are expressed as the mean normalized to Gapdh ±S .D. (N = 3). **= P value < 0.01 as calculated using Student's T test.
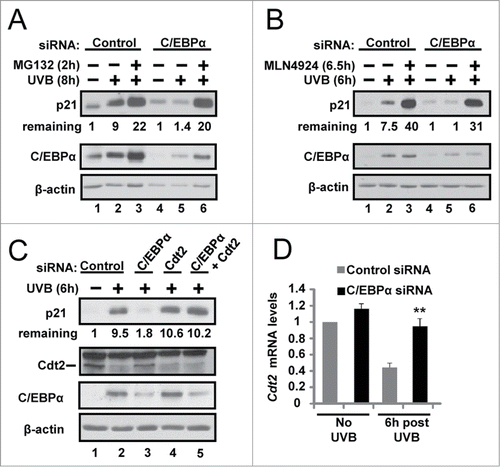
Failure to up-regulate nuclear p21 protein levels in UVB-exposed C/EBPα-deficient keratinocytes has functional consequences on cell cycle protein complexes, CDK2 activity and the G1/S checkpoint
CDK2 is required for completion of G1 and entry/progression through S phase and is a critical target of p21 in the G1/S DNA damage checkpoint response. We examined whether UVB-treated C/EBPα-deficient keratinocytes display reduced CDK2-p21 complex formation and altered CDK2 kinase activity. CDK2 formed a complex with p21 in UVB-treated control siRNA cells, however the CDK2-p21 complex formation was barely detectable in the C/EBPα siRNA treated keratinocytes (). Measurement of CDK2 activity in control and C/EBPα-depleted keratinocytes revealed that control cells displayed ∼75% decrease in CDK2 kinase activity following UVB treatment (6 h) while C/EBPα-deficient keratinocytes displayed only ∼30% decrease in CDK2 kinase activity, with C/EBPα-deficient keratinocytes displaying ∼4-fold higher CDK2 activity 12 h post UVB (). To further investigate the functional consequences of C/EBPα deficiency on p21 protein interactions following UVB treatment, we conducted LC MS/MS analysis of proteins that co-immunoprecipitated with p21 from UVB exposed control and C/EBPα knockdown keratinocytes. UVB-treated C/EBPα-deficient keratinocytes displayed diminished p21 interactions with numerous key cell cycle regulator proteins that are critical in inducing p21-mediated cell cycle arrest (; Fig. S3 and Table S1).
Figure 4. Failure to up-regulate nuclear p21 protein levels in UVB-exposed C/EBPα-deficient keratinocytes leads to functional consequences on cell cycle protein complexes, CDK2 activity and the G1/S checkpoint. (A) Immunoblot analysis of CDK2 co-IP from control and C/EBPα siRNA nuclear extracts 6 h following 5 mJ/cm2 UVB. (B) Phosphorylation of H1 by CDK2 immunoprecipitated from control and C/EBPα siRNA nuclear extracts 5 mJ/cm2 after UVB exposure. (C) Bar plot of peptide abundance for unique peptides for the proteins of interest across the 2 technical p21 IP replicates. Peptide abundances are expressed as a percent of the most abundant for a specific protein. **For PCNA, we could not confidently integrate a peptide signal in the siRNA replicates. A second peptide for each protein shows similar results (Fig. S3). (D)3H-thymidine incorporation in synchronized control and C/EBPα siRNA treated keratinocytes after 5 mJ/cm2 UVB exposure. Data represents the mean ± S.D., * = P value < 0.01 as calculated using Student's T test (N = 3) and are representative of 3 independent experiments.
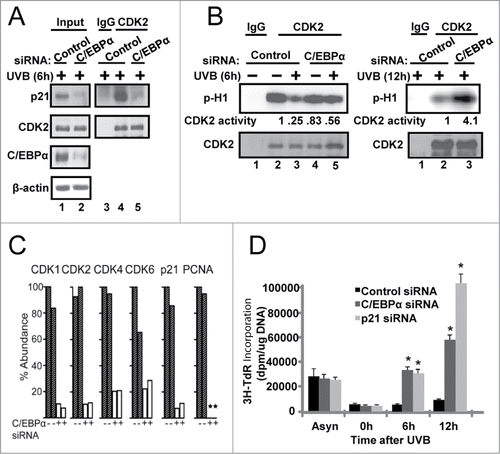
Finally, we compared the effect of knockdown of p21 and C/EBPα on UVB-induced G1/S checkpoint response. Control siRNA, C/EBPα siRNA and p21 siRNA keratinocytes were synchronized by serum and EGF deprivation, released from starvation, irradiated with UVB 6 h after release from starvation, and pulsed with 3H-thymidine 1 h prior to collection. As shown in , UVB-treated control siRNA keratinocytes exposed to UVB did not enter S-phase as determined by the lack of increase in 3H-thymidine incorporation into DNA demonstrating a functional G1/S checkpoint. In contrast, both C/EBPα- and p21-defcient cells displayed an inappropriate entry into S phase and DNA replication following UVB as measured by the increase in 3H-thymidine incorporation. Collectively our results indicate that failure of C/EBPα-deficient keratinocytes to increase p21 protein levels in response to UVB results in diminished p21 interactions with numerous key cell cycle regulator proteins and that these molecular defects define the failed G1/S DNA damage response in C/EBPα-deficient keratinocytes.
Discussion
Normally, cells respond to DNA damage with the transcriptional upregulation of the cyclin-dependent kinase inhibitor, p21, which results in increased p21 protein levels and the subsequent inhibition of numerous CDKs, including CDK2, which has a critical role in the DNA damage-induced G1/S checkpoint.Citation7,8 We observed that this canonical DNA damage G1/S checkpoint pathway involving p21 is inactivated in C/EBPα-deficient cells due to diminished p21 protein stability due to the altered regulation of Cdt2 and the cullin RING ubiquitin ligase CRL4Cdt2. Moreover, this DNA damage pathway is inactivated despite the normal transcriptional up-regulation of Cdkn1a which points to the overall power of C/EBPα on p21 protein stability. The inability of C/EBPα-deficient keratinocytes to accumulate p21 in response to UVB-induced DNA damage results in diminished interactions with CDK6, CDK4, CDK2 and PCNA that are critical in the p21-mediated regulation of G1 and the G1/S checkpoint. We also observed that a decreased p21-CDK2 interaction was accompanied by the incomplete inhibition of CDK2 kinase activity and inappropriate entry into S phase and DNA synthesis following UVB treatment. These findings reveal C/EBPα regulates G1/S cell cycle arrest in response to DNA damage via the control of CRL4Cdt2 mediated degradation of p21.
Cdt2 is a DCAF (DDB1/CUL4-associated factor) protein and is the substrate binding subunit of the CRL4 that is responsible for targeting p21 for proteasomal degradation during S phase and following UVC exposure.Citation38-40 Unlike many substrates of CRLs, no specific posttranslational modifications, such as phosphorylation, are reported to be essential for ubiquitination of substrates by CRL4Cdt2 and the mere overexpression of Cdt2 is sufficient to decrease the stability of p21.Citation38,41 Ctd2 is a critical regulator of cell-cycle progression and genome stability and in addition to p21, CRL4Cdt2 also targets the replication initiation factor Cdt1Citation42-44, the histone methyltransferase Set8Citation45,46, and DNA polymerase δ for proteasomal degradation.Citation47 Degradation of these substrates is important for cell-cycle progression, DNA repair, gene expression, and prevention of aberrant DNA replication.Citation41 The activity of CRL4Cdt2 is unique in that it requires its substrates be bound to chromatin-bound PCNA, a condition met during S phase and during the cellular response to DNA damage.Citation41 This requirement for CRL4Cdt2substrate ubiquitination/degradation is consistent with our findings demonstrating that the loss of C/EBPα preferentially affects nuclear p21 and only during a DNA damage response. Given that we only observe C/EBPα-mediated regulation of Cdt2 following DNA damage and that C/EBPα is highly inducible by DNA damage suggests C/EBPα protein must accumulate and/or be modified in some way after UVB exposure to affect the down-regulation of Cdt2.
Little is known about the regulation of CRL4Cdt2 activity or the factors involved in its assembly or disassembly. Cdt2 undergoes auto-ubiquitination via CRL4A and is ubiquitinated by CRL1FBXO11.Citation48 The downregulation of Cdt2 serves to restrain CRL4Cdt2 activity on its substrates. Following UVB exposure, we observed that Cdt2 is downregulated at both the protein and mRNA level in control keratinocytes, however, in C/EBPα-deficient keratinocytes, Cdt2 mRNA and protein levels are not down-regulated and these cells display elevated Cdt2 levels and decreased p21 protein levels after UVB exposure. Our findings suggest that the C/EBPα, which is highly inducible by UVB-induced DNA damage, is a transcriptional repressor of Cdt2 during a DNA damage response. Previously, Cdt2 had been shown to be a transcriptional target of E2F1.Citation49 C/EBPα has been reported to directly interact with and repress E2F-mediated transcription activity.Citation50,51 While further studies are required to identify the downstream pathway through which C/EBPα controls Cdt2 gene expression (directly or indirectly) our study provides new insights into the function of C/EBPα.
p21 is a universal CDK inhibitorCitation16, and in addition to regulating G1 progression and the G1/S checkpoint, p21 also has a crucial role in regulating S-phase progression and the G2/M checkpoint response, through p21's ability to bind PCNA and block activation of the principal replicative DNA polymerase δ18,19 and to inhibit the activity of cyclin A/CDK2 and cyclin B/CDK1 complexes.Citation52-54 C/EBPα-deficient cells display significant reductions in p21-PCNA, p21-CDK2, and p21-CDK1 interactions indicating that the C/EBPα-mediated regulation of p21 protein stability may also be important in checkpoint responses regardless of cell cycle position. It is possible the silencing of C/EBPα that occurs in numerous epithelial cancers results in defective checkpoint responses to endogenous and exogenous DNA damage and this contributes to the acquisition of somatic mutations and accelerates cancer progression.
In summary, our findings reveal a previously unidentified role for C/EBPα in the canonical DNA damage response involving p21 and in the regulation of Cdt2 levels and CRL4Cdt2 E3 ubiquitin ligase activity.
Materials and Methods
Cells and mice
Balb/MK2 keratinocytes were a gift from B. E. Weissman (UNC) are maintained in Ca2+-free EMEM (06–174 G, Lonza), 8% Chelax-treated FBS (F2442, Sigma Aldrich), 4 ng/ml hEGF (PHG60311, Life Technologies), and 0.05 mM CaCl2.Citation27 Detailed information on control (K5Cretg/+) and epidermal-specific C/EBPα (K5Cretg/+;C/EBPαflox/flox) knockout SKH-1 hairless mice have been described.Citation25,28 All aspects of animal care and experimentation described in this study were conducted according to the NIH guidelines and approved by the Institutional Animal Care and Use Committee of NCSU.
UVB treatment
Balb/MK2 keratinocytes were exposed at ∼40%–50% confluence to 5 mJ/cm2 UVB with a calibrated UVB lamp as previously described.Citation26 SKH-1 mice were treated with a single dose of 50 mJ/cm2 or 100 mJ/cm2 as described.Citation25
Small Interfering RNA
siRNA targeting mC/EBPα (5′-AAAGCCAAACAACGCAACGUGdTdT-3′), mp21 (5′-AACGGUGGAACUUUGACUUCGdTdT-3′), hC/EBPα (5′-CGACGAGUUCCUGGCCGACdTdT-3′), mSkp2 (5′-UUUGUCACUCCCUUUGCCCdTdT-3′), and the negative control (GFP, 5′-GGCUACGUCCAGGAGCGCACCdTdT-3′) were synthesized by Life Technologies and transfected at the final concentration of 100 nM. Pre-designed siRNA targeting Cdt2 was purchased from Life Technologies (DTLM55233769) and used at 50 nM. All transfections were performed using DharmaFECT Reagent 1 (T-2001, ThermoFisher) according to manufacturer's recommendations. Cells were exposed to UVB 40 h post siRNA transfection.
Antibodies and chemicals
Antibodies against C/EBPα (sc-61), p21 M-19 (sc-471), p21 H-164 (sc-756),CDK2 (sc-163), and α tubulin (sc-8035) were purchased from Santa Cruz Biotechnology. Human C/EBPα antibody (NB110) was purchased from Novus Biologicals. p53 antibody (1C12) was purchased from Cell Signaling. β-actin antibody (A5441) was purchased from Sigma-Aldrich. Antibody against Cdt2 was generously provided by Dr. Anindya Dutta (UVA). Cycloheximide (C7698) was purchased from Sigma-Aldrich and MG132 (13697) was purchased from Cayman Chemical. MLN4924 was kindly provided by Dr. Jean Cook (UNC).
RNA and quantitative PCR
Total RNA was isolated using QiaZOL lysis (79306, Qiagen), and further purified using Qiagen RNeasy® columns (74104). cDNA was prepared from RNA by ImProm-II Reverse Transcription System (A3802, Promega). Quantitative PCR using mouse Cdkn1a Mm00432448_m1, Cdt2 Mm00712787_m1, and Gapdh Mm99999915_g1 TaqMan Gene Expression Assays (Life Technologies) in combination with FastStart Universal Probe Master Mix (14001200, Roche). Data were analyzed using the comparative ΔΔCT method.
Immunohistochemistry
Mouse paraffinized skin section slides were subjected to antigen retrieval by incubating in citrate buffer (pH 6.0) at 95°C for 30 min. Slides were incubated with anti-p21 M-19, followed by incubation with biotin labeled secondary antibody at room temperature for 30 min. Detection was made with the ABC kit (PK-6101, Vector Labs) and 3–3′ diaminobenzidine (HK153–5 K, Biogenex) as the chromagen as previously described.Citation25
Preparation of epidermal lysates and nuclear cell extracts
Mice were euthanized by cervical dislocation and dorsal skin was removed and subjected to 6 sec heat shock in 60°C dH20 followed by 15 sec in ice water. Epidermis was isolated and lysates were prepared as previously described.Citation25 Balb/MK2 cells were fractionated using hypotonic lysis buffer and nuclei were lysed in RIPA buffer as previously described.Citation29
Co-immunoprecipitations
Keratinocytes were lysed in p21 IP buffer (50 mM HEPES pH 7.4, 250 mM NaCl, 2 mM EDTA, 2 mM NaF, 0.1% Tween20, 0.1 mM sodium orthovanadate, 1 mM AEBSF (4-(2-Aminoethyl)benzenesulfonyl fluoride hydrochloride), 1 mM DTT, and 1× protease inhibitor cocktail) or CDK2 IP buffer (50 mM Tris pH 7.5, 150 mM NaCl, 0.5% NP-40, 50 mM NaF, 0.1 mM sodium orthovanadate, 1 mM AEBSF, 1 mM DTT, and 1× protease inhibitor cocktail). Equal protein amount of each clarified lysate was incubated with 1 μg normal rabbit IgG (sc-2027, Santa Cruz Biotechnology), p21 or CDK2 antibody for 2 h at 4°C, followed by 1 h with Protein A/G agarose (Santa Cruz Biotechnology, sc-2003). Immune complexes were washed with corresponding IP buffer, solubilized in 2× SDS-sample buffer and resolved by SDS-PAGE followed by immunoblot analysis.
LC MS/MS methods and data analysis
Preparation of the co-IP using a modified filter aided sample preparation and is described.Citation30 Briefly, samples were prepared in technical duplicate and reduced with DTT at 60°C for 30 min, centrifuged, and alkylated with iodoacetamide in the dark for 20 min. Iodoacetamide was removed via centrifugation and samples were digested for 4 h with trypsin at 37°C. Five μL of purified sample were injected on to a microcolumn packed in house using an Easy nanoLC system 1000 (Thermo Scientific). Mass spectrometric analyses were performed using a Q Exactive Plus (Thermo Scientific) operated in the top 12 data dependent mode. Raw files were searched against the SwissProt Reference Mus Musculus database (16657 entries, downloaded 12/2013) appended with common protein contaminants using the Sequest HT algorithm in Proteome Discoverer (version 1.4.0.288, Thermo Scientific). Data were searched against a target and decoy database (reversed) within Proteome Discoverer and PercolatorCitation31 was used as a post processor to enforce a q value threshold of <0 .01. The law of parsimony was used for protein inference and protein identifications were filtered to those having a least 2 unique peptides among the sample set. Two label free methods were used to evaluate differential abundance among proteins of interest including raw spectral counts and the integration of extracted ion chromatograms (EIC) of unique peptides in Skyline. Further details on the LC MS/MS settings, search parameters, and data analysis are available in Supplemental Materials and Methods.
CDK2 IP kinase
CDK2 was immunoprecipitated as described above using 500 μg fresh Balb/MK2 nuclear protein extract. CDK2 kinase assay was performed as described.Citation32 Detection and quantification of phosphorylated H1 was performed with a Storm™ 865 imager and quantified using Image Quant™ TL software (GE Healthcare Life Sciences).
Thymidine incorporation assay
Cells were incubated for 28 h in media deprived of growth factors (0.1% FBS, no EGF, and 0.05 mM CaCl2) to synchronize in G0 (t = 0). Proliferation was re-stimulated by the addition normal growth media. Cells were pulse-labeled with 3 μCi/ml (20 Ci/mmol) 3H-methyl thymidine (NET027E, PerkinElmer) for 1 h before collection, and detection of incorporated3H-methyl thymidine was performed as described.Citation33
p21 stability assay
Cycloheximide (40 μg/ml) was added 5 h post 5 mJ/cm2 UVB (t = 0). Nuclear extracts were resolved by SDS-PAGE followed by immunoblotting and quantified by densitometry using ImageJ normalized to β-actin.
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Acknowledgments
We are grateful to Dr. Marcelo Rodriguez-Puebla (NCSU) for kinase assay reagents and technical advice, Dr. Anindya Dutta (UVA) for providing the Cdt2 antibody, and Dr. Jean Cook (UNC) for providing the MLN4924 drug and for helpful discussions.
962957_Supplementary_Materials.zip
Download Zip (412.1 KB)Funding
This research was supported by grants from the National Institute of Environmental Health Sciences (ES017734 to J.R.H, ES12473 and CA046637 awarded to RCS).
References
- American Cancer Society. Cancer Facts and Figures 2013. Atlanta: American Cancer Society. http://cancer.org
- Brash DE. Sunlight and the onset of skin cancer. Trends Genet 1997; 13:410-4; PMID:9351343; http://dx.doi.org/10.1016/S0168-9525(97)01246-8
- de Gruijl FR, van Kranen HJ, Mullenders LH. UV-induced DNA damage, repair, mutations and oncogenic pathways in skin cancer. J Photochem Photobiol 2001; 63:19-27; PMID:11684448; http://dx.doi.org/10.1016/S1011-1344(01)00199-3
- Brash DE, Rudolph JA, Simon JA, Lin A, McKenna GJ, Baden HP, Halperin AJ, Ponten J. A role for sunlight in skin cancer: UV-induced p53 mutations in squamous cell carcinoma. Proc Natl Acad Sci U S A 1991; 88:10124-8; PMID:1946433; http://dx.doi.org/10.1073/pnas.88.22.10124
- Kastan MB, Bartek J. Cell-cycle checkpoints and cancer. Nature 2004; 432:316-23; PMID:15549093; http://dx.doi.org/10.1038/nature03097
- Ikehata H, Ono T. The mechanisms of UV mutagenesis. J Radiat Res 2011; 52:115-25; PMID:21436607; http://dx.doi.org/10.1269/jrr.10175
- Bartek J, Lukas J. Mammalian G1- and S-phase checkpoints in response to DNA damage. Curr Opin Cell Bio 2001; 13:738-47; PMID:11698191; http://dx.doi.org/10.1016/S0955-0674(00)00280-5
- Niida H, Nakanishi M. DNA damage checkpoints in mammals. Mutagenesis 2006; 21:3-9; PMID:16314342; http://dx.doi.org/10.1093/mutage/gei063
- Macleod KF, Sherry N, Hannon G, Beach D, Tokino T, Kinzler K, Vogelstein B, Jacks T. p53-dependent and independent expression of p21 during cell growth, differentiation, and DNA damage. Genes Dev 1995; 9:935-44; PMID:7774811; http://dx.doi.org/10.1101/gad.9.8.935
- Jung YS, Qian Y, Chen X. Examination of the expanding pathways for the regulation of p21 expression and activity. Cell Signal 2010; 22:1003-12; PMID:20100570; http://dx.doi.org/10.1016/j.cellsig.2010.01.013
- el-Deiry WS, Tokino T, Velculescu VE, Levy DB, Parsons R, Trent JM, Lin D, Mercer WE, Kinzler KW, Vogelstein B. WAF1, a potential mediator of p53 tumor suppression. Cell 1993; 75:817-25; PMID:8242752; http://dx.doi.org/10.1016/0092-8674(93)90500-P
- Gartel AL, Tyner AL. Transcriptional regulation of the p21(WAF1/CIP1) gene. Expl Cell Res 1999; 246:280-9; PMID:9925742; http://dx.doi.org/10.1006/excr.1998.4319
- Gu Y, Turck CW, Morgan DO. Inhibition of CDK2 activity in vivo by an associated 20K regulatory subunit. Nature 1993; 366:707-10; PMID:8259216; http://dx.doi.org/10.1038/366707a0
- Harper JW, Adami GR, Wei N, Keyomarsi K, Elledge SJ. The p21 Cdk-interacting protein Cip1 is a potent inhibitor of G1 cyclin-dependent kinases. Cell 1993; 75:805-16; PMID:8242751; http://dx.doi.org/10.1016/0092-8674(93)90499-G
- Poon RY, Jiang W, Toyoshima H, Hunter T. Cyclin-dependent kinases are inactivated by a combination of p21 and Thr-14/Tyr-15 phosphorylation after UV-induced DNA damage. J Biol Chem 1996; 271:13283-91; PMID:8662825; http://dx.doi.org/10.1074/jbc.271.23.13706
- Xiong Y, Hannon GJ, Zhang H, Casso D, Kobayashi R, Beach D. p21 is a universal inhibitor of cyclin kinases. Nature 1993; 366:701-4; PMID:8259214; http://dx.doi.org/10.1038/366701a0
- Gottifredi V, McKinney K, Poyurovsky MV, Prives C. Decreased p21 levels are required for efficient restart of DNA synthesis after S phase block. J Biol Chem 2004; 279:5802-10; PMID:14597617; http://dx.doi.org/10.1074/jbc.M310373200
- Shivji MK, Grey SJ, Strausfeld UP, Wood RD, Blow JJ. Cip1 inhibits DNA replication but not PCNA-dependent nucleotide excision-repair. Curr Biol 1994; 4:1062-8; PMID:7704570; http://dx.doi.org/10.1016/S0960-9822(00)00244-X
- Waga S, Hannon GJ, Beach D, Stillman B. The p21 inhibitor of cyclin-dependent kinases controls DNA replication by interaction with PCNA. Nature 1994; 369:574-8; PMID:7911228; http://dx.doi.org/10.1038/369574a0
- Deng C, Zhang P, Harper JW, Elledge SJ, Leder P. Mice lacking p21CIP1/WAF1 undergo normal development, but are defective in G1 checkpoint control. Cell 1995; 82:675-84; PMID:7664346; http://dx.doi.org/10.1016/0092-8674(95)90039-X
- Fotedar R, Bendjennat M, Fotedar A. Role of p21WAF1 in the cellular response to UV. Cell Cycle 2004; 3:134-7; PMID:14712074; http://dx.doi.org/10.4161/cc.3.2.658
- Maeda T, Chong MT, Espino RA, Chua PP, Cao JQ, Chomey EG, Luong L, Tron VA. Role of p21(Waf-1) in regulating the G1 and G2/M checkpoints in ultraviolet-irradiated keratinocytes. J Invest Dermatol 2002; 119:513-21; PMID:12190878; http://dx.doi.org/10.1046/j.1523-1747.2002.01828.x
- Oh HS, Smart RC. Expression of CCAAT/enhancer binding proteins (C/EBP) is associated with squamous differentiation in epidermis and isolated primary keratinocytes and is altered in skin neoplasms. J Invest Dermatol 1998; 110:939-45; PMID:9620302; http://dx.doi.org/10.1046/j.1523-1747.1998.00199.x
- Swart GW, van Groningen JJ, van Ruissen F, Bergers M, Schalkwijk J. Transcription factor C/EBPalpha: novel sites of expression and cloning of the human gene. Biol Chem 1997; 378:373-9; PMID:9191024; http://dx.doi.org/10.1515/bchm.1997.378.5.373
- Thompson EA, Zhu S, Hall JR, House JS, Ranjan R, Burr JA, He YY, Owens DM, Smart RC. C/EBPalpha expression is downregulated in human nonmelanoma skin cancers and inactivation of C/EBPalpha confers susceptibility to UVB-induced skin squamous cell carcinomas. J Invest Dermatol 2011; 131:1339-46; PMID:21346772; http://dx.doi.org/10.1038/jid.2011.31
- Yoon K, Smart RC. C/EBPalpha is a DNA damage-inducible p53-regulated mediator of the G1 checkpoint in keratinocytes. Mol Cell Biol 2004; 24:10650-60; PMID:15572670; http://dx.doi.org/10.1128/MCB.24.24.10650-10660.2004
- Weissman BE, Aaronson SA. BALB and Kirsten murine sarcoma viruses alter growth and differentiation of EGF-dependent balb/c mouse epidermal keratinocyte lines. Cell 1983; 32:599-606; PMID:6297803; http://dx.doi.org/10.1016/0092-8674(83)90479-8
- Ramirez A, Page A, Gandarillas A, Zanet J, Pibre S, Vidal M, Tusell L, Genesca A, Whitaker DA, Melton DW, et al. A keratin K5Cre transgenic line appropriate for tissue-specific or generalized Cre-mediated recombination. Genesis 2004; 39:52-7; PMID:15124227; http://dx.doi.org/10.1002/gene.20025
- Hall JR, Kow E, Nevis KR, Lu CK, Luce KS, Zhong Q, Cook JG. Cdc6 stability is regulated by the Huwe1 ubiquitin ligase after DNA damage. Mol Biol Cell 2007; 18:3340-50; PMID:17567951; http://dx.doi.org/10.1091/mbc.E07-02-0173
- Gokce E, Franck WL, Oh Y, Dean RA, Muddiman DC. In-Depth Analysis of the Magnaporthe oryzae Conidial Proteome. J Proteome Res 2012; 11:5827-35; PMID:23039028
- Kall L, Canterbury JD, Weston J, Noble WS, MacCoss MJ. Semi-supervised learning for peptide identification from shotgun proteomics datasets. Nat Methods 2007; 4:923-5; PMID:17952086; http://dx.doi.org/10.1038/nmeth1113
- Sistrunk C, Kim SH, Wang X, Lee SH, Kim Y, Macias E, Rodriguez-Puebla ML. Skp2 deficiency inhibits chemical skin tumorigenesis independent of p27(Kip1) accumulation. Am J Pathol 2013; 182:1854-64; PMID:23474082; http://dx.doi.org/10.1016/j.ajpath.2013.01.016
- Zhu S, Oh HS, Shim M, Sterneck E, Johnson PF, Smart RC. C/EBPbeta modulates the early events of keratinocyte differentiation involving growth arrest and keratin 1 and keratin 10 expression. Mol Cell Biol 1999; 19:7181-90; PMID:10490653
- Cmielova J, Rezacova M. p21Cip1/Waf1 protein and its function based on a subcellular localization; corrected. J Cell Bioch 2011; 112:3502-6; PMID:21815189; http://dx.doi.org/10.1002/jcb.23296
- Timchenko NA, Wilde M, Nakanishi M, Smith JR, Darlington GJ. CCAAT/enhancer-binding protein alpha (C/EBP alpha) inhibits cell proliferation through the p21 (WAF-1/CIP-1/SDI-1) protein. Genes Dev 1996; 10:804-15; PMID:8846917; http://dx.doi.org/10.1101/gad.10.7.804
- Blank JL, Liu XJ, Cosmopoulos K, Bouck DC, Garcia K, Bernard H, Tayber O, Hather G, Liu R, Narayanan U, et al. Novel DNA damage checkpoints mediating cell death induced by the NEDD8-activating enzyme inhibitor MLN4924. Cancer Res 2013; 73:225-34; PMID:23100467; http://dx.doi.org/10.1158/0008-5472.CAN-12-1729
- Jia L, Li H, Sun Y. Induction of p21-dependent senescence by an NAE inhibitor, MLN4924, as a mechanism of growth suppression. Neoplasia 2011; 13:561-9; PMID:21677879
- Abbas T, Sivaprasad U, Terai K, Amador V, Pagano M, Dutta A. PCNA-dependent regulation of p21 ubiquitylation and degradation via the CRL4Cdt2 ubiquitin ligase complex. Genes dev 2008; 22:2496-506; PMID:18794347; http://dx.doi.org/10.1101/gad.1676108
- Kim Y, Starostina NG, Kipreos ET. The CRL4Cdt2 ubiquitin ligase targets the degradation of p21Cip1 to control replication licensing. Genes Dev 2008; 22:2507-19; PMID:18794348; http://dx.doi.org/10.1101/gad.1703708
- Nishitani H, Shiomi Y, Iida H, Michishita M, Takami T, Tsurimoto T. CDK inhibitor p21 is degraded by a proliferating cell nuclear antigen-coupled Cul4-DDB1Cdt2 pathway during S phase and after UV irradiation. J Biol Chem 2008; 283:29045-52; PMID:18703516; http://dx.doi.org/10.1074/jbc.M806045200
- Abbas T, Dutta A. CRL4Cdt2: master coordinator of cell cycle progression and genome stability. Cell Cycle 2011; 10:241-9; PMID:21212733; http://dx.doi.org/10.4161/cc.10.2.14530
- Higa LA, Banks D, Wu M, Kobayashi R, Sun H, Zhang H. L2DTL/CDT2 interacts with the CUL4/DDB1 complex and PCNA and regulates CDT1 proteolysis in response to DNA damage. Cell Cycle 2006; 5:1675-80; PMID:16861906; http://dx.doi.org/10.4161/cc.5.15.3149
- Jin J, Arias EE, Chen J, Harper JW, Walter JC. A family of diverse Cul4-Ddb1-interacting proteins includes Cdt2, which is required for S phase destruction of the replication factor Cdt1. Mol Cell 2006; 23:709-21; PMID:16949367; http://dx.doi.org/10.1016/j.molcel.2006.08.010
- Senga T, Sivaprasad U, Zhu W, Park JH, Arias EE, Walter JC, Dutta A. PCNA is a cofactor for Cdt1 degradation by CUL4/DDB1-mediated N-terminal ubiquitination. J Biol Chem 2006; 281:6246-52; PMID:16407252; http://dx.doi.org/10.1074/jbc.M512705200
- Abbas T, Shibata E, Park J, Jha S, Karnani N, Dutta A. CRL4(Cdt2) regulates cell proliferation and histone gene expression by targeting PR-Set7/Set8 for degradation. Mol Cell 2010; 40:9-21; PMID:20932471; http://dx.doi.org/10.1016/j.molcel.2010.09.014
- Jorgensen S, Eskildsen M, Fugger K, Hansen L, Larsen MS, Kousholt AN, Syljuasen RG, Trelle MB, Jensen ON, Helin K, et al. SET8 is degraded via PCNA-coupled CRL4(CDT2) ubiquitylation in S phase and after UV irradiation. The J Cell Biol 2011; 192:43-54; PMID:21220508; http://dx.doi.org/10.1083/jcb.201009076
- Terai K, Shibata E, Abbas T, Dutta A. Degradation of p12 subunit by CRL4Cdt2 E3 ligase inhibits fork progression after DNA damage. J Biol Chem 2013; 288:30509-14; PMID:24022480; http://dx.doi.org/10.1074/jbc.C113.505586
- Abbas T, Mueller AC, Shibata E, Keaton M, Rossi M, Dutta A. CRL1-FBXO11 promotes Cdt2 ubiquitylation and degradation and regulates Pr-Set7/Set8-mediated cellular migration. Mol Cell 2013; 49:1147-58; PMID:23478445; http://dx.doi.org/10.1016/j.molcel.2013.02.003
- Nakagawa H, Tategu M, Yamauchi R, Sasaki K, Sekimachi S, Yoshida K. Transcriptional regulation of an evolutionary conserved intergenic region of CDT2-INTS7. PloS One 2008; 3:e1484; PMID:18213392; http://dx.doi.org/10.1371/journal.pone.0001484
- Porse BT, Pedersen TA, Xu X, Lindberg B, Wewer UM, Friis-Hansen L, Nerlov C. E2F repression by C/EBPalpha is required for adipogenesis and granulopoiesis in vivo. Cell 2001; 107:247-58; PMID:11672531; http://dx.doi.org/10.1016/S0092-8674(01)00516-5
- Slomiany BA, D'Arigo KL, Kelly MM, Kurtz DT. C/EBPalpha inhibits cell growth via direct repression of E2F-DP-mediated transcription. Mol Cell Biol 2000; 20:5986-97; PMID:10913181; http://dx.doi.org/10.1128/MCB.20.16.5986-5997.2000
- Ando T, Kawabe T, Ohara H, Ducommun B, Itoh M, Okamoto T. Involvement of the interaction between p21 and proliferating cell nuclear antigen for the maintenance of G2/M arrest after DNA damage. J Biol Chem 2001; 276:42971-7; PMID:11559705; http://dx.doi.org/10.1074/jbc.M106460200
- Bunz F, Dutriaux A, Lengauer C, Waldman T, Zhou S, Brown JP, Sedivy JM, Kinzler KW, Vogelstein B. Requirement for p53 and p21 to sustain G2 arrest after DNA damage. Science 1998; 282:1497-501; PMID:9822382; http://dx.doi.org/10.1126/science.282.5393.1497
- Chen J, Jackson PK, Kirschner MW, Dutta A. Separate domains of p21 involved in the inhibition of Cdk kinase and PCNA. Nature 1995; 374:386-8; PMID:7885482; http://dx.doi.org/10.1038/374386a0
