Abstract
The recent finding that the Retinoblastoma protein (Rb) is able to regulate apoptosis in a non-transcriptional manner directly at the mitochondria by interaction with the pro-apoptotic protein Bax prompted this investigation of the complex formed between Rb and Bax. Because the function of Rb in the cellular processes of proliferation, apoptosis, senescence and differentiation is regulated by phosphorylation we endeavored to elucidate the phosphorylation status of Rb with respect to its association with Bax and its role in apoptosis. In this study we found that Rb phosphorylated on at least 4 C-terminal phosphorylation sites (S608, S795, S807/S811, and T821) is present at the mitochondria under non-stressed cellular conditions. An in vitro binding assay showed that Bax binds to Rb phosphorylated at S807/S811 in 3 cancer cell types. Physiologically relevant association between Bax and Rb phosphorylated on S807/S811 was demonstrated by reciprocal co-immunoprecipitation experiments using antibodies specific for Rb phosphorylated on S807/S811 and Bax. Mutant Rb proteins expressed in Rb-null C33A cells showed that phosphorylation of S807 of Rb promotes association with Bax and that mimicking phosphorylation at S807 of Rb can block the induction of apoptosis due to PNUTS downregulation. Finally using siRNA to activate phosphatase activity in MCF7 cells, Rb is dephosphorylated at several sites including S807/S811, dissociates from Bax and apoptosis is triggered. These studies show that phosphorylation of Rb regulates its association with Bax and its role in apoptosis.
Introduction
The Retinoblastoma (Rb) tumor suppressor protein is regulated by phosphorylation and plays a role in several important cellular processes such as proliferation, differentiation, senescence and apoptosis.Citation1 It is known that alterations in the Rb pathway consisting of cyclin dependent kinase 4/6, D-type cyclins, cdk inhibitors, (such as p16ink4a), and Rb itself, that lead to excessive phosphorylation of Rb (hyperphosphorylation) have been observed in almost all cancer types.Citation2,3 Alterations that lead to increased cdk activity toward Rb are more common than mutations to the Rb gene itself, a finding that has prompted investigations into the development of clinical treatments that inhibit the activity of cdks toward Rb.Citation4-6 Thus understanding the role of specific phosphorylation sites of Rb is clinically relevant.
While the role of Rb in the control of proliferation is well understood,Citation7 the role of Rb in apoptosis appears to depend on cellular context and apoptotic stimulus. There is evidence to suggest that hyperphosphorylated Rb protects cells from apoptosis. Rb dephosphorylation has been widely observed during apoptosis.Citation8-11 In various studies, the specific induction of Rb-directed phosphatase activity has been shown to be required for apoptosis to occur.Citation12-14 These studies suggest that hyperphosphorylation of Rb protects against apoptosis, and dephosphorylation of Rb is involved in triggering cell death. In addition, the identification of specific amino acids of Rb that must be dephosphorylated to allow apoptotic death supports this notion.Citation15 These studies further suggest that unphosphorylated Rb is involved in triggering cell death. In fact, a phosphorylation site mutated Rb protein (PSM-RB) which lacks 9 phosphorylation sites (in the c-terminus) can stimulate the apoptotic response.Citation16 Because Rb in mammalian cells has 15 known phosphorylation sites, the regulation of Rb function by phosphorylation is not well understood and is likely to be highly complex. However, from the available evidence, it seems likely that phosphorylation at specific sites on Rb is required for the ability of Rb to regulate apoptosis.
Rb binds to a large number of cellular proteins although the functional significance of many of these interactions is unknown.Citation17 Various studies have shown that phosphorylation of Rb on certain amino acids is capable of regulating the interaction with Rb binding proteins.Citation18-23 One such interaction is the formation of the complex between hyperphosphorylated Rb and a pro-apoptotic factor pp32 (ANP32A). Hyperphosphorylated Rb in this context inhibits the apoptotic activity of pp32 and stimulates proliferation through yet undefined mechanisms.Citation24 This study further supports the notion that Rb phosphorylation status is important for the regulation of apoptosis by Rb.
The role of Rb phosphorylation in apoptosis has been the focus of our recent studies.Citation12,15 Several studies have supported the idea that Rb may act in a non-transcriptional manner to regulate cell death. For example, the finding that Rb is localized to the mitochondria in non-stressed cells is suggestive of a extra-nuclear role for Rb in tumor suppression.Citation25 The current study was initiated due to the finding that Rb interacts directly with the Bax protein, a pro-apoptotic member of the Bcl2 family.Citation26 Bax is an essential regulator of mitochondrial apoptosis, as it is responsible for mitochondrial outer membrane permeabilization (MOMP), that occurs in apoptosis.Citation27 Subsequent release of cytochrome c causes caspase activation to accomplish cell death. In this study we sought to determine the phosphorylation status of Rb when it associates with the Bax protein. We report that Rb is phosphorylated on several C-terminal sites at the mitochondria under non-apoptotic conditions and that phosphorylation of Rb at S807 promotes the association with Bax. Furthermore, in cells where S807 of Rb is dephosphorylated, Bax dissociates from Rb and apoptosis is stimulated.
Results
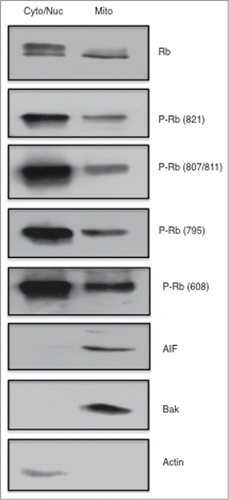
It was recently demonstrated that association of Rb with Bax regulates apoptosis at the mitochondria.Citation26 Our previous work and other studies have shown that the modification of specific phosphorylation sites of Rb play a role in regulating apoptosis.Citation15,19 Thus we sought to evaluate the phosphorylation status of Rb that is localized to the mitochondria. We subjected MCF7 breast cancer cells to subcellular fractionation using a Mitochondria Isolation Kit and performed immunoblotting experiments on equivalent amounts of protein from the combined cytoplasmic and nuclear fraction and compared this to cell lysates from the mitochondrial fraction. As shown in , accurate cell fractionation was verified by immunoblotting with Actin antibodies (cytoplasmic) and AIF and Bak (mitochondria-localized proteins). Phosphorylation site specific antibodies were used to show that cytoplasmic/nuclear fractions contained abundant levels of Rb phosphorylated on T821, S807/S811, S795, and S608, but also that strong signals were detected in mitochondrial fractions, indicating that Rb phosphorylated on these specific sites is found at the mitochondria in proliferating, non-stressed cells.
Figure 1. Phosphorylated Rb is localized to the mitochondria. Asynchronously growing MCF7 breast cancer cells were fractionated using a Mitochondria Isolation kit (Thermo Scientific) according to the manufacturers directions. Equal amounts of protein (15 μg) taken from the cytoplasmic/nuclear fraction and mitochondrial fractions were subjected to immunoblotting analysis as described in the Materials and Methods section. Immunodetection of Actin was used as a marker of the cytoplasmic/nuclear fraction, and detection of AIF and Bak are shown as mitochondrial localized proteins. Antibodies to total Rb and Rb phosphorylated at specific phosphorylation sites are listed to the right of the figure. Data shown is representative of two independent experiments.
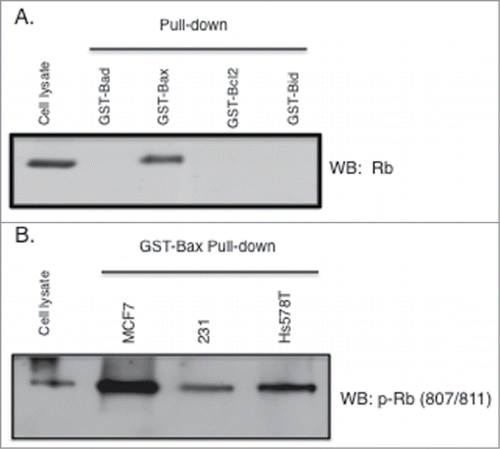
Next we performed Glutathione-S-Transferase (GST) fusion binding assays (pull down) to investigate the phosphorylation state of Rb when bound to Bax. Utilizing several Bcl2 family member proteins fused to GST we show that in this assay, Rb in MCF7 cellular lysates specifically binds to Bax and not the related proteins Bad, Bcl2, and Bid (). Subsequently we used 3 breast cancer cell types (MCF7, MDA-MB-231, and Hs578T) and performed pull-down assays using only GST-Bax. Proteins associated with GST-Bax were analyzed by immunoblotting with Rb phosphorylation site specific antibodies. As shown in , the antibody recognizing phosphorylation of Rb at S807/S811 recognizes Rb that associated with Bax in all 3 cell types. Two additional Rb phosphorylation site specific antibodies recognized Rb that associated with Bax (antibodies raised to Rb phosphorylated on S608, and S795) although at much higher exposures of the film (data not shown). Thus in further experiments we focused our efforts on the significance of Rb phosphorylation on S807/S811 with regard to the interaction with Bax.
Figure 2. Rb phosphorylated on S807/S811 binds to GST-Bax. GST fusion proteins (Sigma) were used in pull down/binding assays performed as previously described (38). (A) Pull down binding assays were performed using 4 bcl2 family proteins individually fused to GST (GST-Bad, GST-Bax, GST-bcl2, and GST-Bid). Two micrograms of fusion protein was incubated with MCF7 cell lysates (500 ug) followed by analysis of GST-fusion protein associated proteins by immunoblotting with antibodies to Rb. (B) Lysates from 3 cancer cell types (MCF7, MDA-MB-231, Hs578T) were utilized in GST-Bax fusion protein binding assays followed by immunoblotting with antibodies to Rb phosphorylated at S807/S811. Data shown is representative of three independent experiments.
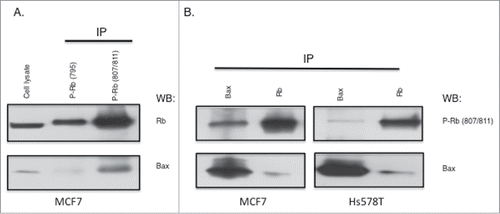
The in vivo association of Rb phosphorylated on S807/S811 with Bax was further analyzed using co-immunoprecipitation. In MCF7 cells, immunoprecipitation experiments using Rb site specific phosphorylation antibodies S795 and S807/S811 show that Bax associates with Rb phosphorylated on S807/S811 (). Additional experiments using both MCF7 and Hs578T cells demonstrate that Rb co-purified with Bax by co-immunoprecipitation is phosphorylated at the S807/S811 site (). As controls, Rb immunoprecipitation is shown to also co-immunopurify Bax. Thus in 2 cell types, Rb that associates with Bax is phosphorylated on the S807 and/or S811 amino acid phosphorylation site.
Figure 3. Association of Bax with Rb phosphorylated on S807/S811. (A) MCF7 cell lysates (1 mg) were used in immunoprecipitations with antibodies to Rb phosphorylated on S795 or S807/S811. Immunoprecipitates were analyzed by immunoblotting with antibodies to Rb or Bax. (B) MCF7 and Hs578T cell lysates were used in Bax or Rb immunoprecipitation experiments, followed by immunoblotting analysis with antibodies to phosphorylated Rb (S807/811) and Bax. Data shown is representative of three independent experiments.
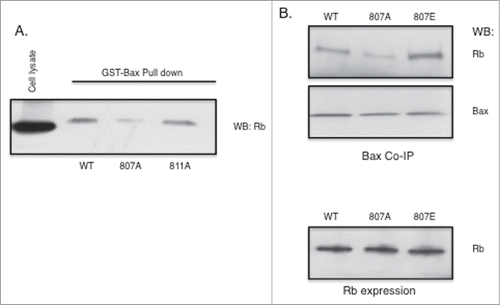
Because the antibody used in the experiments up to this point recognizes phosphorylation at either S807 or S811 or both of the sites, we attempted to identify whether one or both of the phosphorylation sites was responsible for regulating the association of Rb with Bax using expression plasmids encoding Rb mutant proteins. For these studies, C33A cervix carcinoma cells that express a truncated unstable Rb protein were utilized. We performed GST-Bax pull down experiments using lysates that expressed wild type (WT), or alanine mutants S807A and S811A of Rb and determined that while blocking phosphorylation of S807 inhibited the ability of Rb to bind to Bax, the alanine mutant of S811 was fully capable of binding to Bax at an affinity equivalent to that of WT Rb. (). This suggests that phosphorylation of S807 promotes or is required for Rb binding to Bax. We next performed co-immunoprecipitation of Rb with Bax antibodies using C33A cells expressing mutant forms of the Rb protein. In these experiments we used both alanine and glutamic acid mutants of the S807 phosphorylation site of Rb. Whereas alanine blocks the phosphorylation of the amino acid, glutamic acid substitution mimics phosphorylation at the mutated amino acid. We show in that while expression of Rb (WT, S807A, S807E) in cells is equivalent (lower panel), association with Bax is blocked by the alanine mutant and enhanced by the glutamic acid mutant compared to the association of the WT control. These results show that phosphorylation of S807 of Rb is important for association of Rb with Bax.
Figure 4. Rb phosphorylation at S807 is required for association with Bax. Rb mutant plasmids were generated as described in the Materials and Methods section. Rb-negative C33A cells were transfected using Fugene (Promega). (A) Rb plasmids expressing either S807A or S811A alanine mutants were transfected into cells and 48 hours later cell lysates were utilized in GST-Bax fusion protein pull down assays. Proteins associated with GST-Bax were analyzed by immunoblotting with Rb antibodies. (B) Rb plasmids expressing either S807A or S807E mutants were transfected into C33A cells and 48 hours later co-immunoprecipitation with Bax antibodies was performed. Immunoprecipitates were analyzed by immunoblotting with Rb and Bax antibodies. Equivalent expression of the Rb mutant proteins in the C33A cells is verified by immunoblotting (lower panel). Data shown is representative of three independent experiments.
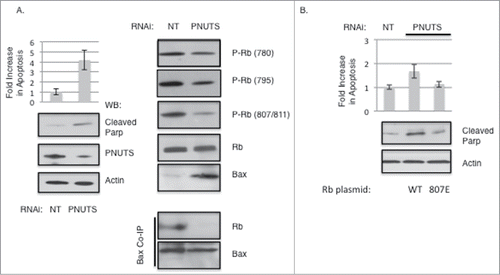
Next we postulated that the change in Rb phosphorylation observed during apoptosis might effect the association between Bax and Rb. We have previously established that by activating phosphatase activity toward Rb using siRNA mediated knockdown of the PP1 regulator PNUTS (Phosphatase Nuclear Targeting Subunit), Rb is dephosphorylated, and apoptosis is triggered in transformed cells in a Rb-dependent manner.Citation12,28 Here, in agreement with previous data, siRNA mediated knockdown of PNUTS expression in MCF7 cells leads to a four-fold increase in apoptosis, as measured by the Cell Death ELISA assay and by the increase in the cleavage of PARP (Poly ADP ribose polymerase) as detected by immunoblotting (). Efficient knockdown of PNUTS is verified by immunoblotting with PNUTS and Actin antibodies. PNUTS knockdown induces dephosphorylation of Rb at specific sites: S780, S795, and S807/S811. The induction of apoptosis is accompanied by an increase in the expression of Bax, in accordance with previous findings.Citation27 However, although Bax expression is increased in apoptosis, its association with Rb is disrupted as shown by co-immunoprecipitation (). These results further support the notion that phosphorylation of S807 of Rb is required for association of Rb with Bax. Finally to determine whether phosphorylation at S807 of Rb could effect apoptosis induction, we performed PNUTS knockdown experiments in C33A cells in which WT Rb or the S807E mutant of Rb was expressed. As shown in , mimicking phosphorylation at S807 blocks apoptosis as measured by the Cell Death ELISA assay and by immunoblotting of cleaved PARP. This result is consistent with the notion that Rb phosphorylated on S807 blocks apoptosis induced by PNUTS downregulation.
Figure 5. Dephosphorylation of Rb on S807/S811 causes Bax dissociation and stimulates Apoptosis. Knockdown of PNUTS (Phosphatase Nuclear Targeting Subunit) was performed in MCF7 cells as described in the Materials and Methods section. (A) Cells were subjected to PNUTS knockdown (PNUTS) or transfected with nontargeting RNA (NT) and 48 hours later apoptosis was measured using the Cell Death Detection ELISA (Roche Diagnostics) which detects degraded DNA released from the nucleus into the cytoplasm. The amount of apoptosis (degraded DNA) detected in control cells (NT) was normalized to one. Graph depicts the fold increase in degraded DNA observed due to PNUTS knockdown. Error bars represent standard deviation of the mean of triplicate samples and data shown is representative of three independent experiments. Apoptosis was also measured by immunoblot analysis of the cleavage of Parp as an indicator of apoptosis. Immunoblotting of Actin and PNUTS verify PNUTS knockdown. Co-immunoprecipitation experiment results and immunoblotting with site-specific Rb phosphorylation antibodies are shown on the right panel. Data shown is representative of three independent experiments. (B) C33A cells were transfected with WT or S807E Rb mutant expressing plasmids and 48 hours later PNUTS knockdown was performed. Apoptosis was measured and quantified as described in (A). Error bars represent standard deviation of the mean of triplicate samples and data shown is representative of three independent experiments.
Discussion
Apoptosis is a complex biological process in multicellular organisms that eliminates damaged cells that would otherwise be deleterious to the individual as a whole.Citation29 Cancer cells often develop mechanisms to evade apoptosis, leading to unchecked proliferation of damaged or mutation-containing cells.Citation30 In addition to its role in cell proliferation, Rb is important in apoptosis. The function of Rb in apoptosis has been addressed in various studies. A role for Rb in apoptosis was initially identified using the Rb-null cell line, Saos-2. In these cells, overexpression of Rb inhibits ionizing radiation-induced apoptosis.Citation31 Additional studies showed that Rb inhibits cell death by interaction with apoptosis-promoting molecules. The c-abl tyrosine kinase is activated by DNA damage or Tumor Necrosis Factor–α (TNF-α) but interaction with Rb inhibits apoptosis.Citation32 In addition, the c-Jun N terminal kinase/stress-activated protein (JNK/SAPK) is involved in UV radiation mediated cell death, however Rb association with JNK/SAPK inhibits apoptosis.Citation33
The phosphorylation status of Rb regulates its function. Thus several studies have investigated the role of phosphorylation on the ability of Rb to regulate apoptosis. There is evidence to suggest that hyperphosphorylated Rb exerts protection against apoptotic cell death. First, sporadic cancers that contain hyperphosphorylated Rb due to upregulation of cdk activity in response to cyclin overexpression or cdki loss are often resistant to apoptosis induced by chemotherapy or radiation.Citation34 In various cell types, dephosphorylation of Rb is observed in apoptosisCitation8-11 and phosphatase activity has been shown to be required for apoptosis to occur.Citation12-14 Because Rb can be phosphorylated on 15 amino acids, the regulation and functions of each site have not yet been elucidated. Therefore the data on the subject of how Rb phosphorylation regulates apoptosis is still conflicting. In one study, Rb that could not undergo phosphorylation due to mutation of all the phosphorylation sites (PSM-RB) could stimulate the apoptotic response to TNFα in Rat-16 cells. However, in the same study, PSM-RB inhibited apoptosis initiated by doxorubicin.Citation16 Thus it is clear that the role of Rb phosphorylation in apoptosis may vary depending on the apoptotic stimulus.
Our study focused on specific phosphorylation sites of Rb with respect to the regulation of binding to Bax in apoptosis. The finding that Rb residing at the mitochondria is phosphorylated on specific sites (T821, S807/S811, S795, and S608) is evidence that phosphorylated Rb has non-nuclear functions in addition to its role in controlling proliferation. These particular phosphorylation sites were chosen for analysis due to the availability of reliable specific antibodies that recognize phosphorylation of Rb at these sites, and not that other phosphorylation sites are insignificant for the regulation of Rb function at the mitochondria. In fact we predict that as the ability to detect phosphorylation at additional sites becomes available, our understanding of the contribution of modification at each site will greatly increase. Cancer cells have elevated levels of phosphorylated Rb, and thus we were able to show that Rb phosphorylated at S807/S811 associates with Bax in an in vitro assay in 3 cell types, and by co-immunoprecipitation of endogenous proteins in 2 cell types. As described in the Results section, Rb phosphorylated on 2 additional sites (S608, S795) was found to associate with Bax in the in vitro assay however at much lower affinity. This observation shows that additional sites in addition to S807/S811 may be phosphorylated when Rb associates with Bax. This result is in agreement with the notion that phosphorylation of S807/S811 acts as a priming mechanism to promote phosphorylation of other sites of Rb.Citation35 In studies of the structural changes that occur due to phosphorylation of specific sites on Rb, it has been shown that certain phosphorylation events exert diverse effects upon the global conformation of Rb. However, phosphorylation of Rb on S807/S811 has little effect on the structure of Rb, instead it regulates phosphorylation at other sites, resulting in hyperphosphorylation. We next showed that blocking phosphorylation of Rb at S807 specifically inhibits complex formation with Bax. This finding is in agreement with previous studies that showed that phosphorylation at this site on Rb regulates binding to other Rb interacting proteins such as E2F and the c-abl tyrosine kinase.Citation20,21 In these studies, association of E2F or c-abl was inhibited by phosphorylation of Rb at S807, whereas in the current study phosphorylation of the site appears to promote binding to Bax. These differences are likely related to the differing functions of each of these Rb binding partners. Our results show that dephosphorylation of Rb on S807/S811 among other sites causes dissociation of the Rb-Bax complex and a four-fold increase in apoptosis. It may be that the specific collection of Rb sites that become dephosphorylated due to the increase in phosphatase activity utilized in our studies is required for and the signal to induce apoptosis in cancer cells. It has been previously shown that activation of phosphatase activity induces apoptosis, and that the effect is dependent on Rb.Citation36,37 Thus our study shows that in cells where Rb is dephosphorylated, the apoptosis that ensues may be due to loss of the Rb-Bax interaction that releases Bax to cause MOMP. Finally mimicking phosphorylation at S807 is able to block apoptosis due to PNUTS downregulation suggesting dephosphorylation of Rb on certain sites such as S807 leads to Bax dissociation which frees Bax to induce apoptosis.
A recent study clearly established that Rb regulates apoptosis by direct binding to pro-apoptotic Bax at the mitochondria.Citation26 The effect was verified to be independent of Rb transcriptional regulation using a mutant Rb tagged to the mitochondria. In addition, it was shown that phosphorylated Rb could stimulate apoptosis, which is incongruous with the conclusion of our present study. We believe this disparity in findings may be due to 2 possibilities. First, the previous study utilized mouse embryonic fibroblasts (MEFs) whereas we performed our analysis of the role of Rb phosphorylation in apoptosis using human breast cancer cells. Second, there were differences in how Rb phosphorylation was manipulated in the 2 studies. In Hilgendorf et al,Citation26 Rb phosphorylation was induced by siRNA mediated knockdown of p16, which allows unrestrained cdk activity. However, in our study of the effect Rb phosphorylation on apoptosis, we utilized a siRNA method that activates PP1 phosphatase activity that causes dephosphorylation of Rb. These 2 strategies may result in differences in site-specific phosphorylation of Rb, which may account for the disparate results. For example, in a previous study, we found that cdk inhibition using Roscovitine targets the S795 site of Rb, but not S807/S811 and S780, whereas upregulation of phosphatase activity using PNUTS siRNA targets S807/S811 and S780 as well as S795 in the same cells.Citation28 The outcome of these studies supports the highly complex nature of Rb regulation by phosphorylation. It could be envisioned that once specific functions for each phosphorylation site of Rb are completely understood, targeted therapies designed to activate the tumor suppressor function of Rb may be developed.Citation4,5
Materials and Methods
Cell culture
All cell lines utilized were obtained from ATCC unless otherwise specified. Cell culture materials were obtained from Invitrogen unless otherwise indicated. MCF7 and C33A cells were grown in Dulbecco's modified Eagle's media (DMEM), supplemented with 10% fetal bovine serum (FBS), 100 U/ml Penicillin, 100 μg/ml Streptomycin (Pen/Strep) and 2 mM glutamine. Hs578T cells were grown in DMEM media with 10% FBS, 0.01 mg/ml Insulin and Pen/Strep. Cells were routinely maintained at 37°C in a humidified, 5% CO2-containing atmosphere and were split 2–3 times weekly to maintain subconfluent cultures. Cell fractionation experiments were performed according to the manufacturers guidelines using the Mitochondria Isolation kit (Thermo Scientific).
Transfections
The CMV/Rb plasmid was obtained from Addgene. It was used to generate S/T to A or E mutations at the following amino acid sites: S807, S811 (Genewiz, Inc.). Plasmids were introduced into C33A cells using Fugene (Promega) performed according to the manufacturer's directions. Cells were harvested 48–72 h post-transfection. All transfections were performed using 10 μg of total DNA/60 mm dish. Expression of wild-type Rb and Rb mutant proteins was routinely verified by immunoblotting.
Pull down assay, immunoprecipitation, immunoblotting
Pull down assays, Immunoprecipitations, and Immunoblotting were performed as described elsewhere.Citation12,38 In this study, we utilized the following primary antibodies: Rb-phospho-821 (Pierce), Rb-phospho-780, Rb-phospho-608, Rb- phospho-795, Rb-phospho-807/811, AIF, Bak, Bax, cleaved PARP, (Cell Signaling Technology); or Rb (IF8, Santa Cruz Biotech); PNUTS (Transduction Labs), β –Actin (Sigma).
siRNA and apoptosis assays
Knockdown of PNUTS by siRNA has been previously described.Citation12 Cells were treated with either Non-targeting RNA or RNA targeting the PNUTS mRNA. The Cell Death Detection ELISA (Roche Diagnostics) was performed as directed by the manufacturer. Briefly, 104 cells from each condition were lysed and subjected to a slow spin centrifugation to pellet nuclei. Extracts from the cytoplasmic fraction were used to detect fragmented DNA, which was measured on a BioRad microplate reader, and experiments were conducted in triplicate.
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Funding
This work was supported by the National Cancer Institute of the NIH Award Numbers R15CA143390 and R15CA182723 awarded to NAK.
References
- Burkhart DL, Sage J. Cellular mechanism of tumor suppression by the retinoblastoma gene. Nat Rev Cancer 2008; 8:671-82; PMID:18650841; http://dx.doi.org/10.1038/nrc2399
- Mittnacht S. The retinoblastoma protein- from bench to bedside. Eur J Cell Biol 2005; 84:97-107; PMID:15819393; http://dx.doi.org/10.1016/j.ejcb.2004.12.012
- Sherr CJ, McCormick F. The RB and p53 pathways in cancer. Cancer Cell 2002; 2:103-12; http://dx.doi.org/10.1016/S1535-6108(02)00102-2
- Knudsen ES, Knudsen KE. Tailoring to Rb: tumour suppressor status and therapeutic response. Nat Rev Cancer 2008; 8:714-24; PMID:19143056; http://dx.doi.org/10.1038/nrc2401
- Knudsen ES, Wang JYJ. Targeting the RB-pathway in cancer therapy. Clin Cancer Res 2110; 16:1094-9; PMID:20145169; http://dx.doi.org/10.1158/1078-0432.CCR-09-0787
- Stone A, Sutherland RL, Musgrove EA. Inhibitors of cell cycle kinases: recent advances and future prospects as cancer therapeutics. Crit Rev in Oncog 2012; 17:175-98; PMID:22471707; http://dx.doi.org/10.1615/CritRevOncog.v17.i2.40
- Sherr CJ. Cancer Cell cycles. Science 1996; 274:1672-7; http://dx.doi.org/10.1126/science.274.5293.1672
- Jeon HS, Dracheva T, Tang SH, Meerzaman D, Fukuoka J, Shakoori A, Shilo K, Travis WD, Jen J. SMAD6 contributes to patient survival in non-small cell lung cancer and its knockdown reestablishes TGF-beta homeostasis in lung cancer cells. Cancer Res 2008; 68:9686-92; PMID:19047146; http://dx.doi.org/10.1158/0008-5472.CAN-08-1083
- Knudsen KE, Booth D, Naderi S, Sever-Chroneos Z, Fribourg AF, Hunton IC, Feramisco JR, Wang JYJ, Knudsen ES. Rb-dependent S-phase response to DNA damage. Mol Cell Biol 2000; 20:7751-63; PMID:11003670; http://dx.doi.org/10.1128/MCB.20.20.7751-7763.2000
- Payton M, Chung G, Yakowec P, Wong A, Powers D, Xiong L, Zhang N, Leal J, Bush TL, Santora V, et al. Discovery and evaluation of dual cdk1 and cdk2 inhibitors. Cancer Res 2006; 66:4299-308; PMID:16618755; http://dx.doi.org/10.1158/0008-5472.CAN-05-2507
- Plastaras JP, Kim SH, Liu YY, Dicker DT, Dorsey JF, McDonough J, Cerniglia G, Rajendran RR, Gupta A, Rustgi AK, et al. Cell Cycle dependent and schedule dependent antitumor effects of sorafenib combined with radiation. Cancer Res 2007; 67:9443-54; PMID:17909054; http://dx.doi.org/10.1158/0008-5472.CAN-07-1473
- De Leon G, Sherry TC, Krucher NA. Reduced expression of PNUTS leads to activation of Rb-phosphatase and caspase-mediated apoptosis. Cancer Bio Ther 2008; 7: 833-9; PMID:18360108; http://dx.doi.org/10.4161/cbt.7.6.5839
- Dou QP, An B, Will PL. Induction of a retinoblastoma phosphatase by anticancer drugs accompanies p53-independent G1 arrest and apoptosis. PNAS USA 1995; 92:9019-23; PMID:7568064; http://dx.doi.org/10.1073/pnas.92.20.9019
- Popowski M, Ferguson HA, Sion AM, Koller E, Knudsen E, Van Den Berg CL. Stress and IGF-1 differentially control cell fate through mammalian target of rapamycin (mTOR. and retinoblastoma protein (pRb). J Biol Chem 2008; 283:28265-73; PMID:18697743; http://dx.doi.org/10.1074/jbc.M805724200
- Lentine B, Antonucci L, Hunce R, Edwards J, Marallano V, Krucher NA. Dephosphorylation of threonine-821 of the retinoblastoma tumor suppressor protein (Rb) is required for apoptosis induced by UV and Cdk inhibition. Cell Cycle 2012; 11:3324-30; PMID:22895174; http://dx.doi.org/10.4161/cc.21693
- Masselli A, Wang JYJ. Phosphorylation site mutated RB exerts contrasting effects on apoptotic response to different stimuli. Oncogene 2006; 25:1290-8; PMID:16205627; http://dx.doi.org/10.1038/sj.onc.1209161
- Taya Y. Rb kinases and Rb binding proteins: new points of view. Trends Biochem Sci 1997; 22:14-7; PMID:9020586; http://dx.doi.org/10.1016/S0968-0004(96)10070-0
- Burke JR, Deshong AJ, Pelton JG, Rubin SM. Phosphorylation-induced conformational changes in the retinoblastoma protein inhibit E2F transactivation domain binding. J Biol Chem 2010; 285:16286-93; PMID:20223825; http://dx.doi.org/10.1074/jbc.M110.108167
- Delston RB, Matatall KA, Sun Y, Onken MD, Harbour JW. p38 phosphorylates Rb on Ser567 by a novel, cell cycle-independent mechanism that triggers Rb-Hdm2 interaction and apoptosis. Oncogene 2011; 30:588-99; PMID:20871633; http://dx.doi.org/10.1038/onc.2010.442
- Knudsen ES, Wang JYJ. Differential regulation of retinoblastoma protein function by specific cdk phosphorylation sites. J Biol Chem 1996; 271:8313-20; PMID:8626527; http://dx.doi.org/10.1074/jbc.271.14.8313
- Knudsen ES, Wang JYJ. Dual mechanisms for the inhibition of E2F binding to Rb by cyclin-dependent kinase mediated Rb phosphorylation. Mol Cell Bio 1997; 17:5771-83; PMID:9315635
- Rubin SM, Deciphering the retinoblastoma protein phosphorylation code. Trends Biochem Sci 2013; 38:12-9; PMID:23218751; http://dx.doi.org/10.1016/j.tibs.2012.10.007
- Ma D, Zhou P, Harbour JW. Distinct mechanisms for regulating the tumor suppressor and antiapoptotic functions of Rb. J Biol Chem 2003; 278:19358-66; PMID:12646568; http://dx.doi.org/10.1074/jbc.M301761200
- Agdebola O, Pasternack GR. Phosphorylated retinoblastoma protein complexes with pp32 and inhibits pp32-mediated apoptosis. J Biol Chem 2005; 280:15497-502; PMID:15716273; http://dx.doi.org/10.1074/jbc.M411382200
- Ferecatu I, Le Floch N, Bergeaud M, Rodríguez-Enfedaque A, Rincheval V, Oliver L, Vallette FM, Mignotte B, Vayssière J-L. Evidence for a mitochondrial localization of the retinoblastoma Protein. BMC Cell Biol 2009; 10:50; http://dx.doi.org/10.1186/1471-2121-10-50
- Hilgendorf KI, Leshchiner ES, Nedelcu S, Maynard MA, Calo E, Ianari A, Walensky LD, Lees JA. The retinoblastoma protein induces apoptosis directly at the mitochondria. Genes Dev 2013; 27:1003-15; http://dx.doi.org/10.1101/gad.211326.112
- Walensky LD, Gavathiotis E. Bax unleashed: the biochemical transformation of an inactive cytosolic monomer into a mitochondrial pore. Trends Biochem Sci 2011; 36:642-52; PMID:21978892; http://dx.doi.org/10.1016/j.tibs.2011.08.009
- De Leon G, Cavino M, D’Angelo M, Krucher NA. PNUTS knockdown potentiates the apoptotic effect of roscovitine in breast and colon cancer cells. Int J Oncol 2010; 36:1269-75; PMID:20372802
- Taylor RC, Cullen SP, Martin SJ. Apoptosis: controlled demolition at the cellular level Nat Rev Mol Cell Biol 2008; 9:231-41; PMID:18073771; http://dx.doi.org/10.1038/nrm2312
- Hanahan D, Weinberg RA. The hallmarks of cancer, Cell 2000; 100:57-70.
- Haas-Kogan D, Kogan SC, Levi D, Dazin P, T’Ang A, Fung Y-KT, Isreal MA. Inhibition of apoptosis by retinoblastoma gene product. EMBO J 1995;14: 461-72.
- Chau BN, Chen T-T, Wan YY, DeGregori J, Wang JYJ. Tumor necrosis factor alpha-induced apoptosis requires p73 and c-ABL activation downstream of RB degradation. Mol Cel Biol 2004; 24:4438-47; PMID:15121862; http://dx.doi.org/10.1128/MCB.24.10.4438-4447.2004
- Shim J, Park H-S, Kim MJ, Park J, Park E, Cho S-G, Eom S-J, Lee H-W, Joe CO, Choi E-J. Rb protein down-regulates the stress-activated signals through inhibiting c-Jun N-terminal kinasestress-activated protein kinase. J Biol Chem 2000; 275:14107-11; PMID:10799486; http://dx.doi.org/10.1074/jbc.275.19.14107
- Shackney SE, Shankey TV. Cell cycle models for molecular biology and molecular oncology: exploring new dimensions. Cytometry 1999; 35:97-116; PMID:10554165; http://dx.doi.org/10.1002/(SICI)1097-0320(19990201)35:2<97::AID-CYTO1>3.0.CO;2-5
- Rubin SM. Deciphering the retinoblastoma protein phosphorylation code. TIBS 2013; 38:12-9; PMID:23218751; http://dx.doi.org/10.1016/j.tibs.2012.10.007
- Berndt N, Dohadwala M, Liu CWY. Constitutively active protein phosphatase 1α causes Rb-dependent G1 arrest in human cancer cells. Curr Biol 1997; 7:375-86; PMID:9197238; http://dx.doi.org/10.1016/S0960-9822(06)00185-0
- Wang R-H, Liu CWY, Avramis VI, Berndt N. Protein phosphatase 1α-mediated stimulation of apoptosis is associated with dephosphorylation of the retinoblastoma protein. Oncogene 2001; 20:6111-22; PMID:11593419; http://dx.doi.org/10.1038/sj.onc.1204829
- Krucher NA, Zygmunt A, Mazloum N, Tamrakar S, Ludlow JW, Lee YMWT. Interaction of the retinoblastoma protein (pRb) with the catalytic subunit of DNA polymerase δ (p125). Oncogene 2000; 19:5464-70; PMID:11114723; http://dx.doi.org/10.1038/sj.onc.1203930
