Abstract
Survivin is a multitasking protein that can inhibit cell death and that is essential for mitosis. Due to these prosurvival activities and the correlation of its expression with tumor resistance to conventional cancer treatments, survivin has received much attention as a potential oncotherapeutic target. Nevertheless, many questions regarding its exact role at the molecular level remain to be elucidated. In this study we ask whether the extreme C- and NH2 termini of survivin are required for it to carry out its cytoprotective and mitotic duties. When assayed for their ability to act as a cytoprotectant, both survivin1–120 and survivin11–142 were able to protect cells against TRAIL-mediated apoptosis, but when challenged with irradiation cells expressing survivin11–142 had no survival advantage. During mitosis, however, removing the NH2 terminal 10 amino acids (survivin11–142) had no apparent effect but truncating 22 amino acids from the C-terminus (survivin1–120) prevented survivin from transferring to the midzone microtubules during anaphase. Collectively the data herein presented suggest that the C-terminus is required for cell division, and that the NH2 terminus is dispensable for apoptosis and mitosis but required for protection from irradiation.
Abbreviations
| CPC | = | chromosomal passenger complex |
| CPP | = | chromosomal passenger protein |
| IAP | = | inhibitor of apoptosis |
| IR | = | irradiation |
| NES | = | nuclear exportation signal |
| SVN | = | survivin |
| WT | = | wild type |
| TRAIL | = | Tumor-necrosis factor Responsive Apoptosis Inducing Ligand |
Introduction
Survivin is a prosurvival factor that inhibits cell death and is essential for mitosis. It is overexpressed in all cancers Citation1 and its abundance correlates with tumor resistance to conventional therapies including chemo and radiation.Citation2-4 Thus it has received much attention scientifically and as a potential oncotherapeutic target.
The crystal structure of human survivin has been solved in 2 forms, as a homodimer,Citation5,6 and in complex with its mitotic partners, borealin and INCENP.Citation7 These structural analyses have revealed that survivin is essentially a globular protein with an outward projecting C-terminal α helix of approximately 40 amino acids (). It homodimerises via a short linker positioned centrally between the globular domain and the extended helical tail, and engages leucine 6 and tryptophan 10 from the NH2 terminus at this interface to secure the interaction.Citation5,6,8 The central linker also contains a nuclear export signal (NES), which actively shuttles survivin into the cytoplasm in a CRM1 dependent manner.Citation9-11 If the NES is mutated or survivin is artificially relocated to the nucleus, it no longer protects cells from apoptosis or radiation.Citation9 This loss of cytoprotection might be due to more rapid turnover of survivin in the nucleus.Citation12 However, there is also evidence that nuclear survivin may play a protective role by reducing DNA damage or facilitating DNA repair as cells expressing survivin have fewer DNA lesions when compared with control cells.Citation13 Reconciling this apparent paradox, when cells are exposed to irradiation (IR), survivin relocates from the cytoplasm to the nucleus.Citation13,14 Given that forced nuclear relocation eliminates cytoprotection, these data suggest that survivin may be post-translationally modified when retained in the nucleus post-IR, indeed acetylation at K129, has been implicated in nuclear enrichment.Citation15
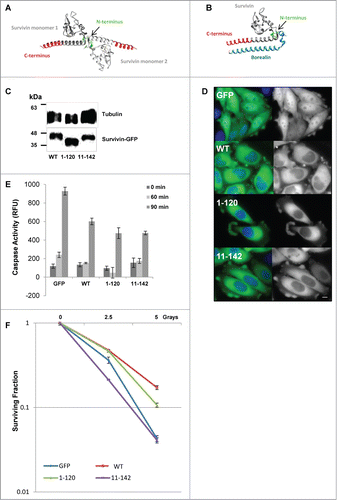
Figure 1. Expression of survivin truncations and their ability to protect cells from death threats. (A and B) 3D models of the crystal structure of (A) the survivin homodimer (1F3H), and (B) survivin bound to the NH2 terminus of borealin (2RAW). Models were created using UCSF Chimera Modeling software Citation29. (A) Survivin monomers are shown in light and dark gray. The terminal 22 amino acids removed in survivin1–120 are indicated in red, the NH2 terminus of survivin is shown in green. Note that only amino acids 6–10 are highlighted as residues 1–5 remain unresolved. (B) The NH2 terminus of borealin is indicated in blue. (C) An immunoblot showing expression of the ectopic forms of survivin in each stable line; tubulin indicates loading. (D) Fluorescence imaging reveals that all forms examined are predominantly cytoplasmic. Scale bar 5 μm. (E) Caspase activity assay in response to TRAIL treatment over 90 minutes expressed in relative fluorescence units (RFU). Error bars indicate standard deviation from the mean. (F) Clonogenic survival in response to X-irradiation plotted on a logarithmic scale. Experiments were performed in triplicate, 3 independent times.
During mitosis survivin is a chromosomal passenger protein (CPP), which operates in the chromosomal passenger complex (CPC) with aurora-B, borealin and INCENP. The CPC facilitates the correction of maloriented chromosomes during prometaphase congression, and the execution of cytokinesis.Citation16,17 When bound to its mitotic partners, the C-terminus becomes part of a 3 helix bundle comprised of the NH2 terminus of borealin (see ), and the coiled coil region of INCENP.Citation7 Survivin targets the CPC to the centromere, via two acidic residues D71D72, in the BIR domain, which bind to threonine 3 of Histone H3, when it has been phosphorylated by the mitotic kinase, haspin.Citation18
The aim of this study was to determine whether the C- and NH2-termini of survivin are required for its known roles in cytoprotection and/or mitosis.
Results and Discussion
In this report whether the extreme C- and NH2-terminal ends of survivin are required for its ability to inhibit cell death, or for its essential function during mitosis was investigated. To do this 2 truncation mutants were generated, one in which the C-terminal 22 amino acids were removed (see ), referred to as survivin1–120; and the other lacking the first 10 amino acids, survivin11–142. Expression constructs encoding these truncations were transfected into HeLa cells and cell lines stably expressing these forms were generated. GFP and survivin-GFP expressing cells were used as controls, see.Citation19 Immunoblotting was used to verify the size of ectopic proteins and compare their levels of expression (). Fluorescence imaging revealed that 90% of all cells were expressing these constructs (data not shown), and like wild type (WT) both survivin1–120 and survivin11–142 were predominantly localized within the cytoplasm of interphase cells.()
Survivin terminal truncations can inhibit apoptosis
To test whether these truncation mutants could protect cells against apoptosis, asynchronous cells expressing GFP, or the GFP tagged survivin forms indicated, were treated with TRAIL for 0, 60, or 90 minutes, whole cell lysates prepared, and apoptosis assessed using a caspase-3 tetrapeptide cleavage assay (). In this assay lysates from cells overexpressing WT reduced caspase-3 activity compared with cells expressing GFP alone, as expected. Lysates prepared from cells expressing survivin1–120 and survivin11–142 were also able to protect cells against this apoptotic challenge, demonstrating that neither end of the protein is required to inhibit TRAIL- mediated apoptosis.
NH2 truncation eliminates protection from X-irradiation
Next to test the ability of survivin mutants to protect cells against a second death-signaling pathway, cells were exposed to increasing doses of X-rays (0, 2.5 and 5 Gy). Survival was monitored using a clonogenic assay in which colonies of 50 cells or more were counted 7 d post-treatment, and plotted on a logarithmic scale as the “surviving fraction.” As shown in , when compared to control cells expressing only GFP, WT protected cells against this challenge. Surprisingly, despite effectively protecting cells against TRAIL, cells expressing survivin11–142 exhibited the same sensitivity to IR as GFP control cells, while survivin1–120, conferred a similar level of protection against this insult as cells expressing the WT form. These data suggest that the NH2 terminus is required for survivin to mediate protection from IR.
This finding is intriguing as survivin11–142 is the first form that we have encountered that can protect cells from TRAIL-mediated apoptosis but not IR-mediated death, and suggests that survivin may operate in distinct survival pathways in response to TRAIL and IR. Moreover, it suggests that although exclusion from the nucleus is necessary for survivin to protect cells against IR,Citation9,12 it is not sufficient: the NH2 terminus is also required. X-irradiation induces double strand DNA breaks and survivin expression has been shown to reduce DNA lesions post IR,Citation13 thus it is possible that the NH2 terminus is engaged in some aspect of DNA repair, see also.Citation20,21 However, in addition to damaging DNA, recent evidence has demonstrated that irradiation induces autophagy,Citation22 an intracellular recycling program that is activated in times of stress. At its outset autophagy is prosurvival, but if it persists excessive recycling can ultimately kill the cell, hence it is often considered to be a second form of programmed cell death, reviewed in.Citation23 Whether survivin is part of the autophagic response remains to be determined but could explain the dichotomous responses of survivin11–142 cells to TRAIL and IR. Clearly, as survivin is overexpressed in all cancers, and its abundance correlates with tumor resistance to radiotherapy, the modus operandi of survivin in protecting cells from IR warrants further investigation.
Localization of survivin truncations during mitosis
To investigate whether the ends of survivin are required for cell division the localization of these truncation mutants during mitosis was examined. Survivin has a very distinct pattern of localization during mitosis.Citation24, 25 It is centromeric until the metaphase-anaphase transition, after which it transfers to the central anaphase spindle, decorates the equatorial cortex at the site where the cell will form the cleavage furrow, and finally it is discarded from the cell during cytokinesis via midbody externalisation (). Interestingly, although the C-terminal truncation, survivin1–120 localized to the centromeres during early mitosis, it was not specifically confined to these foci, instead it was distributed all along the chromosome arms (, upper panel). Most strikingly instead of transferring to the midzone during anaphase, survivin1–120 remained associated with the chromosome arms and appeared to become enriched at the ends of the separating chromosomes (, middle panels). The NH2-terminal truncation, survivin11–142 also mislocalised but in contrast to survivin1–120, it was simply found diffusely localized at all stages (). Neither truncation concentrated at the midbody ( and , lower panels). The inability of these mutants to localize to the central anaphase spindle was not due to a defect in this structure itself as intact midzone microtubules were clearly evident in fixed anaphase cells immunoprobed with anti-tubulin antibodies (). We also noted that the chromosomal localization of survivin1–120 witnessed in live cells was compromised when cells were fixed, compare middle panels in and
Figure 2. Survivin truncations mislocalise during mitosis. (A–C) Exponentially growing HeLa cells expressing (A) survivin-GFP (WT); (B) survivin1–120-GFP and (C) survivin 11–142-GFP as indicated, were stained with NucBlue and imaged live. (D) Anaphase cells were fixed with formaldehyde and immunoprobed with anti-tubulin antibodies to show the integrity of the central spindle in the different lines. Scale bars 5 μm. (E) Analysis of mitotic stages of cells 120 minutes post-release from DMA-induced mitotic arrest.
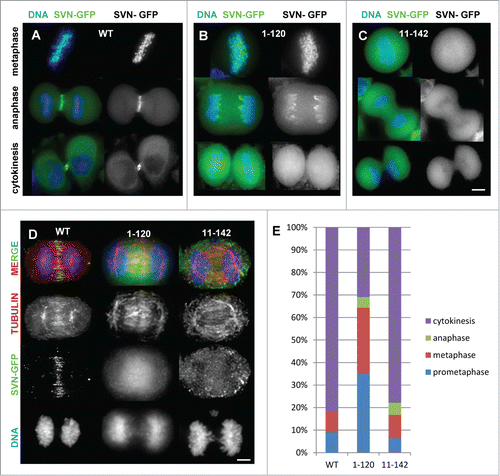
The competence of these versions of survivin to correct maloriented chromosomes was then assessed using an error correction assay. Briefly, cells were arrested in mitosis with monopolar spindles and syntelically attached chromosomes using the Eg5 inhibitor dimethylenastron (DMA), harvested by mitotic shake-off, then released for 120 minutes before fixing and immunoprobing with anti-tubulin antibodies. The percentage of cells in each mitotic stage was then assessed by fluorescence microscopy and quantified (). As judged by the percentage of cells persisting in prometaphase at 120 minutes (35%), survivin1–120 was less efficient at correcting maloriented chromosomes compared to either WT or survivin11–142 which both had a majority (approx. 80%) of their populations in cytokinesis. In addition we noted that while only 10.9% (N = 92; WT) and 10% (N = 70; survivin11–142) cells exhibited abnormalities during mitotic exit, 92.3% (N = 26) of the survivin1–120 population were aberrant, clearly demonstrating that this form causes genomic instability during mitosis.
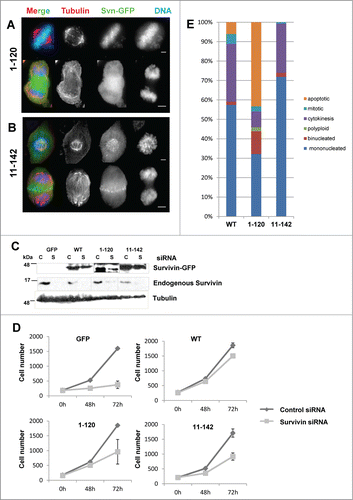
In all cells examined in the native protein as well as the ectopic form was present. Thus to ascertain whether the localization of the ectopic forms was influenced by the endogenous protein the distribution of the siRNA resistant truncation mutants was re-examined after depletion of the native protein. Surprisingly, although the localization of survivin1–120 remained unchanged (), removing native survivin enabled survivin11–142 to localize normally, gaining access to the centromeres and the midzone during prometaphase and anaphase respectively (). Removal of endogenous survivin and resistance of the ectopic forms to siRNA was verified by immunoblotting (). Mislocalisation only in the presence of the endogenous protein suggests competition rather than interaction and raises the question as to whether the NH2 terminus is really involved in survivin dimerization, as has been suggested for L6 and W10.Citation5 Note also that removal of the endogenous protein from survivin11–142 cells does not impact on the outcome of the TRAIL assay, survivin11–142 remains protective in its absence (data not shown). On the flipside, loss of this end is not expected to interfere with the essential mitotic borealin-INCENP helix interactionCitation7 (see ). These experiments suggest that the NH2 terminus is dispensable for mitosis, but that the C-terminus facilitates centromere targeting and is required for transfer to the central spindle during anaphase, data that concur with.Citation26
Figure 3. Mitotic competency of survivin truncations. Endogenous survivin was removed by siRNA from cells expressing survivin1–120-GFP (A) or survivin 11–142-GFP. (B) 72 h later cells were fixed and immunoprobed with anti-tubulin antibodies (red) and counterstained with DAPI (blue). Scale bars 5 μm. (C) Immunoblotting with anti-survivin antibodies confirming removal of the endogenous protein, and resistance of the ectopic form. Tubulin is shown to indicate equality in loading. C = control, S = survivin specific siRNA. (D) Cell proliferation over 72h was assessed using a metabolic (resazurin) assay. Average and standard deviation of cell number is plotted on the Y-axis. (E) Analysis of cell condition 72h post-siRNA treatment in each population, viewed in real time by fluorescence imaging using GFP and NucBlu signals.
Truncation of the C-terminus inhibits mitosis.
To determine whether these mutants can support mitosis alone, cell proliferation was monitored at 0, 48 and 72h post-siRNA. As shown in survivin depletion inhibited proliferation of cells expressing GFP alone (control), while those expressing the siRNA resistant WT form, continued to grow. By contrast, although populations of both survivin1–120 and survivin11–142 -expressing cells expanded, they did not rescue proliferation as efficiently as WT suggesting a partial, but incomplete rescue. Next the DMA arrest and release assay was repeated in cell populations that had been depleted of endogenous survivin for 48h, and their ability to exit mitosis analyzed by fluorescence imaging. As shown in , while the majority of WT and survivin11–142 -expressing cells were either completing a normal cytokinetic event, or were in G1 (mononucleated) 2h post-release from DMA, the sur- vivin1–120 experienced many difficulties, which resulted in 14% with multiple nuclei, 43% undergoing apoptosis.
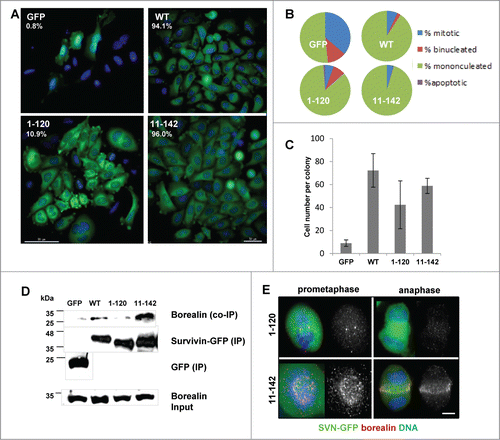
As survivin1–120 and survivin11–142 clearly were different in their mitotic competency, yet the proliferation assay in suggested that they were equally effective at (partially) restoring cell growth, the siRNA experiment was extended to a clonogenic analysis to assess survival more specifically. Briefly, cells that had been exposed to control or survivin specific siRNA for 72 h were seeded at low density onto live imaging chambers and petri dishes. The number of colonies that formed in the survivin siRNA treated populations was counted and expressed as a percentage of the control siRNA treated population (top left corner of each panel, ). In addition, fluorescence imaging of colonies seeded onto live imaging chambers revealed that the GFP control cells were unable to form viable colonies (), while those expressing WT survivin grew into uniform colonies with cells of regular size. This assay clearly demonstrated that survivin11–142 expressing cells form a similar number of highly regular exponentially growing colonies to WT, however, consistent with the mitotic defects observed in previous experiments, the colonies that developed from cells expressing survivin1–120, contained many binucleated and multinucleated cells () further confirming that mitosis and cytokinesis was impaired in this population. The average number of cells in each colony was also counted (), and was found to be highest in the WT (72.3+/−14.6) population and lowest and most variable in the population expressing survivin1–120 (42.4+/−20.8). Interestingly, survivin11–142 expressing colonies had on average 58.8+/−6.6 cells, suggesting that the less efficient rescue observed for this cell line in the proliferation assay may be due to a reduced rate of cell growth, rather than any reduction in viability.
Figure 4. C-terminal truncation inhibits borealin interaction and inhibits mitosis. (A) To determine viability over a longer period, cells treated for 72 h with siRNA were re-seeded at low density and the resultant clonal morphologies assessed 7 d later. Scale bar 50 μm. The number of colonies that formed as expressed in the top left corner as a percentage of colonies on control siRNA treated plates. (B) Quantitation of the percentage of cells that are mononucleated, binucleated, apoptotic or mitotic within individual colonies. (C) Average number of cells in each colony and the standard deviation from the mean. (D) Immunoprecipitation using anti-GFP antibodies and immunoblot analysis to assess interaction with endogenous borealin. Equality of borealin expression is indicated in lower panel which is an immunoblot of whole cell extracts. (E) Immunofluorescence of fixed cells in prometaphase or anaphase, probed with anti-borealin antibodies, and visualised with Cy5 anti-rabbit secondary antibodies. Bar, 5 μm.
Truncation of the C-terminus disrupts interaction with borealin
The observation that survivin1–120 expressing cells fail to divide is consistent with previous studies that have reported that the C-terminal helix is responsible for binding to borealin (see ). Indeed, in a parallel paper, we recently reported that substitution of a lysine at position 129, for a glutamic acid, K129E, is sufficient to inhibit survivin from interacting with borealin.Citation27 Thus to confirm that C-terminal truncation abrogates interaction with borealin, an immunoprecipitation assay was performed with each of the cell lines. Immunoblotting analysis after immunoprecipitation with anti-GFP antibodies revealed that borealin does indeed co-immunoprecipitate with WT survivin during mitosis, as does survivin11–142, but not with survivin1–120 (). Finally, immunolocalisation in fixed cells revealed that borealin was misplaced in prometaphase and anaphase cells expressing only survivin1–120, but localized normally in cells expressing survivin11-142 ().
In summary, the data herein presented suggest that the last 22 amino acids of survivin are required to bind to borealin and support mitosis and cytokinesis, whereas the first 10 amino acids are needed to protect cells from irradiation. How this short sequence contributes to protection from IR remains to be determined but initial data suggest that its removal may influence the rate of cell growth.
Materials and Methods
Unless otherwise stated, tissue culture reagents were obtained from www.lifetechnologies.com, and all other reagents were from www.sigmaaldrich.com.
Molecular biology
Survivin truncations were generated by PCR using VENT polymerase (www.neb.com), with wild type human survivin (accession number NM001168) in pBluescript as template, see.Citation24,28 Once generated, mutants were subcloned into the mammalian expression vector pcDNA3.1 (Invitrogen) such that the protein was fused at its C-terminus to GFP. All final constructs were sequence verified prior to use.
Cell Lines, transfection and proliferation
HeLa cell lines stably expressing GFP, survivin-GFP, survivin1–120-GFP or survivin11–142-GFP were established by transfection with the relevant pcDNA3.1 constructs using Transit LT1 (ww. mirusbio.com) diluted in Optimem, and selection post-transfection with 500 μg/ml G418, followed by clonal identification, pooling and FACS sorting on the GFP signal. Cells were cultured at 37˚C and 5% CO2 in DMEM with 10% Hyclone Bovine Growth Serum (www.thermoscientific.com), 1% penicillin-streptomycin; 1% fungizome and 500 μg/ml G418. Cell proliferation was assessed in triplicate using a resazurin assay in which cells grown in 96 welled plates were incubated for 1 h at 37˚C in 10 μg/ml resazurin (Sigma R7017) prepared in complete medium. Resazurin signal was measured using a spectrophotometer (FLUOstar Galaxy, BMG Labtechnologies) with excitation set at 530 nm and emission at 590 nm.
Fluorescence imaging
Cells were either grown on poly-l-lysine coated 4-chambered slides (nunc Lab-TekII, www.thermoscientific.com) or 13 mm2 glass coverslips. To image cells live, the growth medium was replaced with CO2 independent, phenol red free medium containing NucBlue (www.lifetechnologies.com) to visualize the DNA. To fix cells for immunofluorescence, coverslips were incubated with 4% formaldehyde (37˚C, 5 min) and permeabilised in 0.15% triton in PBS (37˚C, 2 min). Before immunoprobing, cells were blocked with 1% BSA in PBS, then incubated sequentially for 1 h at room temperature with anti-tubulin (B512, 1/2000, Sigma, T5168) or anti-borealin (1/500, in-house), then texas-red conjugated anti-mouse secondary antibodies (1/200, www.vectorlabs.com) or Cy5-anti-rabbit antbodies (1/1000, www.abcam.com). All fixed samples were counterstained with DAPI and mounted in 1% propyl gallate/ glycerol mounting medium. Images were acquired using an Olympus inverted microscope fitted with a x 20, or a x 63 (NA 1.4, oil) objective and operated with Deltavision Software (api.gehealthcare.com). High resolution images are presented as 2D projections of 0.3 μm stepped Z-sections and processed for presentation using Adobe Photoshop, adhering to image manipulation regulations.
RNAi
Exponentially growing HeLa cells were seeded immediately before RNAi transfection at a density of 5×104 per well of a 24-welled plate and cultured in antibiotic free DMEM with 10% FCS. Control and survivin specific oligonucleotides (Ambion, www.lifetechnologies.com) diluted in OptiMEM were transfected into cells at a final concentration of 10 nM, using HiPerfect (www.qiagen.com), and cells allowed a minimum of 48 h to grow in antibiotic free medium before analysis. RNAi insensitive versions of survivin were made resistant to siRNA knockdown by a base substitution C54G (Wheatley et al., 2004).
Immunoblotting and immunoprecipitation
Cells were lysed in RIPA buffer (20mM Tris, pH8.0, 137 mM NaCl, 0.5 mM EDTA, 10% glycerol, 1% Non-Idet P40, and 1% SDS) with protease and phosphatase inhibitors. Cell lysates were then boiled in Laemmli sample buffer, and 50 μg protein lysate loaded per lane. Proteins were separated by SDS-PAGE (12%) and transferred to 0.22 μm nitrocellulose (www.pall.com) using standard Tris/glycine based methods. The following antibodies were used: anti-survivin (in-house; 1/1000) and anti-tubulin (B512, 1/2000, Sigma, T5168). HRP-conjugated secondary antibodies (diluted 1/2000) were from www.dako.com. All antibodies were prepared in 5% milk (Marvel) in PBS with 0.1% Tween 20. Signals were detected using enhanced chemiluminescence and hyperfilm (www.gehealthcare.com).
Immunoprecipitation was carried out as described in,Citation27 except that protein G Dynabeads (www.lifetechnologies.com) were used.
Caspase assay
Cells were seeded at 1×105 per well in 24-welled plates, and cultured for 16 h before induction of apoptosis. Cells were treated with 250 ng/ml recombinant TRAIL (www.peprotech.com cat no. 310–04) for the times indicated, then lysed in 200 μl of mammalian protein extraction reagent (MPER; www.pierce.com) containing 1 mM EDTA, 1 μg/μl pepstatin A, 1 mM AEBSF for 45 minutes. Caspase activity assays were performed in a 96-well microtiter plate: 4 μg of the caspase-3 substrate Ac-DEVD-AMC (Biomol cat no. P-411, www.enzolifesciences.com) was incubated with 40 μl of cell lysate and 200 μl of reaction buffer (20 mM HEPES, 10% glycerol, 2 mM DTT, pH 7.5) at 37°C for 1 h. Fluorescent emission (450 nm) was measured using a Spectrofluorometer (Fluostar Galaxy, BMG Labtechnologies) with excitation wavelength set at 390 nm.
X-Irradiation and clonogenic survival Aasays
Cells were seeded at low density in 10 cm2 Petri dishes and irradiated 2 h later at 0, 2.5, or 5 Grays using an Hs-X-ray System (A.G.O Installations Ltd). Seven days post-irradiation cultures were stained with 1% methylene blue in absolute ethanol (1 h, 25°C), and colonies of >50 cells counted.
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Acknowledgements
I would like to thank Mr. Jamie Webster and Ms. Elisa Marelli for technical support. Thanks also to Jessica Thomas, Yanisa Jarusyingdumrong and Katrina Garcha for their assistance with experiments during their undergraduate dissertations.
Funding
Imaging was performed in the Advanced Microscopy Unit, at the University of Nottingham with a DeltaVision Elite system funded by the Wellcome Trust (Award number WT094233MA).
References
- Velculescu VE, Madden SL, Zhang L, Lash AE, Yu J, Rago C, Lal A, Wang CJ, Beaudry GA, Ciriello KM, et al. Analysis of human transcriptomes. Nat Gen 1999; 23:387-8; PMID:10581018; http://dx.doi.org/10.1038/70487
- Altieri DC. Survivin, cancer networks and pathway-directed drug discovery. Nature Reviews Cancer 2008; 8:61-70; PMID:18075512; http://dx.doi.org/10.1038/nrc2293
- Guha M, Altieri DC. Survivin as a global target of intrinsic tumor suppression. Cell Cycle 2009; 8:2708-10; PMID:19717980; http://dx.doi.org/10.4161/cc.8.17.9457
- Zaffaroni N, Pennati M, Daidone MG. Survivin as a target for new anticancer interventions. J Cell Mol Med 2005; 9:360-72; PMID:15963255; http://dx.doi.org/10.1111/j.1582-4934.2005.tb00361.x
- Verdecia MA, Huang H, Dutil E, Kaiser DA, Hunter T, Noel JP. Structure of the human anti-apoptotic protein survivin reveals a dimeric arrangement. Nat Struct Biol 2000; 7:602-8; PMID:10876248; http://dx.doi.org/10.1038/77929
- Chantalat L, Skoufias DA, Kleman JP, Jung B, Dideberg O, Margolis RL. Crystal structure of human survivin reveals a bow tie-shaped dimer with two unusual α-helical extensions. Mol Cell 2000; 6:183-9; PMID:10949039; http://dx.doi.org/10.1016/S1097-2765(00)00019-8
- Jeyaprakash AA, Klein UR, Lindner D, Ebert J, Nigg EA, Conti E. Structure of a survivin-borealin-INCENP core complex reveals how chromosomal passengers travel together. Cell 2007; 131:271-85; PMID:17956729; http://dx.doi.org/10.1016/j.cell.2007.07.045
- Sun C, Nettesheim D, Liu Z, Olejniczak ET. Solution structure of human survivin and its binding interface with Smac/DIABLO. Biochem 2005; 44:11-7; PMID:15628841; http://dx.doi.org/10.1021/bi0485171
- Colnaghi R, Connell CM, Barrett RMA, Wheatley SP. Separating the anti-apoptotic and mitotic roles of survivin. J Biol Chem 2006; 281:33450-6; PMID:16950794; http://dx.doi.org/10.1074/jbc.C600164200
- Knauer SK, Bier C, Habtemichael N, Stauber RH. The survivin-Crm1 interaction is essential for chromosomal passenger complex localization and function. EMBO Reports 2006; 12:1259-65; PMID:17099693; http://dx.doi.org/10.1038/sj.embor.7400824
- Engelsma D, Rodriguez JA, Fish A, Giaccone G, Fornerod M. Homodimerization antagonizes nuclear export of survivin. Traffic 2007; 11:1495-502; PMID:17714426; http://dx.doi.org/10.1111/j.1600-0854.2007.00629.x
- Connell CM, Colnaghi R, Wheatley SP. Nuclear survivin has reduced stability and is not cytoprotective. J Biological Chemistry 2008; 238:3289-96; PMID:16950794; http://dx.doi.org/10.1074/jbc.M704461200
- Chakravarti A, Zhai GG, Zhang M, Malhotra R, Lathman DE, Delaney MA, Robe P, Nestler U, Song Q, Loeffler J. Survivin enhances radiation resistance in primary human glioblastoma cells via caspase-independent mechanisms. Oncogene 2004; 23:7494-506; PMID:15326475; http://dx.doi.org/10.1038/sj.onc.1208049
- Wheatley SP. Nuclear survivin: consequences and therapeutic implications. Advances in Cancer Therapy, InTech open 2011; Chapter 15:331-42
- Wang HJ, Holloway MP, Ma L, Cooper ZA, Riolo M, Samkari A, Elenitoba-Johnson KSJ, Chin YE, Altura RA. Acetylation directs survivin nuclear localization to repress STAT3 oncogenic activity. J Biol Chem 2010; 285:36129-37; PMID:20826784; http://dx.doi.org/10.1074/jbc.M110.152777
- Ruchaud S, Carmena M, Earnshaw WC. Chromosomal passengers: conducting cell division. Nat Rev Mol Cell Biol 2007; 8:798-812; PMID:17848966; http://dx.doi.org/10.1038/nrm2257
- Vader G, Medema R, Lens SMA. The chromosomal passenger complex: guiding Aurora-B through mitosis. J Cell Biol 2006; 173:833-7; PMID:16769825; http://dx.doi.org/10.1083/jcb.200604032
- Kelly AE, Ghenoiu C, Xue JZ, Zierhut C, Kimura H, Funabiki H. Survivin Reads Phosphorylated Histone H3 Threonine 3 to Activate the Mitotic Kinase Aurora B. Sci 2010; 330:235-9; PMID:20705815; http://dx.doi.org/10.1126/science.1189505
- Barrett RMA, Osborne TP, Wheatley SP. Phosphorylation of survivin at threonine 34 inhibits its mitotic function and enhances its cytoprotective activity. Cell Cycle 2009; 8:278-83; PMID:19158485; http://dx.doi.org/10.4161/cc.8.2.7587
- Asumen MG, Ifeacho TV, Cockerham L, Pfandl C, Wall NR. Dynamic changes to survivin subcellular localization are initiated by DNA damage. OncoTargets and Therapy 2010; 3:129-37; PMID:20856848
- Capalbo G, Dittman K, Weiss C, Reichert S, Hausmann E, Rodel C, Rodel F. Radiation-induced survivin nuclear accumulation is linked to DNA damage repair. Int J Radiat Onc Biol Phys 2010; 77:226-34; PMID:20394854; http://dx.doi.org/10.1016/j.ijrobp.2009.12.001
- Chaachouay H, Ohneseit P, Toulany M, Kehlbach R, Multhoff G, H.P.R. Autophagy contributes to resistance of tumor cells to ionizing radiation. Radiother Oncol 2011; 99:287-92; PMID:21722986; http://dx.doi.org/10.1016/j.radonc.2011.06.002
- Rosenfeldt MT, Ryan KM. The multiple roles of autophagy in cancer. Carcinogenesis 2011; 32:955-63; PMID:21317301; http://dx.doi.org/10.1093/carcin/bgr031
- Carvalho A, Carmena M, Sambade C, Earnshaw WC, Wheatley SP. Survivin is required for stable checkpoint activation in response to loss of spindle tension in HeLa cells. J Cell Sci 2003; 116:2987-98; PMID:12783991; http://dx.doi.org/10.1242/jcs.00612
- Lens SM, Medema RH. The survivin/Aurora B complex: its role in coordinating tension and attachment. Cell Cycle 2003; 2:507-10; PMID:14504461; http://dx.doi.org/10.4161/cc.2.6.559
- Lens SMA, Rodriguez JA, Vader G, Span SW, Giaccone G, Medema RH. Uncoupling the central spindle-associated function of the chromosomal passenger complex from its role at centromeres. Mol Biol Cell 2006; 17:1897-909; http://dx.doi.org/10.1091/mbc.E05-08-0727
- Aljaberi A, Webster J, Wheatley S. Analysis of the functional repertoire of a mutant form of survivin, K129E, which has been linked to lung cancer. Cancer Cell Internl 2014; 14:78-85.
- Wheatley SP, Carvalho A, Vagnarelli P, Earnshaw WC. INCENP is required for proper targeting of survivin to the centromeres and the anaphase spindle during mitosis. Curr Biol 2001; 11:886-90; PMID:11516652; http://dx.doi.org/10.1016/S0960-9822(01)00238-X
- Pettersen EF, Goddard TD, Huang CC, Couch GS, Greenblatt DM, Meng EC, Ferrin TE. A visualization system for exploratory research and analysis. J Comput Chem 2004; 25:1605-12; PMID:15264254
