Eukaryotic DNA replication is controlled by a 2-step mechanism, which is crucial for genome stability. In the first step, during late M-phase of the cell cycle, the core of replicative helicase becomes loaded onto DNA and in the second step, during the G1-S transition, the helicase holo-enzyme becomes assembled and activated. Crucially, helicase loading is inhibited during S-phase, so that DNA can be only replicated once. A consequence of this regulatory principle is that the loading of the replicative DNA helicase needs to be very efficient, since no helicase reloading can occur once DNA replication has initiated. The mini-chromosome-maintenance proteins 2-7 (MCM2-7) form the core of the helicase. The hexameric MCM2-7 proteins associate at replication origins with the origin-recognition complex (ORC), Cdc6 and Cdt1 to form a pre-replicative complex (pre-RC).Citation1 During pre-RC formation, 2 MCM2-7 hexamers become loaded into a double-hexamer encircling double-stranded DNA. This reaction occurs in a step-wise manner, with the initial formation of an ORC/Cdc6/Cdt1/MCM2-7 (OCCM) complex, ATP-hydrolysis dependent Cdt1 release, which is followed by the ORC/Cdc6/MCM2-7 (OCM) dependent recruitment of a second MCM2-7 hexamer.Citation2 To understand the process of helicase loading, one has to consider that this reaction must involve an opening of the MCM2-7 complex and insertion of the DNA, before the ring closes around the DNA. The structural organization of the MCM2-7 is crucial in this context. Moreover, at which point during the multi-step assembly reaction DNA is inserted into the MCM2-7 ring, and what interface of MCM2-7 represents the DNA entry gate for helicase loading, are crucial questions that need to be addressed to understand the mechanical alterations in the core-helicase during DNA loading.
The structural basis of MCM2-7 and its organization has been unclear for the longest time. Archaeal homologues of the MCM helicase adopt ring-shaped or spiral hexamer structures, but only the ring-shaped complex is active in DNA unwinding. If eukaryotic MCM2-7 would be organized in an open ring (spiral),Citation3 then ORC, Cdc6 and Cdt1 independent helicase loading could occur, which could lead to DNA re-replication and genome instability. Alternatively, MCM2-7 could be organized as a closed ring,Citation4,5 which would facilitate regulated helicase loading, but current knowledge is insufficient to proof a ring shaped structure. To contribute to the resolution of this important question we recently investigated the organization of the Saccharomyces cerevisiae MCM2-7 complex by electron microscopy (EM).Citation6 Our 3-dimensional reconstruction of the EM images revealed that the complex adopts a closed ring-shape, arguing for regulated MCM2-7 loading onto DNA. To address the all-important helicase opening mechanism, in particular to identify the subunits that serve as the DNA entry gate, we engineered a small-molecule dependent linkage between neighboring Mcm subunits. Introduction of the linkage at 5 out of 6 possible MCM2-7 subunit-pairs had no effect on pre-RC formation. However, linking Mcm2 with Mcm5 inhibited MCM2-7 double-hexamer formation completely (). When the DNA entry gate was blocked, initial ORC/Cdc6/Cdt1/MCM2-7 complex formation could occur, but ATP-hydrolysis dependent OCM formation failed in large and the complex disassembled (). This demonstrates that DNA loading is surveyed by a quality control mechanism, which promotes active disassembly of a failed helicase loading intermediate. Consistent with these in vitro experiments, the in vivo analysis revealed that linkage of Mcm2 and Mcm5 blocks cell cycle progression and causes a defect in MCM2-7 association with chromatin. Multiple different constructs, that added the linkage near the N- or C-terminal section of Mcm2 and Mcm5, behaved similarly, highlighting that the observed effects are highly specific.
Figure 1. (A) Rapamycin dependent interactions between Mcm2 and Mcm5 reveal the DNA entry gate of the MCM2-7 helicase. (B) The loading of the replicative helicase onto DNA itself is regulated by a quality control mechanism that leads to complex disassembly if the reaction fails.
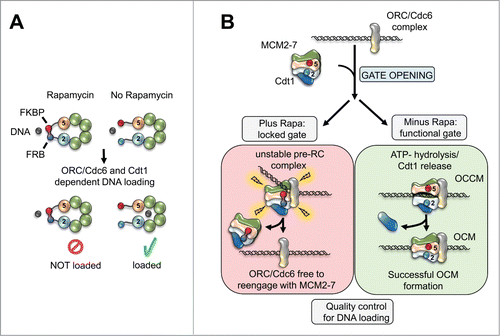
Up to now, it has not been known how DNA insertion is coordinated with MCM2-7 double-hexamer formation. Initial models suggested that this is a concerted reaction. Then a cryo-EM structure of the OCCM complexCitation7 hinted the presence of DNA inside this early pre‑RC intermediate. We wanted to resolve this issue by asking if MCM2-7 already encircles DNA when the OCCM is formed. We performed high salt washes of the OCCM complex and observed that the small molecule induced linkage specifically stabilised a hexameric MCM2‑7 complex on DNA, while ORC, Cdc6 and Cdt1 were washed away. Therefore our experiment proved that helicase loading onto DNA occurs already at the stage of OCCM formation - very early during pre-RC formation. Consequently, eukaryotic helicase loading could function similar to bacterial helicase loading, as both helicases are loaded as single-hexamers onto DNA. Indeed, it is known that in some bacteria the replication factor DnaC acts as a DnaB helicase ring breaker by adopting a spiral shape, which forces the opening of the hexameric DnaB complex. The recent cryo-EM structure of the OCCM provided some evidence for a related mechanism, as ORC/Cdc6 also adopt a spiral shape, but more work will be required until we fully understand the mechanism of MCM2-7 ring opening. What is clear now is that helicase loading occurs through a unique DNA entry gate – Mcm2 and Mcm5. This fits with biochemical evidence, which showed that this interface becomes destabilised in the absence of ATP.Citation5 Whether this or another interface functions as the DNA exit gate during helicase activation or MCM2-7 de-loading is an open question that we are eager to address.
In summary, our recent publication describes critical mechanisms that together detail crucial structural and mechanical principles in helicase loading and reveal important similarities between bacteria and eukaryotes. Nevertheless, the eukaryotic hetero-hexameric helicase structure offers a multitude of regulatory, functional and structural opportunities, which cannot be achieved by the simpler prokaryotic homo-hexamer.
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
References
- Riera A, Tognetti S, Speck C. Helicase loading: how to build a MCM2-7 double-hexamer. Semin Cell Dev Biol 2014; 30:104-109; PMID:24637008; http://dx.doi.org/10.1016/j.semcdb.2014.03.008
- Fernandez-Cid A, Riera A, Tognetti S, Herrera MC, Samel S, Evrin C, Winkler C, Gardenal E, Uhle S, Speck C. An ORC/Cdc6/MCM2-7 complex is formed in a multistep reaction to serve as a platform for MCM double-hexamer assembly. Mol Cell 2013; 50:577-588; PMID:23603117; http://dx.doi.org/10.1016/j.molcel.2013.03.026
- Costa A, Ilves I, Tamberg N, Petojevic T, Nogales E, Botchan MR, Berger JM. The structural basis for MCM2-7 helicase activation by GINS and Cdc45. Nat Struct Mol Biol 2011; 18:471-477; PMID:21378962; http://dx.doi.org/10.1038/nsmb.2004
- Remus D, Beuron F, Tolun G, Griffith JD, Morris EP, Diffley JF. Concerted loading of Mcm2-7 double hexamers around DNA during DNA replication origin licensing. Cell 2009; 139:719-730; PMID:19896182; http://dx.doi.org/10.1016/j.cell.2009.10.015
- Bochman ML, Schwacha A. Differences in the single-stranded DNA binding activities of MCM2-7 and MCM467: MCM2 and MCM5 define a slow ATP-dependent step. J Biol Chem 2007; 282:33795-33804; PMID:17895243; http://dx.doi.org/10.1074/jbc.M703824200
- Samel SA, Fernández-Cid A, Sun J, Riera A, Tognetti S, Herrera MC, Li H, Speck C. A unique DNA entry gate serves for regulated loading of the eukaryotic replicative helicase MCM2-7 onto DNA. Genes Dev 2014; 28:1653-1666; PMID:25085418; http://dx.doi.org/10.1101/gad.242404.114
- Sun J, Evrin C, Samel SA, Fernández-Cid A, Riera A, Kawakami H, Stillman B, Speck C, Li H. Cryo-EM structure of a helicase loading intermediate containing ORC-Cdc6-Cdt1-MCM2-7 bound to DNA. Nat Struct Mol Biol 2013; 20:944-951; PMID:23851460; http://dx.doi.org/10.1038/nsmb.2629
