Abstract
During meiosis, crossover recombination is tightly regulated. A spatial patterning phenomenon known as interference ensures that crossovers are well-spaced along the chromosomes. Additionally, every pair of homologs acquires at least one crossover. A third feature, crossover homeostasis, buffers the system such that the number of crossovers remains steady despite decreases or increases in the number of earlier recombinational interactions. Here we summarize recent work from our laboratory supporting the idea that all 3 of these aspects are intrinsic consequences of a single basic process and suggesting that the underlying logic of this process corresponds to that embodied in a particular (beam-film) model.
Abbreviations
| CO | = | crossover |
| BF | = | beam-film |
| STUbL | = | SUMO-targeted ubiquitin ligase |
| NCO | = | noncrossover |
| DSBs | = | double-strand breaks |
| DDF | = | designation driving force |
| SC | = | synaptonemal complex |
Background: Three CO Patterning Phenomena
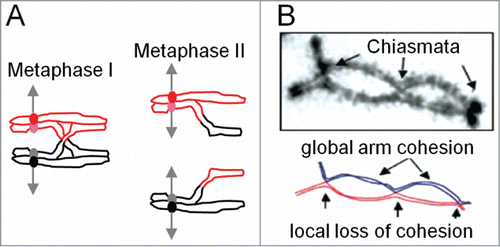
During meiosis, DNA crossovers (COs) increase genetic diversity. COs are also essential for proper segregation of homologous chromosomes (homologs) at the first division of meiosis (Meiosis I). In brief, homologs must be connected so that tension arises on their centromeres when they are properly oriented and thus being pulled toward opposite poles (). This tension is sensed by regulatory surveillance mechanisms. When all homolog pairs are correctly aligned and under tension, anaphase is allowed to proceed. The required inter-homolog connection arises by the combined effects of COs (seen cytologically as chiasmata) and links between sister chromatids along chromosome arms ().
Figure 1. Meiotic crossover (chiasma) patterns. (A) Meiosis comprises 2 successive rounds of chromosome segregation. At Meiosis I, homologous chromosomes (red and black) segregate to opposite poles. The segregating chromosomes are connected by the combined effects of a CO between one sister of each homolog and links between sister chromatids along their arms. Because of this connection, when homologous chromosomes are connected to opposite poles, tension arises on their corresponding centromeres (red and black circles). When all pairs are under such tension, cell cycle regulatory mechanisms license onset of Anaphase I. At Meiosis II, sister chromatids segregate to opposite poles, this time guided by connections between sister centromeres.(B) In many organisms, the CO-generated links between homologs at Meiosis I (A) are seen cytologically as "chiasmata." Note that in favorable cases, sister cohesion is disrupted around chiasma sites (arrows) in accord with the connection of non-sister chromatids by COs at those positions (from ref.Citation4).
The number and positions of COs are tightly regulated by events that occur well before Meiosis I, during an extended prophase period. Three distinct aspects of this regulation are known. Classical studies of CO/chiasma patterns identified 2 hallmark features: CO interference and the obligatory CO.Citation1-5 Modern studies have identified a third feature: CO homeostasis.Citation6-12
Crossover interference
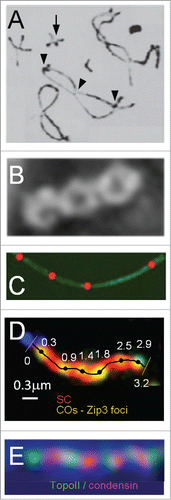
COs occur stochastically at different positions in different meiotic nuclei. Nonetheless, along each given chromosome, they tend to be evenly spaced. This tendency is visually apparent in the array of chiasmata or of CO-specific recombination complexes along mid/late prophase chromosomes (). It is also reflected in the fact that the distribution of inter-CO distances has the general shape of a gamma distribution (e.g. refs.Citation13-16). This phenomenon reflects the century-old phenomenon of “crossover interference,” where occurrence of a CO at one position disfavors the occurrence of a CO nearby.Citation17,18
Figure 2. Even spacing of chiasmata and pachytene CO-correlated recombination complexes. (A) Pairs of homologous grasshopper chromosomes ("bivalents") linked by chiasmata (from ref.Citation5). Note that each bivalent has at least one chiasma, sometimes only one (arrow) and that multiple chiasmata (arrow heads) on a single bivalent are spaced far away from one another. (B) A diplotene bivalent of Sordaria macrospora showing evenly-spaced chiasmata (D.Z.). (C) A pachytene bivalent of Sordaria macrospora. SC illuminated with Sme4-GFP (green) and decorated with evenly-spaced CO-correlated Hei10-T3-mCherry foci (red) (D.Z. unpublished) (D) A pachytene bivalent of budding yeast. SC (red) and CO-correlated Zip3 foci (green) illuminated with antibodies against Zip1 and Myc (for detection of Myc tagged Zip3) respectively (from ref.Citation11). (E) Patterning along mammalian mitotic metaphase chromosomes as manifested in evenly-spaced alternating domains of TopoII and condensin I (from ref.Citation76).
The nature this CO pattern is revealed in more detail by a measure known as the Coefficient of Coincidence (CoC), originally defined to describe interference observed genetically (; ref.Citation17,19; further discussion in ref.Citation10). By this approach, the test chromosome is divided into a convenient number of intervals. For each pair of intervals, in all combinations, the observed frequency of double COs (i.e., the frequency of chromosomes with a CO in both intervals), is compared with the frequency of double COs predicted if they occurred independently (i.e., without interference) as given by the product of the CO frequencies in each of the 2 intervals. The ratio of these 2 frequencies is the CoC. In the absence of interference, CoC = 1. If CoC < 1, the frequency of double COs is less than expected for independent occurrence and the presence of interference is thus inferred. Universally, interference is strong for pairs of intervals that are close together. Essentially no double COs are observed for intervals that are very close together, i.e. CoC = 0. Interference then weakens progressively as the 2 intervals are farther and farther apart, finally reaching unity (e.g., refs.Citation10,19; ).
Figure 3. Crossover Interference in wild-type and mutant budding yeast. CoC relationships for positions of CO-correlated Zip3 foci along pachytene bivalents of budding yeast in wild type and mutant situations (from ref.Citation11). (A) Experimental data for Chromosome XV (black) and BF best-fit simulation (green). Left: CoC relationships, plotted as function of inter-interval distance in μm axis length. Right: the distribution of Zip3 foci (COs) per bivalent. A convenient measure of the apparent strength of CO interference is provided by the inter-interval distance at which CoC = 0.5. In wild type, this "interference distance" is ∼300 nm. This value corresponds approximately to the average distance between adjacent COs. (B) Meiotic depletion of TopoII increases the CoC at smaller inter-interval distances and shifts the CoC curve to the left (pink) as compared to wild type [black; from (A)], with corresponding BF best fit simulation (right, green). Interference distance in the mutant is ∼200 nm. (C) A yeast condensin mutant has longer chromosome axes. CoC relationships in the mutant (red) are the same as in wild type (black) when the metric of inter-interval distance is physical chromosome length (μm) (top) and are different when the metric is genomic distance (Kb) (bottom). (D) Model for the role of TopoII in crossover interference. Chromatin expansion puts stress on the protein/protein/DNA meshwork of chromosome axes (top). Local CO-designation gives local relaxation, which then spreads along the axes; TopoII is required to adjust the relationships among DNA segments in response to this relaxation. All images are from ref.Citation11
![Figure 3. Crossover Interference in wild-type and mutant budding yeast. CoC relationships for positions of CO-correlated Zip3 foci along pachytene bivalents of budding yeast in wild type and mutant situations (from ref.Citation11). (A) Experimental data for Chromosome XV (black) and BF best-fit simulation (green). Left: CoC relationships, plotted as function of inter-interval distance in μm axis length. Right: the distribution of Zip3 foci (COs) per bivalent. A convenient measure of the apparent strength of CO interference is provided by the inter-interval distance at which CoC = 0.5. In wild type, this "interference distance" is ∼300 nm. This value corresponds approximately to the average distance between adjacent COs. (B) Meiotic depletion of TopoII increases the CoC at smaller inter-interval distances and shifts the CoC curve to the left (pink) as compared to wild type [black; from (A)], with corresponding BF best fit simulation (right, green). Interference distance in the mutant is ∼200 nm. (C) A yeast condensin mutant has longer chromosome axes. CoC relationships in the mutant (red) are the same as in wild type (black) when the metric of inter-interval distance is physical chromosome length (μm) (top) and are different when the metric is genomic distance (Kb) (bottom). (D) Model for the role of TopoII in crossover interference. Chromatin expansion puts stress on the protein/protein/DNA meshwork of chromosome axes (top). Local CO-designation gives local relaxation, which then spreads along the axes; TopoII is required to adjust the relationships among DNA segments in response to this relaxation. All images are from ref.Citation11](/cms/asset/d0964fce-eb1f-477e-9d60-e2558e2a17ca/kccy_a_991185_f0003_c.gif)
Interference and even spacing are particularly interesting because they imply the existence of communication along the chromosomes, which occurs over distances of tenths to tens of microns among different organisms. The same type of patterning occurs in many other systems, in both the biological and physical worlds. For chromosomes, it applies to mammalian DNA replication origin firings and metaphase chromosomal domains in somatic cells () and, in bacteria, the positions of extra-chromosomal plasmids and Mu transposon insertion sites.Citation20-25
The obligatory CO
Essentially every chromosome acquires at least one CO, irrespective of chromosome length. Among different organisms, or among different chromosomes in the same organism, the frequency of zero-CO chromosomes is usually much less than 1%, but can be as high as several percent.Citation10,19,26 This “obligatory CO rule” reflects the fact that at least one crossover is required to ensure correct chromosome segregation at meiosis I (). The obligatory CO is not achieved by having a high average number of COs per bivalent. In many cases, that average number is ∼2; and in certain cases/organisms, a chromosome always acquires one and only one CO (e.g. ).Citation1,2,4,5,10,27
CO homeostasis
Meiotic recombination is initiated from programmed double-strand breaks (DSBs),Citation28 which then mediate establishment of cytologically-observable links between homolog partner chromosomes (below). A subset of these interactions (so-called “precursors”) are designated to be COs. When the number of DSBs is decreased (or increased), the number of COs is decreased (or increased) less than proportionally. This phenomenon, “crossover homeostasis,” buffers the system against deficits (or excesses) of DSBs or precursor interactions. CO homeostasis was identified in budding yeast and subsequently found in Drosophila, mouse and C. elegans.Citation6-12
A central question emerges: what is the relationship among these 3 features?
Experimental Evidence
Our recent workCitation10,11,20 suggests that all 3 aspects of CO regulation can be explained as direct manifestations of a single patterning process. We identified a genetic pathway involved in CO interference in budding yeast.Citation11 Topoisomerase II (Top2) is a key player. Protein SUMOylation (by Ubc9) and SUMO-targeted ubiquitin ligase- (STUbL-) mediated protein removal (by Slx5/8-Sir2) are also required. In mutants defective in this pathway, CO interference is “less strong” than in wild type, as defined by CoC analysis ().
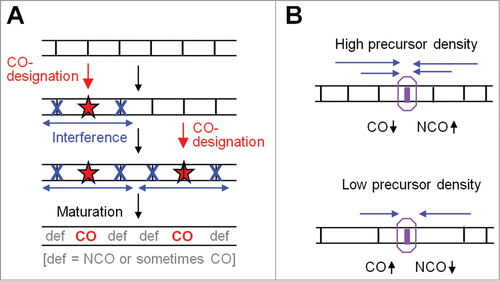
Detailed interpretation of this phenotype requires consideration of the overall nature of CO patterning. Current models (; below) envision a process that acts on an array of undifferentiated precursor interactions via 2 effects: (i) CO-designation, in which particular precursors become molecularly-fated to mature as COs, and (ii) an inhibitory signal, triggered at each CO-designation site, which spreads outward in both directions to disfavor the subsequent occurrence of further CO-designations in the vicinity. The latter can be thought of as a “spreading interference signal.” The identified yeast pathway could be important for either or both of these features; however, mutant defects are best explained by a decrease in the distance over which the interference signal spreads, implying a primary role for the pathway in communication along the chromosomes.Citation11
Figure 4. The logic of beam-film model and CO homeostasis. (A) The general logic of the beam-film model (from ref.Citation10). The "designation driving force" works on an array of precursors (vertical black lines) and promotes CO designation (red stars) with resulting spreading of the interference signal outward in both directions, dissipating with distance. Sequential CO designations with ensuing spreading interference signals lead to COs that tend to be evenly spaced (text). (B) Crossover homeostasis from the perspective of an individual DSB-mediated recombinational interaction according to the logic of the BF model. Crossover homeostasis results from interplay between precursor density and spreading interference.Citation6,10,11 At high (low) precursor (vertical black lines) density, a precursor will be more (less) affected by the spreading interference signal (blue arrows) from nearby crossover-designations and thus will be less (more) likely to be a crossover.
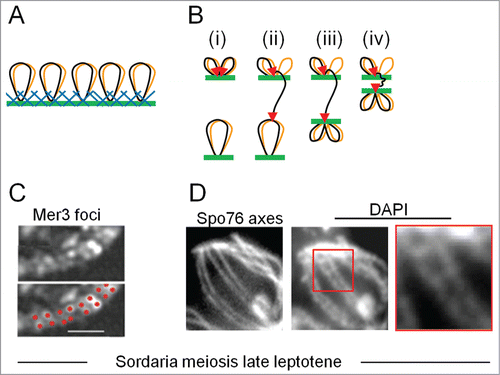
Mutant analysis provides additional insights. At the stage when interference is imposed, chromosomal DNA is organized into linear arrays of loops whose bases are decorated with a meshwork of structural proteinsCitation29 (). Meiotic recombination occurs in the context of these axesCitation29-34 (). Correspondingly, we find that normal axes are required for interference.Citation11 Since Topoisomerase II occurs along chromosome axes, we suggest that it might exert its effects by mediating adjustment of DNA segments within the axis meshwork, first locally and then as the interference signal spreads along the chromosomes ().Citation11
Figure 5. Recombination occurs in the context of chromosome structural axes. (A) Meiotic prophase chromosomes comprise co-oriented sister linear arrays of chromatin loops (black and brown), the bases of which are decorated by structural components in a protein/DNA meshwork (blue and green) (ref.Citation29) These proteins (e.g., Topoisomerase II, condensins, cohesins and meiotic specific Red1 and Hop1) bind to axis association sites, which are usually regularly distributed, locally AT-rich regions. (B) A model for recombinosome-mediated homolog juxtaposition (ref.Citation1). (i) One dual loop (black and brown) from one homolog is tethered onto axis (green; axial meshwork not shown) by recombinosome complex (not shown) and a DSB forms (2 red arrow heads) within one chromatin loop (black). (ii) The "leading" end of the DSB is released and searches for a homologous sequence on its homolog partner while the "lagging" DSB end is retained on the axis. (iii) The homolog is caught and bought into closer proximity, to ∼400 nm (the equivalent stage in Panels C and D). (iv) Finally the 2 axes move closer, to a distance of ∼100 nm, and synaptonemal complex forms (not shown). (C, D) DSB-mediated inter-axis bridges along coaligned late leptotene axes in Sordaria macrospora. (C) In late leptotene nuclei, pairs of foci of meiotic helicase Mer3 occur along the axes of homologous chromosomes as illuminated with fluorescent axis component Spo76/Pds5 (from ref.Citation35). Foci of each pair mark the 2 ends of a single DSB, implying that single DSB-mediated recombinational interaction is bridging the 2 axes. (D) Left: coaligned axes as in (C). Middle and Right: DAPI staining reveals inter-axis DNA bridges that presumptively correspond to the recombination-mediated bridges in (C) (D.Z.)
In the fungus Sordaria macrospora, where events are particularly well-defined,Citation35,36 CO patterning is likely imposed at the “late leptotene” stage, on recombination-mediated bridges that link homolog axes at ∼400 nm (), concomitant with installation of synaptonemal complex (SC), the tripartite meiosis-specific structure then links axes along their lengths at ∼100 nm.Citation29,35-40 The same timing occurs in budding yeast and, potentially, in mammals and plants.Citation36,41-47 In budding yeast, SC is not required for CO interference.Citation11,42,43 In fact, we have suggested that SC might initially be installed in the wake of the spreading interference signal.Citation42,48 Interestingly, in C. elegans, CO interference can arise after SC formationCitation49-51 and axial structures are again critical, but this time with involvement of the SC.Citation51 It has also been shown in C. elegans that transmission of the CO interference signal along a chromosome requires physical integrity along both involved homologs.Citation27
Our mutant analysis further revealed that in budding yeast, as seen also in Arabidopsis, mouse and human, the distance metric for spreading of the interference signal is physical chromosome length (microns) rather than genomic distance (kb/Mb) or genetic distance (cM).Citation10,11,52-54 This conclusion emerges from the finding that when genetically identical chromosomes are of different lengths (e.g., in different sexes or in mutants), CoC values are different when plotted as a function of inter-interval distance in Mb but the same when plotted as a function of distance in microns (e.g. ).
Our above-described findings arose from analysis of Zip3 foci along pachytene chromosomes of budding yeast. Zip3 foci provide a uniquely early cytological marker of CO-designationCitation10,43 (). Given this assay, and the existence of bona fide interference-defective mutants, we could quantitatively evaluate relationships among CO interference, the obligatory CO and CO homeostasis.Citation11 Two new findings emerged. (1) Reduced CO interference is accompanied by reduced CO homeostasis. Thus, these 2 features are intrinsically linked. The simplest possibility is that CO homeostasis is dependent upon CO interference as proposedCitation6 and supported also by other recent studies, including ours.Citation7-11 (2) Mutants with reduced interference (and reduced CO homeostasis) still show the obligatory CO. Thus, this feature does not require (strong) CO interference and accompanying homeostasis.
Identification of the Top2/SUMO/STUbL pathway is an important advance. However, thus far, only 2 targets of this pathway have been identified: TopoII and meiotic axis component Red1. Many more likely remain to be discovered. Moreover, the detailed consequences of this pathway remain to be elucidated. Even more importantly, since ablation of this pathway reduces, but does not eliminate, CO interference, additional components and their roles, likely even more fundamental, remain to be elucidated. It seems likely that the initial consequence of CO-designation is a local change at the site of an axis-associated recombination complex. A general question of interest is whether the primary target of interest for CO-designation is the recombination complex itself, the underlying structural components and/or other unsuspected players.
Insights from the Beam-Film Model
We have proposed a model for CO patterning, with corresponding quantitative expressions, that allows simulation of predicted outcomes.Citation10,20 This model has been useful in thinking about the nature of CO patterning. In addition, it has suggested a basic logic, different from that of previously-proposed models. Finally, formulation of this model has brought under consideration several features that had not been previously considered.
The stress hypothesis
The “beam-film” (BF) model involves accumulation, local relief and redistribution of mechanical stress. The process operates on an array of undifferentiated “precursor interactions.” The entire ensemble comes under stress until one interaction “goes critical,” undergoing a molecular change that commits it to becoming a CO. This event is “CO-designation.” By its nature, this stress-promoted event involves local relief of stress at the affected position. Moreover, that relief of stress will automatically spread outward in both directions, dissipating with distance, thus disfavoring subsequent stress-promoted events in the affected area. Redistribution of stress comprises the “spreading inhibitory interference signal.” As multiple CO designations occur, they tend to “fill in the holes” between prior events, ultimately giving a relatively evenly-spaced array, as depicted generically in .
Precursors appear to be recombinational interactions that bridge homolog axes at a distance of ∼400 nm (; ref.Citation2,37). Thus, stress might accumulate along chromosome axes. Since bridges would comprise “stress-sensitive” weak points along the axes, accumulating stress could cause a bridge ensemble to buckle, giving “CO-designation,” with ensuing redistribution of stress relief outward from that position giving the “spreading interference signal.”Citation42 Involvement of Topoisomerase II and chromosome axes (above) would be consistent with such a mechanism.
The question then arises: what is the source of the stress? We have suggested one viable possibility.Citation10,11,20,42 In brief: along the axis meshwork, the DNA/chromatin fiber is constrained into a too-small volume and thus is in a high energy (“stressed”) state. Such stress could be alleviated locally at CO sites by a change in the state of the fiber and/or by removal of constraining tethers. Indeed, in accord with this scenario, C. elegans chromosomes exhibit local axis expansion specifically at CO sites.Citation51 The interference signal would then comprise redistribution of this local stress relief along the axis.
Implications of logic and mathematical expressions
Importantly, the underlying logic and the mathematical expressions of the BF model can equivalently apply to mechanisms that are not based on stress and stress relief. In this broader context, the BF model accurately explains CO patterns in diverse organisms.Citation10,11 It also explains the basic features of CO patterning revealed by our recent experimental analysis as described above.
The obligatory CO
By the BF model, the obligatory CO is ensured primarily by making the driving force for CO-designation (i.e., the level of accumulated stress) strong enough to ensure that at least one precursor will undergo CO-designation. Since this effect ensures occurrence of the first CO-designation, it does not require the spreading interference signal. Secondarily, occurrence of the obligatory CO is favored by an appropriate array of precursor interactions. This is important to ensure that each chromosome in the nucleus always exhibits at least one sufficiently-reactive precursor, without which it could not acquire even a single CO. Additionally, occurrence of the obligatory CO requires that CO-designated interactions must mature efficiently into final CO products; otherwise, the effects of efficient CO-designation will be lost. The precise balance among these various features is likely achieved somewhat differently in different organisms.Citation10 However, most importantly, neither CO interference nor CO homeostasis is required to ensure the obligatory CO, in accord with observation (above). Also, it has been suggested that a particular process, “CO assurance,” is involved in ensuring the obligatory CO (e.g., refs.Citation45,55). The BF model suggests that there is no one specific process; instead, the obligatory CO is ensured by the appropriate constellation of all of the involved features.Citation10
CO homeostasis
The BF model also predicts an intrinsic functional link between CO interference and CO homeostasis, as pointed out previously by Martini et al., (ref.Citation6) and supported by experimental findings (above) (). The chance that a given undifferentiated precursor will become a CO is reduced when a spreading interference signal emanates from an adjacent position. If there are fewer DSBs/precursors along the chromosome, adjacent precursors will, on average, be farther away and the chance of such reduction is decreased. Thus, the probability of occurrence of a CO at each given position will be reduced less by the effect of interference than otherwise would have been the case. Consequently, overall, the frequency of COs will be reduced less than expected from the reduction in DSBs/precursors. Analogous buffering will occur if there is an increase in DSBs/precursors along the chromosome. Put more generally: CO homeostasis suggests that any tendency for more DSB interactions and thus more COs will be opposed by the occurrence of more spreading inhibitory signals emanating from those COs, which will dampen the effect; conversely, any tendency for fewer COs will result in occurrence of fewer impinging inhibitory signals and thus a commensurately lesser interference-mediated reduction.
In correspondence to these effects, CO homeostasis is predicted to be stronger or weaker depending on either (a) the distance over which the spreading interference signal acts; or (b) the probability of CO-designation per precursor, i.e. the strength of the CO designation driving force as given by the level of stress in the stress hypothesis.
This scenario is supported quantitatively by BF analysis of Zip3 focus distributions in budding yeast. In strains with different DSB levels, Zip3 focus (CO) levels vary as predicted from independently-defined parameters for interference.Citation10 The same was found to be true for Drosophila.Citation10 Furthermore, in conditions of Top2 depletion, decreased CO interference not only results in decreased CO homeostasis but does so with exactly the quantitative effect predicted. Specifically: in the top2 background, observed variations in Zip3 focus (CO) levels at altered DSB levels are explained quantitatively by assuming that the interference signal spreads over the specifically reduced distance inferred from analysis at the normal DSB level.Citation11 These results confirm homeostasis/interference interplay and provide strong experimental evidence that the mathematical formulation of the BF model can quantitatively describe CO patterning.
Phenomenologically, the distance over which CO interference extends, as a fraction of total chromosome length, is greater in other organisms as compared to budding yeast. The BF model further predicts that CO homeostasis should be stronger in those organisms.Citation10,20 This is qualitatively true in mouse and C. elegans.Citation7,9
Models for CO Patterning: Logic vs. Mechanism
As a general prospect: how could CO patterning work? The answer to this question can be divided into 2 categories: What is the basic logic of the process? And what is the molecular mechanism? The distinction between these 2 issues is illustrated by comparisons of our BF model (above)Citation10,20,42 with the seminal early model of King and MortimerCitation56 and another (also earlier-proposed) scenario, the “counting model” from Stahl and colleagues.Citation57
With respect to mechanism, the BF model envisions a stress and stress-relief process in which communication along the chromosomes results from redistribution of mechanical stress (above). In contrast, the King and Mortimer model envisions that the interference signal involves molecular polymerization of a key component along the chromosomes.
The logic of the 2 models is also significantly different. In the BF model, CO-designations are sequential. Each CO-designation triggers a spreading interference signal that automatically dissipates with distance, with this effect implemented prior to the next CO-designation (e.g. ). In a more generic formulation, CO-designation would be promoted by a “designation driving force” (DDF) and would involve a spreading inhibitory interference signal that (i) disfavors event-designation and (ii) dissipates exponentially with distance from its nucleation site.Citation10
The King and Mortimer model also involves precursor interactions, CO-designation and an interference signal that spreads in both directions from each designation site. However, contrary to the BF model, the interference signal does not dissipate with distance; instead it continues unabated until it runs into another interference signal approaching in the opposite direction from another CO-designation site. As a result, the number and distribution of COs will be governed by the kinetics of the system, i.e., the relative rates of CO-designation and interference signal spreading.
The King/Mortimer model further posits that interference results in the release of encountered precursor interactions, which then rebind to sites that have not yet experienced interference. This feature was required by the model's assumption that precursor interactions are randomly distributed along each given chromosome and that, among different nuclei, the number along a given chromosome is Poisson-distributed. A consequence of these assumptions is that some chromosomes will fail to acquire even one precursor and thus could never acquire a first (obligatory) CO. Release and rebinding of precursors rectifies this deficit. Importantly, this feature implies that, by the King/Mortimer model, the obligatory CO requires spreading of the interference signal. Our data argues against this possibility (above). Cytological evidence also argues against this scenario: all available evidence points to sequential formation and evolution of precursors into COs , without release and rebinding (e.g., refs.Citation36,38), although more complex dynamics could have been missed. Moreover, it is now clear that the hypothetical need for release and rebinding of precursors is eliminated by appropriate specification of the precursor arrayCitation10 (above).
The previous model from Stahl and colleagues proposes that an interference process proceeds progressively from one chromosome end and counts precursor interactions, with a CO-designation occurring once every "N" precursors.Citation57 This model gives a very good match to experimental CO data.Citation57 Nonetheless, it specifically predicts that: (i) any variation in precursor density will result in a proportional variation in CO frequency, which is qualitatively contradicted by the finding of CO homeostasis (ref.Citation6 but also see ref.Citation58); and (ii) interference will act over a longer genetic distance at reduced precursor densities, which is not observed.Citation6,11 Also, this model does not address or ensure the obligatory CO.
Other proposed models for CO interference that specify specific processes have invoked reaction-diffusion random walk collision between precursors,Citation59 dynamic filament fluctuations.Citation60 For the other chromosomal phenomena, both stress-based and reaction-diffusion models are shown or proposed (e.g. refs.Citation3,11,20-25). In the physical world, of course, analogous patterning necessarily occurs by stress and stress relief.
Implications
Given the above considerations, the array of chiasmata seen at diplotene can be understood as the intrinsic consequences of one relatively simple basic patterning process. This single process provides both the obligatory CO and CO interference, which are concomitant but independent effects. The combination of CO-designation and a spreading interference signal, in turn, not only ensure that COs are far apart but, at the same time, as automatic additional consequence, confers CO homeostasis, which buffers the system against early perturbations. All of these effects are well-described, qualitatively and quantitatively by the beam-film model. Thus, the underlying basic logic of this model is robust. The challenge now is to understand specific mechanisms that could underlie this logic, whether involving macroscopic mechanical stress or not.
Extensions
CO patterning is part of a broader structural program
The phenomenon of CO interference and its related effects have always been considered in the narrow context of recombination and chiasma formation. However, another recent study from our laboratory points to a broader perspective.Citation36 In most organisms, installation of SC is nucleated at the sites of DSB-mediated inter-axis bridges. Analysis in Sordaria macrospora suggests that, in that organism, SC nucleations comprise a subset of total DSB-mediated bridges, among which a further subset is concomitantly undergoing CO-designation. Moreover, total SC nucleation sites and CO sites both exhibit interference; however, the interference exhibited by SC nucleations is apparently “weaker” than that exhibited by CO sites, as defined by CoC analysis. Since SC nucleation and CO-designation appear to occur contemporaneously,Citation61 they are likely to be outcomes of a single process. Such a process would therefore yield relatively evenly-spaced SC nucleations, a subset of which are also CO sites. These coordinate effects support the possibility that CO patterning might fundamentally be a structure-based process.
Consideration of these phenomena through the lens of the BF model has provided appreciation of another aspect of CO patterning. Given a process with CO-designation and spreading interference, as more and more CO-designations occur, the resulting COs will tend to be closer and closer together. This will give the appearance, phenomenologically, of “weaker CO interference.” For example, CoC curves will shift "leftward" to give higher frequencies of double COs at smaller inter-interval distances. However, mechanistically, there will have been no change in the actual nature of the spreading inhibitory interference signal. This effect suggests an explanation for the observed pattern of SC nucleations and COs: all “event-designations” would give SC nucleation while only the first designations (which will necessarily occur at the most reactive precursor sites) would also concomitantly give CO-designation. As a result, the same ongoing process would give SC nucleations that give apparently “weaker” interference than CO designations, which would exhibit a classical interference distribution. An intriguing extrapolation is that apparently weaker CO interference could occur in mutant conditions where the relevant cell cycle stage is prolonged by a surveillance checkpoint delay (e.g., refs.Citation62,63).
Biological implications
Meiotic crossover patterns, and their disruption, have implications for efficient chromosome segregation during gametogenesis and thus, for example, in humans, for infertility and aneuploidy-related birth defects. This link is illustrated by 2 phenomena. (i) In human meiosis (and similarly in other organisms), COs are more frequent in female vs. male, with important genetic implications.Citation14,54,64-67 Moreover, differences in COs parallel differences in chromosome axis lengths (e.g. refs.Citation10,52,53 and references therein). Variations in interference are often suggested as the underlying basis.Citation14,54,64-67 Our alternative idea,Citation68 receiving preliminary support,Citation11,52-54 suggests that interference is irrelevant and that differences reflect a constant density of DSB-initiated precursors along longer/shorter axes. (ii) Shorter chromosomes, which are specifically implicated in birth defects,Citation69 often have higher CO density than longer chromosomes (e.g., ref.Citation70; unpublished). Many proposed models remain to be critically evaluated (e.g. refs.Citation14,65-67).
A remaining mystery
This emergent picture further highlights a fundamental remaining issue: what is the evolutionary rationale for CO interference? The evolutionary dictate of the obligatory CO is clear: at least one inter-homolog connection, and thus at least one CO/chiasma is required for regular meiosis I segregation (). But what could be the rationale for the complex, but widely conserved, phenomenon of CO interference and its corollary, CO homeostasis?
One possibility is that even spacing of COs (and thus chiasmata) facilitates regular chromosome segregation at meiosis I, as first proposed by Muller.Citation17 Such models remain tempting, especially now given that CO interference is part of the same process that gives the obligatory CO, which clearly exists to promote homolog segregation. On the other hand, in the plant Arabidopsis, mutations that increase the frequency of COs and chiasmata by as much as 10-fold have no obvious deleterious effect on meiotic chromosome segregation.Citation71,72 Moreover, meiotic chromosome segregation is very faithful in S. pombe and A. nidulans where CO interference is absent and most bivalents acquire many COs that are not spaced out by effects of interference.Citation73-75
The alternative possibility is that there is a genetic advantage to patterned COs. For example, there could be a selective advantage for a relatively low, homeostatically-regulated number of crossover recombination events. However, additional assumptions regarding genome organization would then be required to explain why that low number should tend to be evenly spaced along the chromosomes. Instead, we can now propose that CO patterning might promote the evolution and co-segregation of functionally-related sets of linked genes. This might be critical in light of the fact that CO recombination has the potential to eliminate existing favorable gene combinations as well as to create more favorable new combinations. Even spacing of COs would provide a balance between the 2 effects.
Conclusion
Solving this evolutionary conundrum remains an interesting challenge for the future.
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Acknowledgments
We thank S. Keeney, N. Hunter for discussions and members of Kleckner and Zickler laboratories for comments on the manuscript.
Funding
This work, S.W., L.Z. D.Z. and N.K. were supported by a grant to N.K. from the National Institutes of Health (RO1 GM044794). D.Z. was also supported by grants from the Center National de la Recherche Scientifique (Unité Mixte de Recherche 8621).
References
- Kleckner N, Zhang L, Weiner B, Zickler D. Meiotic chromosome dynamics. In: Rippe, ed. Genome Organization and Function in the Mammalian Cell Nucleus. New York: John Wiley and Sons, 2011:487-534.
- Zickler D, Kleckner N. Recombination, pairing and synapsis of homologs in meiosis. In: Kowalczykowski S, Hunter N, Heyer WD. DNA Replication. Cold Spring Harbor: Cold Spring Harbor Press, 2015; in press.
- Berchowitz LE, Copenhaver GP. Genetic interference: don't stand so close to me. Curr Genomics 2010; 11(2):91-102; PMID:20885817; http://dx.doi.org/10.2174/138920210790886835
- Jones GH, Franklin FC. Meiotic crossing-over: obligation and interference. Cell 2006; 126(2):246-8; PMID:16873056; http://dx.doi.org/10.1016/j.cell.2006.07.010
- Jones GH. The control of chiasma distribution. Symp Soc Exp Biol 1984; 38:293-320; PMID:6545727
- Martini E, Diaz RL, Hunter N, Keeney S. Crossover homeostasis in yeast meiosis. Cell 2006; 126(2):285-95; PMID:16873061; http://dx.doi.org/10.1016/j.cell.2006.05.044
- Cole F, Kauppi L, Lange J, Roig I, Wang R, Keeney S, Jasin M. Homeostatic control of recombination is implemented progressively in mouse meiosis. Nat Cell Biol 2012; 14(4):424-30; PMID:22388890; http://dx.doi.org/10.1038/ncb2451
- Globus ST, Keeney S. The joy of six: how to control your crossovers. Cell 2012; 149(1):11-2; PMID:22464316; http://dx.doi.org/10.1016/j.cell.2012.03.011
- Yokoo R, Zawadzki KA, Nabeshima K, Drake M, Arur S, Villeneuve AM. COSA-1 reveals robust homeostasis and separable licensing and reinforcement steps governing meiotic crossovers. Cell 2012; 149(1):75-87; PMID:22464324; http://dx.doi.org/10.1016/j.cell.2012.01.052
- Zhang L, Liang Z, Hutchinson J, Kleckner N. Crossover patterning by the beam-film model: analysis and implications. PLoS Genet 2014; 10(1):e1004042; PMID:24497834; http://dx.doi.org/10.1371/journal.pgen.1004042
- Zhang L, Wang S, Yin S, Hong S, Kim KP, Kleckner N. Topoisomerase II mediates meiotic crossover interference. Nature 2014; 511(7511):551-6; PMID:25043020; http://dx.doi.org/10.1038/nature13442
- Mehrotra S, McKim KS. Temporal analysis of meiotic DNA double-strand break formation and repair in Drosophila females. PLoS Genet 2006; 2(11):e200; PMID:17166055; http://dx.doi.org/10.1371/journal.pgen.0020200
- de Boer E, Stam P, Dietrich AJ, Pastink A, Heyting C. Two levels of interference in mouse meiotic recombination. Proc Natl Acad Sci U S A 2006; 103(25):9607-12; PMID:16766662; http://dx.doi.org/10.1073/pnas.0600418103
- Broman KW, Weber JL. Characterization of human crossover interference. Am J Hum Genet 2000; 66(6):1911-26; PMID:10801387; http://dx.doi.org/10.1086/302923
- McPeek MS, Speed TP. Modeling interference in genetic recombination. Genetics 1995; 139(2):1031-44; PMID:7713406
- Falque M, Anderson LK, Stack SM, Gauthier F, Martin OC. Two types of meiotic crossovers coexist in maize. Plant Cell 2009; 21(12):3915-25; PMID:20040539; http://dx.doi.org/10.1105/tpc.109.071514
- Muller HJ. The Mechanism of Crossing Over. Am Nat 1916; 50: 193-434; http://dx.doi.org/10.1086/279534
- Sturtevant AH. The behavior of the chromosomes as studied through linkage. Z Indukt Abstamm-u VererbLehre 1915; 13: 234-87.
- Charles DR. The spatial distribution of cross-overs in X-chromosome tetrads of Drosophila melanogaster. J Genetics 1938; 36: 103-26; http://dx.doi.org/10.1007/BF02982376
- Kleckner N, Zickler D, Jones GH, Dekker J, Padmore R, Henle J, Hutchinson J. A mechanical basis for chromosome function. Proc Natl Acad Sci U S A 2004; 101(34):12592-7; PMID:15299144; http://dx.doi.org/10.1073/pnas.0402724101
- Lebofsky R, Heilig R, Sonnleitner M, Weissenbach J, Bensimon A. DNA replication origin interference increases the spacing between initiation events in human cells. Mol Biol Cell 2006; 17(12):5337-45; PMID:17005913; http://dx.doi.org/10.1091/mbc.E06-04-0298
- Maeshima K, Eltsov M, Laemmli UK. Chromosome structure: improved immunolabeling for electron microscopy. Chromosoma 2005; 114(5):365-75; PMID:16175370; http://dx.doi.org/10.1007/s00412-005-0023-7
- Kleckner N. Mesoscale spatial patterning in the Escherichia coli Min system: reaction-diffusion versus mechanical communication. Proc Natl Acad Sci U S A 2010; 107(18):8053-4; PMID:20421460; http://dx.doi.org/10.1073/pnas.1002477107
- Vecchiarelli AG, Hwang LC, Mizuuchi K. Cell-free study of F plasmid partition provides evidence for cargo transport by a diffusion-ratchet mechanism. Proc Natl Acad Sci USA 2013; 110(15):E1390-7; PMID:23479605; http://dx.doi.org/10.1073/pnas.1302745110
- Ivanov V, Mizuuchi K. Multiple modes of interconverting dynamic pattern formation by bacterial cell division proteins. Proc Natl Acad Sci USA 2010; 107(18):8071-8; PMID:20212106; http://dx.doi.org/10.1073/pnas.0911036107
- Lao JP, Cloud V, Huang CC, Grubb J, Thacker D, Lee CY, Dresser ME, Hunter N, Bishop DK. Meiotic crossover control by concerted action of Rad51-Dmc1 in homolog template bias and robust homeostatic regulation. PLoS Genet 2013; 9(12):e1003978; PMID:24367271; http://dx.doi.org/10.1371/journal.pgen.1003978
- Hillers KJ, Villeneuve AM. Chromosome-wide control of meiotic crossing over in C. elegans. Curr Biol 2003; 13(18):1641-7; PMID:13678597; http://dx.doi.org/10.1016/j.cub.2003.08.026
- Keeney S. Spo11 and the formation of DNA double-strand breaks in meiosis. Genome Dyn Stab 2008; 2:81-123; PMID:21927624; http://dx.doi.org/10.1007/7050_2007_026
- Kleckner N. Chiasma formation: chromatin/axis interplay and the role(s) of the synaptonemal complex. Chromosoma 2006; 115(3):175-94; PMID:16555016; http://dx.doi.org/10.1007/s00412-006-0055-7
- Panizza S, Mendoza MA, Berlinger M, Huang L, Nicolas A, Shirahige K, Klein F. Spo11-accessory proteins link double-strand break sites to the chromosome axis in early meiotic recombination. Cell 2011; 146(3):372-83; PMID:21816273; http://dx.doi.org/10.1016/j.cell.2011.07.003
- Lichten M, de Massy B. The impressionistic landscape of meiotic recombination. Cell 2011; 147(2):267-70; PMID:22000007; http://dx.doi.org/10.1016/j.cell.2011.09.038
- Borde V, de Massy B. Programmed induction of DNA double strand breaks during meiosis: setting up communication between DNA and the chromosome structure. Curr Opin Genet Dev 2013; 23(2):147-55; PMID:23313097; http://dx.doi.org/10.1016/j.gde.2012.12.002
- de Massy B. Initiation of meiotic recombination: how and where? Conservation and specificities among eukaryotes. Annu Rev Genet 2013; 47:563-99; PMID:24050176; http://dx.doi.org/10.1146/annurev-genet-110711-155423
- Blat Y, Protacio RU, Hunter N, Kleckner N. Physical and functional interactions among basic chromosome organizational features govern early steps of meiotic chiasma formation. Cell 2002; 111: 791-802; http://dx.doi.org/10.1016/S0092-8674(02)01167-4
- Storlazzi A, Gargano S, Ruprich-Robert G, Falque M, David M, Kleckner N, Zickler D. Recombination proteins mediate meiotic spatial chromosome organization and pairing. Cell 2010; 141(1):94-106; PMID:20371348; http://dx.doi.org/10.1016/j.cell.2010.02.041
- Zhang L, Espagne E, de Muyt A, Zickler D, Kleckner NE. Interference-mediated synaptonemal complex formation with embedded crossover designation. Proc Natl Acad Sci U S A 2014:pii: 201416411. [Epub ahead of print]; PMID:25380597
- Zickler D, Kleckner N. Meiotic chromosomes: integrating structure and function. Annu Rev Genet 1999; 33:603-754; PMID:10690419; http://dx.doi.org/10.1146/annurev.genet.33.1.603
- De Muyt A, Zhang L, Piolot T, Kleckner N, Espagne E, Zickler D. E3 ligase Hei10: a multifaceted structure-based signaling molecule with roles within and beyond meiosis. Genes Dev 2014; 28(10):1111-23; PMID:24831702; http://dx.doi.org/10.1101/gad.240408.114
- Fraune J, Schramm S, Alsheimer M, Benavente R. The mammalian synaptonemal complex: protein components, assembly and role in meiotic recombination. Exp Cell Res 2012; 318(12):1340-6; PMID:22394509; http://dx.doi.org/10.1016/j.yexcr.2012.02.018
- Espagne E, Vasnier C, Storlazzi A, Kleckner NE, Silar P, Zickler D, Malagnac F. Sme4 coiled-coil protein mediates synaptonemal complex assembly, recombinosome relocalization, and spindle pole body morphogenesis. Proc Natl Acad Sci U S A 2011; 108(26):10614-9; PMID:21666097; http://dx.doi.org/10.1073/pnas.1107272108
- Bishop DK, Zickler D. Early decision; meiotic crossover interference prior to stable strand exchange and synapsis. Cell 2004; 117(1):9-15; PMID:15066278; http://dx.doi.org/10.1016/S0092-8674(04)00297-1
- Börner GV, Kleckner N, Hunter N. Crossover/noncrossover differentiation, synaptonemal complex formation, and regulatory surveillance at the leptotene/zygotene transition of meiosis. Cell 2004; 117(1):29-45; PMID:15066280; http://dx.doi.org/10.1016/S0092-8674(04)00292-2
- Fung JC, Rockmill B, Odell M, Roeder GS. Imposition of crossover interference through the nonrandom distribution of synapsis initiation complexes. Cell 2004; 116(6):795-802; PMID:15035982; http://dx.doi.org/10.1016/S0092-8674(04)00249-1
- Reynolds A, Qiao H, Yang Y, Chen JK, Jackson N, Biswas K, Holloway JK, Baudat F, de Massy B, Wang J, et al. RNF212 is a dosage-sensitive regulator of crossing-over during mammalian meiosis. Nat Genet 2013; 45(3):269-78; http://dx.doi.org/10.1038/ng.2541
- Hunter N. Meiotic recombination. In: Aguilera A, Rothstein R, eds. Molecular Genetics of Recombination. Heidelberg: Springer Berlin, 2006:381-442.
- Osman K, Higgins JD, Sanchez-Moran E, Armstrong SJ, Franklin FC. Pathways to meiotic recombination in Arabidopsis thaliana. New Phytol 2011; 190(3):523-44; PMID:21366595; http://dx.doi.org/10.1111/j.1469-8137.2011.03665.x
- Baudat F, de Massy B. Regulating double-stranded DNA break repair towards crossover or non-crossover during mammalian meiosis. Chromosome Res 2007; 15(5):565-77; PMID:17674146; http://dx.doi.org/10.1007/s10577-007-1140-3
- Tesse S, Storlazzi A, Kleckner N, Gargano S, Zickler D. Localization and roles of Ski8p protein in Sordaria meiosis and delineation of three mechanistically distinct steps of meiotic homolog juxtaposition. Proc Natl Acad Sci U S A 2003; 100: 12865-70; http://dx.doi.org/10.1073/pnas.2034282100
- Rog O, Dernburg AF. Chromosome pairing and synapsis during Caenorhabditis elegans meiosis. Curr Opin Cell Biol 2013; 25(3):349-56; PMID:23578368; http://dx.doi.org/10.1016/j.ceb.2013.03.003
- Lui DY, Colaiácovo MP. Meiotic development in Caenorhabditis elegans. Adv Exp Med Biol 2013; 757:133-70; PMID:22872477; http://dx.doi.org/10.1007/978-1-4614-4015-4_6
- Libuda DE, Uzawa S, Meyer BJ, Villeneuve AM. Meiotic chromosome structures constrain and respond to designation of crossover sites. Nature 2013; 502(7473):703-6; PMID:24107990; http://dx.doi.org/10.1038/nature12577
- Drouaud J, Mercier R, Chelysheva L, Bérard A, Falque M, Martin O, Zanni V, Brunel D, Mézard C. Sex-specific crossover distributions and variations in interference level along Arabidopsis thaliana chromosome 4. PLoS Genet 2007; 3(6):e106; PMID:17604455
- Petkov PM, Broman KW, Szatkiewicz JP, Paigen K. Crossover interference underlies sex differences in recombination rates. Trends Genet 2007; 23(11):539-42; PMID:17964681; http://dx.doi.org/10.1016/j.tig.2007.08.015
- Hou Y, Fan W, Yan L, Li R, Lian Y, Huang J, Li J, Xu L, Tang F, Xie XS, et al. Genome analyses of single human oocytes. Cell 2013; 155(7):1492-506; PMID:24360273; http://dx.doi.org/10.1016/j.cell.2013.11.040
- Shinohara M, Oh SD, Hunter N, Shinohara A. Crossover assurance and crossover interference are distinctly regulated by the ZMM proteins during yeast meiosis. Nat Genet 2008; 40(3):299-309; PMID:18297071; http://dx.doi.org/10.1038/ng.83
- King JS, Mortimer RK. A polymerization model of chiasma interference and corresponding computer simulation. Genetics 1990; 126(4):1127-38; PMID:2127577
- Lande R, Stahl FW. Chiasma interference and the distribution of exchanges in Drosophila melanogaster. Cold Spring Harb Symp Quant Biol 1993; 58:543-52; PMID:7956068; http://dx.doi.org/10.1101/SQB.1993.058.01.061
- Getz TJ, Banse SA, Young LS, Banse AV, Swanson J, Wang GM, Browne BL, Foss HM, Stahl FW. Reduced mismatch repair of heteroduplexes reveals “non”-interfering crossing over in wild-type Saccharomyces cerevisiae. Genetics 2008 178(3):1251-69; PMID:18385111; http://dx.doi.org/10.1534/genetics.106.067603
- Fujitani Y, Kawai J, Kobayashi I. Random-walk mechanism in the genetic recombination. Adv Exp Med Biol 2010; 680: 275-82; PMID:20865510; http://dx.doi.org/10.1007/978-1-4419-5913-3_31
- Hulten MA. On the origin of crossover interference: a chromosome oscillatory movement (COM) model. Mol Cytogenet 2011; 4:10; http://dx.doi.org/10.1186/1755-8166-4-10
- Zickler D, Moreau PJ, Huynh AD, Slezec AM. Correlation between pairing initiation sites, recombination nodules and meiotic recombination in Sordaria macrospora. Genetics 1992; 132(1):135-48; PMID:1398050
- Joyce EF, McKim KS. Chromosome axis defects induce a checkpointmediated delay and interchromosomal effect on crossing over during Drosophila meiosis. PLoS Genet 2010; 6: e1001059.
- Joyce EF, McKim KS. Meiotic checkpoints and the interchromosomal effect on crossing over in Drosophila females. Fly (Austin) 2011; 5: 134-40; http://dx.doi.org/10.4161/fly.5.2.14767
- Gruhn JR, Rubio C, Broman KW, Hunt PA, Hassold T. Cytological studies of human meiosis: sex-specific differences in recombination originate at, or prior to, establishment of double-strand breaks. PLoS One 2013; 8(12):e85075; PMID:24376867; http://dx.doi.org/10.1371/journal.pone.0085075
- Lynn A, Kashuk C, Petersen MB, Bailey JA, Cox DR, Antonarakis SE, Chakravarti A. Patterns of meiotic recombination on the long arm of human chromosome 21. Genome Res 2000; 10(9):1319-32; PMID:10984450; http://dx.doi.org/10.1101/gr.138100
- Tease C, Hartshorne GM, Hultén MA. Patterns of meiotic recombination in human fetal oocytes. Am J Hum Genet 2002; 70(6):1469-79; PMID:11992253; http://dx.doi.org/10.1086/340734
- Tease C, Hultén MA. Inter-sex variation in synaptonemal complex lengths largely determine the different recombination rates in male and female germ cells. Cytogenet Genome Res 2004; 107(3-4):208-15; PMID:15467366; http://dx.doi.org/10.1159/000080599
- Kleckner N, Storlazzi A, Zickler D. Coordinate variation in meiotic pachytene SC length and total crossover/chiasma frequency under conditions of constant DNA length. Trends Genet 2003; 19(11):623-8; PMID:14585614; http://dx.doi.org/10.1016/j.tig.2003.09.004
- Risch N, Stein Z, Kline J, Warburton D. The relationship between maternal age and chromosome size in autosomal trisomy. Am J Hum Genet 1986; 39(1):68-78; PMID:3752082
- Sun F, Mikhaail-Philips M, Oliver-Bonet M, Ko E, Rademaker A, Turek P, Martin RH. The relationship between meiotic recombination in human spermatocytes and aneuploidy in sperm. Hum Reprod 2008; 23(8):1691-7; PMID:18482994; http://dx.doi.org/10.1093/humrep/den027
- Crismani W, Girard C, Froger N, Pradillo M, Santos JL, Chelysheva L, Copenhaver GP, Horlow C, Mercier R. FANCM limits meiotic crossovers. Science 2012; 336(6088):1588-90; PMID:22723424; http://dx.doi.org/10.1126/science.1220381
- Girard C, Crismani W, Froger N, Mazel J, Lemhemdi A, Horlow C, Mercier R. FANCM-associated proteins MHF1 and MHF2, but not the other Fanconi anemia factors, limit meiotic crossovers. Nucleic Acids Res 2014; 42(14):9087-95; PMID:25038251; http://dx.doi.org/10.1093/nar/gku614
- Munz P. An analysis of interference in the fission yeast Schizosaccharomyces pombe. Genetics 1994; 137(3):701-7; PMID:8088515
- Strickland WN. An analysis of interference in Aspergillus nidulans. Proc R Soc Lond B Biol Sci 1958; 149(934):82-101; PMID:13554433; http://dx.doi.org/10.1098/rspb.1958.0053
- Egel-Mitani M, Olson LW, Egel R. Meiosis in Aspergillus nidulans: another example for lacking synaptonemal complexes in the absence of crossover interference. Hereditas 1982; 97(2):179-87; PMID:6761318; http://dx.doi.org/10.1111/j.1601-5223.1982.tb00761.x
- Maeshima K, Laemmli UK. A two-step scaffolding model for mitotic chromosome assembly. Dev Cell 2003; 4(4):467-80; PMID:12689587; http://dx.doi.org/10.1016/S1534-5807(03)00092-3
