Abstract
The Atg1 complex, comprising Atg1, Atg13, Atg17, Atg29, and Atg31, is a key initiator of autophagy. The Atg17-Atg31-Atg29 subcomplex is constitutively present at the phagophore assembly site (PAS), while Atg1 and Atg13 join the complex when autophagy is triggered by starvation or other signals. We sought to understand the energetics and dynamics of assembly using isothermal titration calorimetry (ITC), sedimentation velocity analytical ultracentrifugation, and hydrogen-deuterium exchange (HDX). We showed that the membrane and Atg13-binding domain of Atg1, Atg1EAT, is dynamic on its own, but is rigidified in its high-affinity (∼100 nM) complex with Atg13. Atg1EAT and Atg13 form a 2:2 dimeric assembly and together associate with lower affinity (∼10 μM) with the 2:2:2 Atg17-Atg31-Atg29 complex. These results lead to an overall model for the assembly pathway of the Atg1 complex. The model highlights the Atg13-Atg17 binding event as the weakest link in the assembly process and thus as a natural regulatory checkpoint.
Autophagy is a highly conserved catabolic pathway that maintains cellular homeostasis by removing damaged organelles, misfolded proteins, and pathogens, and replenishes biosynthetic precursors during starvation. Autophagy is characterized by a double-membrane structure, the phagophore, that encapsulates cellular components targeted for degradation and delivers them to the vacuole or lysosome. Starvation induces autophagy in part through deactivation of the nutrient-sensing TORC1 kinase. Deactivation of TORC1 promotes the recruitment of the most upstream set of autophagy-regulating proteins to the phagophore assembly site (PAS), the Atg1 complex. This complex consists of 5 proteins, Atg1, Atg13, Atg17, Atg29, and Atg31 in yeast. In human, the first three proteins correspond to ULK1/2, ATG13, and RB1CC1/FIP200, respectively, which assemble with the unique ATG101 protein. Upon starvation, Atg13 is rapidly dephosphorylated and recruited to Atg17 along with Atg1. It is not fully understood if Atg1 and Atg13 are recruited to Atg17 as a preformed complex or sequentially.
To understand the molecular mechanism of the assembly of the Atg1 complex at the PAS, we analyzed a minimal Atg1 complex, consisting of Atg1 EAT domain (Atg1EAT), the central region of Atg13 that binds Atg1EAT and Atg17, and full-length Atg17, Atg29, and Atg31. Using hydrogen-deuterium exchange mass spectrometry (HDX-MS), we found that the C-terminal half of Atg1EAT undergoes cooperative unfolding on an ∼10 s time scale. The N-terminal half of Atg1EAT forms the interface of the Atg1EAT homodimer, and is therefore much less dynamic. When Atg13 binds, Atg1EAT becomes much more rigid, and no longer manifests local unfolding. Thus, in order for Atg1EAT to reach the level of order normally associated with a well-folded domain, it must bind to Atg13.
To identify the domain within Atg13 that binds to Atg1EAT, we coarsely mapped the region by deletional analysis. We then used HDX-MS to more finely map the binding site by assessing regions of Atg13 that were protected from HDX upon binding to Atg1. The mapped residues coincide with 2 regions that were identified in the crystal structure of the Atg1EAT-Atg13 complex. This provides a satisfying mutual validation of the crystallographic and HDX identifications of the binding site. Atg1EAT binds to this small region of Atg13 with high affinity as judged by ITC, which yields an ∼100 nM binding affinity between the 2 domains. The high affinity of the complex is consistent with the rigidification of the Atg1EAT structure upon Atg13 binding. These data argue that the Atg1-Atg13 subcomplex is quite stable and seems likely to encounter the Atg17-Atg31-Atg29 subcomplex as a unit.
Having found that Atg1EAT-Atg13 form a tight complex, we went on to characterize the interaction between Atg1EAT-Atg13 and Atg17-Atg31-Atg29. We found that Atg13 and Atg17 bind via a weak interaction, with ∼10 μM affinity. To understand how Atg1 and Atg13 are recruited to Atg17-Atg29-Atg31, we assembled and characterized the pentamer in solution using analytical ultracentrifugation. We found that the pentameric complex exists predominately as a 2:2:2:2:2 dimer. A model for the assembly of the Atg1-Atg13 and Atg17-Atg31-Atg29 complexes into the pentamer is shown in .
Figure 1. Model for Atg1 complex assembly. Left, the Atg1 EAT domain dimer is partially melted in the absence of Atg13. Middle, the EAT domain become rigidified when Atg13 binds to it. The parts of Atg13 that directly bind to Atg1 also become ordered. The equilibrium strongly favors Atg1-Atg13 complex formation, at least when Atg13 is dephosphorylated. Right, the Atg1-Atg13 subcomplex binds weakly to the tips of the Atg17-Atg31-Atg29 double crescent. A 4:4:2:2:2 complex of Atg1-Atg13-Atg17-Atg31-Atg29 is shown for illustration purposes, but in solution the hydrodynamic data suggest a mixture of 2:2:2:2:2 and 4:4:4:4:4 species.
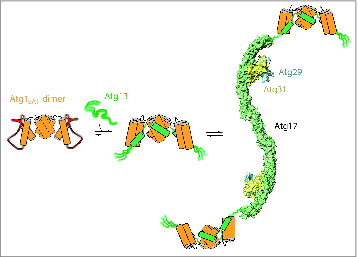
Some tetramer formation is also observed. The nature of the higher order assembly of pentamers is an open question, given that there are several possibilities for different arrangements of the 2:2 Atg1-Atg13 and 2:2:2 Atg17-Atg31-Atg29 dimers with respect to one another in tetramers or larger structures. Given the model that the Atg1 complex is involved in tethering and organizing the vesicles that give rise to the autophagosome, the nature of this arrangement might be pivotal to the initiation of autophagy.
These data have provided the initial framework for understanding the structure of the Atg1 complex at the PAS. One limitation of the experiments is that they made use of truncated constructs that lack the kinase domain of Atg1 and the HORMA domain of Atg13. A closer look at the full-length Atg1 complex will be an important next step. Another outstanding issue is that Atg1-Atg13 recruitment to Atg17 is driven by an interaction whose affinity is 2 orders of magnitude weaker than the concentrations likely to be present in cells. Overcoming this weak affinity maybe the result of increasing the local concentrations of the 5 proteins by targeting them to membranes. Alternatively, other proteins and complexes known to function in early autophagy, including Ypt1 and TRAPPIII, might help stabilize the interaction of the 2 subcomplexes. The spatial alignment of the entire complex with vesicles and downstream proteins will likely be essential to direct membrane remodeling and phagophore biogenesis.
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Funding
This work was supported by the National Institutes of Health GM111730 (J.H.H.) and Ruth Kirschstein NRSA Fellowship GM112301 (C.W.D.).
