Abstract
Plant cells are surrounded by rigid cell walls, and hence, their division is associated with a plant-specific mode of cytokinesis in which the cell plate, a new cell wall, is generated and separates 2 daughter nuclei. The successful execution of cytokinesis requires the timely activation of multiple regulatory pathways, which include the AtNACK1/HINKEL kinesin-induced MAPK cascade and MYB3R1/4-mediated transcriptional activation of G2/M-specific genes. However, it remains unclear whether and how these pathways are functionally interconnected to each other. By analyzing enhancer mutations of myb3r4, here we found a close genetic interaction between the 2 pathways; a mutation in ANP3, which encodes MAPKKK (acting downstream of AtNACK1/HINKEL), strongly enhanced the defective cytokinesis observed in the myb3r4 mutant. This interaction may not be due to the direct activation of MYB3R1/4 by the MAPK cascade; rather, possibly to the downstream targets of these 2 signaling pathways, acting in close proximity. Our results showed that MYB3R1/4 may positively affect cytokinesis via multiple pathways, one of which may act independently from the KNOLLE-dependent pathway defined previously, and affect the downstream events that may also be under the control of the AtNACK1/HINKEL-mediated MAPK cascade.
Unlike animals, plants typically have a rigid cell wall outside of each cell, which provides mechanical strength that sustains their shape and growth. Because of the presence of the cell wall, plants have evolved a unique mode of cytokinesis, which accompanies the generation of a new cell wall, called the cell plate. During cell division, a microtubule-based structure, the phragmoplast, appears between 2 separating daughter nuclei and fulfills functions that are essential for the accumulation and fusion of Golgi-derived vesicles at the equatorial region.Citation1,2 The molecular mechanisms underlying the formation of the cell plate are largely unknown; however, recent genetic studies have revealed components of signaling pathways that are essential for the execution of cytokinesis in Arabidopsis thaliana.Citation2,3
The cascade of the mitogen-activated protein kinase (MAPK) regulates phragmoplast dynamics during plant cytokinesis.Citation4,5,6,7 This pathway comprises Arabidopsis NPK1-RELATED PROTEIN KINASE (ANP) MAPK kinase kinases (MAPKKKs), Arabidopsis NQK1 (ANQ1)/MKK6 MAPKK, and multiple MAPKs that are classified in the group B subfamily. A kinesin-like protein, AtNACK1/HINKEL, is localized at the phragmoplast equator and binds to and activates the ANP MAPKKKs, thus transmitting the signals to a downstream kinase cascade.Citation8,9 This entire signaling cascade is designated the NACK-PQR pathway.Citation3 Mutational analyses demonstrated that the components of each step of this cascade are essential for the completion of cytokinesis in Arabidopsis.Citation4,5,7,8
Transcriptional regulation also plays important roles in the execution of plant cytokinesis. The two R1R2R3-type Myb transcription factors, MYB3R1 and MYB3R4 (hereafter MYB3R1/4), act redundantly and bind to the cis-regulatory element called mitosis-specific activator (MSA), for transcriptional activation.Citation10,11 Through binding to the MSA elements, MYB3R1/4 activates a variety of genes that are expressed specifically during G2 and M phases in the cell cycle.Citation10,11 Loss of these Myb proteins by mutation results in the frequent occurrence of incomplete cytokinesis, mainly because of the reduced expression of the critical target gene, KNOLLE (KN), which encodes a syntaxin-like protein that has an essential function for vesicle fusion at the site of cell-plate formation.Citation12 Although NACK1, a tobacco homolog of AtNACK1/HIK, is known as a downstream target of an R1R2R3-type Myb in tobacco,Citation13,14 it remains unclear if there is a functional interconnection between MYB3R-mediated transcriptional regulation and the NACK-PQR pathway for the promotion of cytokinesis.
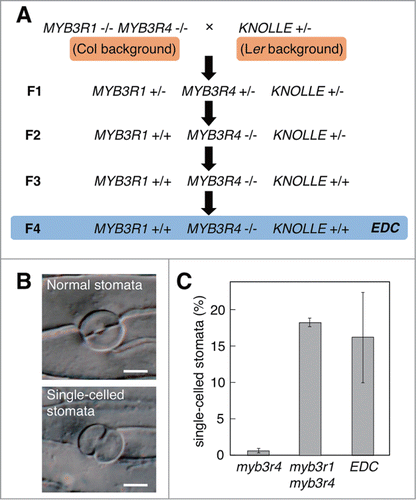
In our previous study, we generated crosses between plants with a heterozygous kn mutation and those with an myb3r1 myb3r4 double mutation, to examine their genetic interactions ().Citation10 As the mutant alleles of MYB3R1 and MYB3R4, we used myb3r1-1 and myb3r4-1,Citation10 respectively, for all experiments described here. We reported that the myb3r4 mutant, which normally has only limited defects in cytokinesis because of the functional redundancy between MYB3R1 and MYB3R4, showed pronounced defects when plants also carried a heterozygous kn mutation.Citation11 Surprisingly, in F3 populations, we occasionally found myb3r4 plants with enhanced defective cytokinesis, even in the absence of mutations in kn and myb3r1. The selfed progenies of such plants reproducibly showed similar enhanced defects, and one such line, which was designated enhanced defective cytokinesis (EDC), was chosen for further analyses (). In the epidermis of the myb3r4 and EDC plants, we observed typical defects of cytokinesis, such as single-celled stomata, which are generated by incomplete cytokinesis of guard mother cells ().Citation11 To compare quantitatively the cytokinesis defects in myb3r4 and EDC plants, we determined the proportion of single-celled stomata in silique valves, and showed that this value was much higher in EDC plants than in myb3r4 plants, despite the fact that these plants have identical genotypes at the MYB3R1, MYB3R4, and KN loci (). The cytokinesis defect observed in EDC plants even reached the levels of that detected in the myb3r1 myb3r4 double mutant ().
Figure 1. Generation of the EDC line and its enhanced defective cytokinesis. (A) Generation of the EDC line after the genetic cross between the kn heterozygote in the Ler background and the myb3r1 myb3r4 double mutant in the Col background. In the selfed F3 progeny, plants were segregated that showed enhanced defective cytokinesis and were homozygous for the mutant allele of myb3r4 and for the wild-type allele of both MYB3R1 and KN. These plants yielded F4 offspring that exhibited constantly enhanced defects; one representative line was termed EDC and analyzed further. The mutant and wild-type alleles are indicated by minus and plus signs, respectively. (B) Representative differential interference contrast (DIC) images of normal (upper) and cytokinesis-defective (lower) stomata in wild-type and myb3r4 silique valves, respectively. Siliques were fixed and cleared for observation using DIC microscopy, as described previously.Citation10 The occurrence of incomplete cytokinesis typically generates single-celled stomata, as shown in the lower panel. Scale bars, 10 μm. (C) Increased frequency of single-celled stomata in EDC plants. Silique valves from the indicated lines were observed using DIC microscopy to count normal and single-celled stomata, as described previously.Citation10 Bars indicate the SD (n ≥ 10).
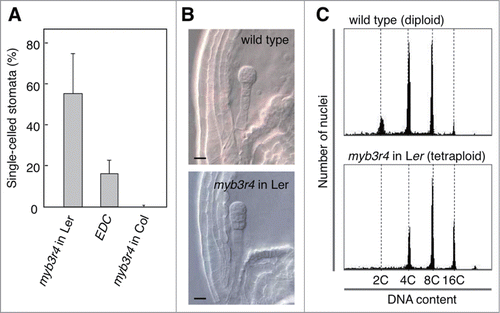
Because the kn mutation (allele X37-2) had been generated by X-ray irradiation,Citation15 the enhanced defects in EDC plants may have been caused by additional mutations introduced by X-ray, which were transmitted to the original kn mutant. Alternatively, the enhanced defects may be attributed to the hybrid genetic background that was achieved by our genetic cross between the kn mutant in the Landsberg erecta (Ler) background and the myb3r1 myb3r4 double mutant in the Columbia (Col) background (). To test these possibilities, we analyzed the F1 and F2 progenies of the cross between EDC plants and the myb3r4 mutant (). Because both parents are homozygous for myb3r4, all F1 and F2 plants carried the same homozygous myb3r4 mutation. In the F1 generation, all plants showed much weaker phenotypes compared with EDC plants, although we still observed a small number of plants with slightly more pronounced defects than that observed in the parental myb3r4 plants (). All F1 plants examined showed defects in less than 8% of stomata, whereas more than 8% of stomata were defective in all the EDC plants examined. In the F2 generation, however, some of the plants examined showed an elevated frequency (>8%) of defective cytokinesis (). We observed 6 such plants among a total of 20 F2 plants analyzed, and this ratio statistically satisfied the theoretical segregation ratio of 3:1 that is expected from a single recessive locus that is responsible for phenotypic enhancement (P = 0.82, chi-squared test). To evaluate the effects of the Ler genetic background, we prepared another cross between the myb3r4 mutant in the Col background and a wild-type plant in the Ler background, to rule out the potential effects of additional mutations introduced by X-ray (). In the F2 progeny, we selected plants that were homozygous for myb3r4 and used them for phenotypic analysis. In this genetic analysis, we again observed plants with enhanced defective cytokinesis. The control cross between myb3r4 and wild-type plants, both in the Col background, did not yield such progeny with enhanced phenotype, demonstrating that the abnormality of myb3r4 is differently affected by the Col and Ler genetic backgrounds. It should also be noted that the F2 segregants of the cross between wild-type (Ler) and myb3r4 (Col) plants exhibited a much less severe phenotype compared with EDC plants (). This is consistent with our interpretation that EDC plants may carry additional mutations that enhance the myb3r4 phenotype, in addition to the half-Ler genome background retained in EDC plants. Moreover, we observed severely enhanced cytokinesis defects in a near isogenic line (NIL) carrying the myb3r4-1 mutation in the Ler genomic background, which had been developed by repeated (5 times) backcrossing with Ler and genotyping. In addition to the dramatically elevated proportion of single-celled stomata, the NIL showed abnormal embryo development and produced tetraploid plants in the selfed progeny (31 tetraploids out of a total of 122 plants examined), which was previously observed in the myb3r1 myb3r4 double mutant,Citation10 but never in the myb3r4 single mutant in the Col background or in EDC plants (). Taken together, our results led us to conclude that the enhanced defects observed in EDC plants may result from the combined effects of additional mutations and the genetic background (Col/Ler hybrid).
Figure 2. Inheritance pattern of enhanced defective cytokinesis after the various genetic crosses. The histograms show the frequency distribution of plants with different severities of defective cytokinesis in the indicated populations. The frequency of single-celled stomata was determined in silique valves of each plant, and the number of plants that fell into the range of each bin is shown for each population. (A) Parental myb3r4 plants in the Col background. (B) Parental EDC plants in the Col/Ler hybrid background. (C) F1 progeny of the cross between EDC and myb3r4 plants. (D) F2 progeny of the cross between EDC and myb3r4 plants. (E) F2 progeny of the cross between the myb3r4 plant and a wild-type plant in the Ler background. (F) F2 progeny of the cross between the myb3r4 plant and a wild-type plant in the Col background. For (E) and (F), plants that were homozygous for myb3r4 were selected from the F2 segregating populations and used for phenotypic analysis.Citation11
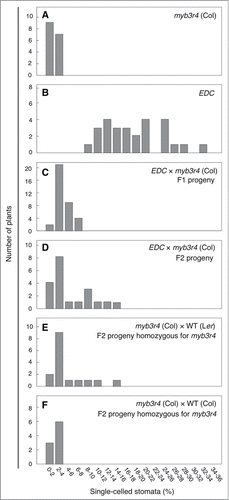
Figure 3. Enhanced defective cytokinesis in a NIL carrying myb3r4 in the Ler background. (A) The proportion of single-celled stomata in silique valves was compared between the NIL with myb3r4 in the Ler background, EDC plants (in the Col/Ler hybrid background), and myb3r4 in the Col background. Bars indicate the SD (n ≥ 10). (B) Malformed embryo in the NIL with myb3r4 in the Ler background (lower panel) compared with the normal embryo that developed in the wild-type ovule (upper panel). For observation of embryos using DIC microscopy, ovules were fixed and cleared as described previously.Citation10 Scale bars, 10 μm. (C) The NIL with myb3r4 in the Ler background generated tetraploid offspring. The lower panel shows the representative DNA ploidy pattern of the tetraploid plants found in the selfed progeny of the NIL, which was characterized by the loss of the 2C peak compared with the DNA ploidy pattern of a wild-type diploid plant (upper panel). Leaf samples were used for ploidy analysis, as described previously.Citation11 Higher levels of DNA ploidy correspond to nuclei that underwent endoreplication during the leaf development, as typically observed in this analysis.
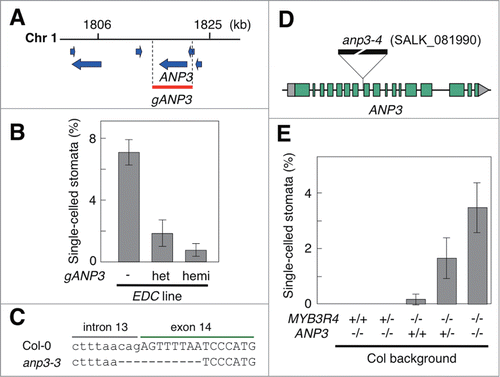
To identify the potential recessive locus in EDC plants, we performed map-based cloning using the F2 segregating population of the cross between EDC and myb3r4 (Col) plants. By analyzing approximately 500 F2 plants using PCR-based polymorphic markers, we localized the potential recessive locus within a 20 kb region located on chromosome 1, which was flanked by the 2 PCR markers at positions 1806 kb and 1825 kb of chromosome 1. This region contains the full coding sequences (CDSs) of 4 annotated genes: AT3G06020, AT3G06030, AT3G06035, and AT3G06040 (). Because AT3G06030 encodes ANP3 MAPKKK, a component of the MAPK pathway regulating cytokinesis, we analyzed this gene in further detail. First, we introduced genomic DNA containing only AT3G06030 as a full CDS (gANP3) into EDC plants, and confirmed that it complemented the enhanced cytokinesis defects (). The effect of phenotypic complementation was dependent on gene dosage, as plants that were hemizygous for gANP3 showed more pronounced complementation than did heterozygous plants (). Next, we sequenced genomic ANP3 and found that EDC plants and the original kn mutant have an 11 bp deletion that spans the junction between intron 13 and exon 14 of ANP3 (). This mutation, anp3-3, results in the retention of intron 13 in mature mRNA (not shown) and presumably represents a null allele of anp3. Finally, to analyze the effects of the ANP3 knockout allele, we identified its T-DNA insertion mutant (SALK_081990) from the SALK collection,Citation16 which was named anp3-4 and used for crossing with the myb3r4 mutant (). As expected, the double myb3r4 anp3-4 mutant in the Col background exhibited an increased number of single-celled stomata compared with the myb3r4 mutant, although no cytokinesis defect was observed in the single anp3-4 mutant (). In addition, we again observed gene dosage effects of ANP3 on the severity of the cytokinesis defects, suggesting that the expression levels of ANP3 may be an important determinant for the promotion of cytokinesis (). All of these results support our conclusion that the enhanced cytokinesis defects observed in EDC plants are caused partially by the loss of function of ANP3, which had been mutated during the generation of mutant populations.Citation15
Figure 4. Mutations in anp3 enhanced the defective cytokinesis observed in myb3r4. (A) Chromosomal location of the putative recessive locus that is responsible for the EDC phenotype. The recessive locus was mapped to the 20 kb region located between 2 polymorphic markers at positions 1806 and 1825 kb on chromosome 1. The blue arrows show the positions and orientations of the CDSs of the genes annotated in this genomic region. The red box indicates the position of the genomic construct, gANP3, which was used for complementation analysis. (B) Complementation analysis of EDC plants. The gANP3 construct was introduced into the EDC plants, and both hemizygous and heterozygous plants in the T2 generation were used for phenotypic analysis. The T2 segregants that had lost the transgene were used as a control. (C) The EDC plants had an 11 bp deletion in the ANP3 gene. The deletion in this mutant allele, anp3-3, eliminated the acceptor site of intron 13 and caused retention of this intron in the mature mRNA. (D) The T-DNA insertion allele of anp3. The position of the T-DNA insertion is indicated for the SALK_081990 line, which was used as a knockout mutant of anp3 (anp3-4). The boxes indicate exons, and the areas in green and gray colors correspond to protein coding and noncoding regions, respectively. (E) Genetic interaction between anp3 and myb3r4. After the genetic cross of the 2 knockout mutants, anp3 and myb3r4, both in the Col background, F2 segregants with the indicated genotypes were selected and used for phenotypic analysis. The mutant and wild-type alleles are indicated by minus and plus signs, respectively. For (B) and (E), the frequency of single-celled stomata was determined in silique valves, to estimate the severity of the cytokinesis defects in each plant. Bars indicate the SD (n ≥ 5).
The genetic interaction observed between myb3r4 and anp3 may reflect functional crosstalk between these 2 proteins. One explanation is that ANP3 may be involved in the activation of MYB3R1/4 by transmitting activatory signals to the downstream MAPK pathway. If ANP3 affects MYB3R1/4 activity, the anp3 mutation may have some impact on the expression of the downstream targets of MYB3R1/4. To clarify this possibility, we performed a real-time RT–PCR analysis and determined the transcript levels of INFLORESCENCE RECEPTOR-LIKE KINASE2 (IMK2)Citation17, KN, and CYCB2;1, which are representative target genes of MYB3R1/4 (). The transcript levels of these genes were reduced significantly upon myb3r4 mutation, as reported previously.Citation10,11 However, an additional anp3 mutation did not further decrease their transcript levels (), suggesting that the phenotypic effect of the anp3 mutation is not due to decreased MYB3R1/4 activity. Because anp3 single mutation had negligible effect on target transcription (Fig. S1), we concluded that the observed transcript abundance in a mutant combination is not due to epistatic interaction between anp3 and myb3r4, but to the absence of the mutational effect of anp3 on transcription of MYB3R1/4 target genes.
Figure 5. Molecular basis of the genetic interaction between myb3r4 and anp3. (A) Transcript levels of MYB3R1/4-target genes. Seedlings (7-day-old plants) with the indicated genotypes were used for real-time RT–PCR analysis, as described previously.Citation10 For each quantification, 3 biological replicates were analyzed. The expression levels of ACT2 were used as an internal standard to normalize the transcript levels of each gene. The CDKA;1 gene, which is not under the control of MYB3R1/4, was similarly analyzed as a control. Bars indicate the SD. Sequences of primers used for RT-PCR are available upon request. (B) KN and MPK13 had a negligible effect of phenotypic rescue in myb3r4 anp3 plants. The full-length cDNAs of KN and MPK13 were placed under the control of the CDKA;1 promoter as described previouslyCitation10 and introduced into the myb3r4 anp3 plants via the floral dip method.Citation20 The frequency of single-celled stomata in silique valves was determined in more than 5 T1 plants for each construct. Bars indicate the SD. The method for the plasmid construction is available upon request. (C) Heat map showing expression changes upon myb3r1 myb3r4 mutation. Fold-change levels in log2 value were calculated for the genes encoding components of the NACK-PQR pathway and downstream targets using published data from a microarray analysis of wild-type and myb3r1 myb3r4 plants.Citation11
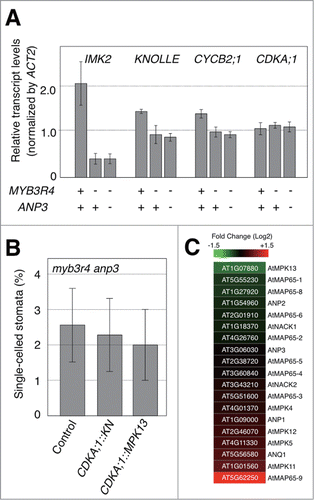
An alternative explanation for the ANP3–MYB3R4 genetic interaction is that MYB3R4 may transcriptionally activate the genes that act as components of the ANP3-mediated MAPK pathway. In this scenario, there may be critical target genes of MYB3R1/4 encoding components of the MAPK pathway, the downregulation of which is responsible for cytokinesis defects in myb3r4 anp3. We previously showed that transgenic expression of KN under the CDKA;1 promoter rescued the cytokinesis defects of the myb3r1 myb3r4 double mutant.Citation10 However, we showed here that the CDKA;1:KN construct did not significantly rescue the cytokinesis defects of myb3r4 anp3 (), suggesting that, unlike what was observed in the myb3r1 myb3r4 double mutant, downstream target genes other than KN may be critical in this double mutant. To identify such critical target genes of MYB3R1/4, we evaluated the effects of a myb3r1/4 double mutation on the expression of the genes involved in the ANP3-mediated MAPK pathway. Because the microtubule-associated proteins of the MAP65 family are known as downstream targets of the NACK-PQR pathway for the execution of cytokinesis,Citation18,19 all 9 genes in the MAP65 family of Arabidopsis were included in this analysis. Using our previous microarray data from the comparison of wild-type and myb3r1/4 double-mutant plants,Citation11 we found that only MPK13 among the genes tested was significantly downregulated in the double mutant (). However, transgenic expression of MPK13 driven by the CDKA;1 promoter did not significantly rescue the cytokinesis defects of the myb3r4 anp3 double mutant (); therefore, we were not able to specify the MYB3R1/4 target genes that are critical for the myb3r4 anp3 phenotype. It remains possible that MYB3R1/4 regulates the MPK cascade by weakly affecting multiple components of this pathway; thus, none of the individual components would be critical.
In summary, we showed that the genetic interaction between myb3r4 and anp3 may not be due to either direct positive regulation of MYB3R1/4 by the NACK-PQR pathway or to MYB3R1/4-mediated activation of specific components of the NACK-PQR pathway. Rather, it is most likely that downstream targets of the MYB3R1/4-mediated pathway and the ANP3-mediated MAPK cascade act in close proximity to promote cytokinesis. Alternatively, common downstream targets that are essential for cytokinesis may be shared by the 2 signaling pathways. Therefore, MYB3R1/4 may positively affect cytokinesis via the activation of multiple pathways, one of which may affect the downstream events that may also be regulated by the ANP3-mediated MAPK cascade. This pathway may be independent of the KN-mediated pathway defined previously, because the genetic interaction between myb3r4 and anp3 may not be due to the downregulation of KN.
Our results also suggest that the isolation of mutations that enhance myb3r4 is an effective strategy to elucidate the novel genetic pathway that connects MYB3R1/4 with the promotion of cytokinesis. Finally, we found that there is a vast difference between the effects of the genetic backgrounds of Col and Ler on the cytokinesis defects caused by the myb3r4 mutation. Further genetic analyses may allow the identification of important quantitative trait loci that affect the myb3r4 phenotype, which may be responsible for the novel regulatory pathways of cytokinesis in Arabidopsis.
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Figure_S1.docx
Download MS Word (3.9 MB)Acknowledgments
We thank K. Komatsu, Y. Sako, C. Kotani, C. Inoue, and A. Yamamoto for technical assistance; and Y. Machida, Y. Yoshioka, and Y. Mizukami for helpful discussion.
Supplemental Material
Supplemental data for this article can be accessed on the publisher's website.
Funding
This work was supported in part by JSPS KAKENHI (grant numbers 25119710 and 26113509), MEXT KAKENHI (grant number 26291058), and JST, CREST.
References
- Lee YR, Liu B. The rise and fall of the phragmoplast microtubule array. Curr Opin Plant Biol 2013; 16:757-63; PMID:24172707; http://dx.doi.org/10.1016/j.pbi.2013.10.008
- Jürgens G. Plant cytokinesis: fission by fusion. Trends Cell Biol 2005; 15:277-83; PMID:15866032; http://dx.doi.org/10.1016/j.tcb.2005.03.005
- Takahashi Y, Soyano T, Sasabe M, Machida Y. A MAP kinase cascade that controls plant cytokinesis. J Biochem 2004; 136:127-32; PMID:15496582; http://dx.doi.org/10.1093/jb/mvh118
- Kosetsu K, Matsunaga S, Nakagami H, Colcombet J, Sasabe M, Soyano T, Takahashi Y, Hirt H, Machida Y. The MAP kinase MPK4 is required for cytokinesis in Arabidopsis thaliana. Plant Cell 2010; 22:3778-90; PMID:21098735; http://dx.doi.org/10.1105/tpc.110.077164
- Soyano T, Nishihama R, Morikiyo K, Ishikawa M, Machida Y. NQK1/NtMEK1 is a MAPKK that acts in the NPK1 MAPKKK-mediated MAPK cascade and is required for plant cytokinesis. Genes Dev 2003; 17:1055-67; PMID:12704083; http://dx.doi.org/10.1101/gad.1071103
- Nishihama R, Ishikawa M, Araki S, Soyano T, Asada T, Machida Y. The NPK1 mitogen-activated protein kinase kinase kinase is a regulator of cell-plate formation in plant cytokinesis. Genes Dev 2001; 15:352-63; PMID:11159915; http://dx.doi.org/10.1101/gad.863701
- Krysan PJ, Jester PJ, Gottwald JR, Sussman MR. An Arabidopsis mitogen-activated protein kinase kinase kinase gene family encodes essential positive regulators of cytokinesis. Plant Cell 2002; 14:1109-20; PMID:12034900
- Nishihama R, Soyano T, Ishikawa M, Araki S, Tanaka H, Asada T, Irie K, Ito M, Terada M, Banno H, Yamazaki Y, Machida Y. Expansion of the cell plate in plant cytokinesis requires a kinesin-like protein/MAPKKK complex. Cell 2002; 109:87-99; PMID:11955449; http://dx.doi.org/10.1016/S0092-8674(02)00691-8
- Takahashi Y, Soyano T, Kosetsu K, Sasabe M, Machida Y. HINKEL kinesin, ANP MAPKKKs and MKK6/ANQ MAPKK, which phosphorylates and activates MPK4 MAPK, constitute a pathway that is required for cytokinesis in Arabidopsis thaliana. Plant Cell Physiol 2010; 51:1766-76; PMID:20802223
- Haga N, Kato K, Murase M, Araki S, Kubo M, Demura T, Suzuki K, Müller I, Voss U, Jürgens G, Ito M. R1R2R3-Myb proteins positively regulate cytokinesis through activation of KNOLLE transcription in Arabidopsis thaliana. Development 2007; 134:1101-10; PMID:17287251
- Haga N, Kobayashi K, Suzuki T, Maeo K, Kubo M, Ohtani M, Mitsuda N, Demura T, Nakamura K, Jürgens G, Ito M. Mutations in MYB3R1and MYB3R4cause pleiotropic developmental defects and preferential down-regulation of multiple G2/M-specific genes in Arabidopsis. Plant Physiol 2011; 157:706-17; PMID:21862669
- Lukowitz W, Mayer U, Jürgens G. Cytokinesis in the Arabidopsis embryo involves the syntaxin-related KNOLLE gene product. Cell 1996; 84:61-71; PMID:8548827
- Ito M, Araki S, Matsunaga S, Itoh T, Nishihama R, Machida Y, Doonan JH, Watanabe A. G2/M-phase-specific transcription during the plant cell cycle is mediated by c-Myb-like transcription factors. Plant Cell 2001; 13:1891-905; PMID:11487700; http://dx.doi.org/10.1105/tpc.13.8.1891
- Araki S, Ito M, Soyano T, Nishihama R, Machida Y. Mitotic cyclins stimulate the activity of c-Myb-like factors for transactivation of G2/M phase-specific genes in tobacco. J Biol Chem 2004; 279:32979-88; PMID:15175336; http://dx.doi.org/10.1074/jbc.M403171200
- Mayer U, Torres Ruiz RA, Berleth T, Miséra S, Jürgens G. Mutations affecting body organization in the Arabidopsis embryo. Nature 1991; 353:402-7.
- Alonso JM, Stepanova AN, Leisse TJ, Kim CJ, Chen H, Shinn P, Stevenson DK, Zimmerman J, Barajas P, Cheuk R, et al. Genome-wide insertional mutagenesis of Arabidopsis thaliana. Science 2003; 301:653-7; PMID:12893945
- ten Hove CA, Bochdanovits Z, Jansweijer VM, Koning FG, Berke L, Sanchez-Perez GF, Scheres B, Heidstra R. Probing the roles of LRR RLK genes in Arabidopsis thaliana roots using a custom T-DNA insertion set. Plant Mol Biol 2011; 76:69-83; PMID:21431781; http://dx.doi.org/10.1007/s11103-011-9769-x
- Sasabe M, Soyano T, Takahashi Y, Sonobe S, Igarashi H, Itoh TJ, Hidaka M, Machida Y. Phosphorylation of NtMAP65-1 by a MAP kinase down-regulates its activity of microtubule bundling and stimulates progression of cytokinesis of tobacco cells. Genes Dev 2006; 20:1004-14; PMID:16598040
- Sasabe M, Kosetsu K, Hidaka M, Murase A, Machida Y. Arabidopsis thaliana MAP65-1 and MAP65-2 function redundantly with MAP65-3/PLEIADE in cytokinesis downstream of MPK4. Plant Signal Behav 2011; 6:743-7; PMID:21455028
- Clough SJ, Bent AF. Floral dip: a simplified method for Agrobacterium -mediated transformation of Arabidopsis thaliana. Plant J 1998; 16:735-43; PMID:10069079
