Abstract
OsPti1a (Pto-interacting protein 1a) has important roles in the regulation of immune responses in rice. Phosphorylation of a conserved threonine in OsPti1a is necessary to activate defense responses; however, the regulatory mechanism of OsPti1a-mediated immune responses is still obscure. Recently, we revealed that OsPti1a forms protein complex(es) at the plasma membrane and this localization is required for its function. Here, we show that membrane-localized OsPti1a was selectively phosphorylated. Additionally, phosphorylation was not required for the localization of OsPti1a at the membrane. These results suggest that OsPti1a protein is selectively regulated by its phosphorylation after OsPti1a localizes to the plasma membrane.
Abbreviations
| HR | = | hypersensitive response |
| Pti1 | = | Pto-interacting protein 1 |
Developing an understanding of the plant immune system can potentially lead to the breeding of crops with enhanced disease resistance against pathogens. Already it is known that basal resistance including pathogen-associated molecular pattern (PAMP)-triggered immunity (PTI) confers resistance against a broad range of pathogens in plants.Citation1 Therefore, unraveling the details of PTI signaling will contribute to disease control for a broad range of pathogen infections.
Previously, we identified OsPti1a, a serine/threonine protein kinase with high sequence similarity to tomato Pti1.Citation2 In the OsPti1a null mutant (ospti1a), a series of defense responses including HR (hypersensitive response)-like spontaneous lesion formation is activated even without pathogen challenge. Overexpressor of OsPti1a has enhanced disease susceptibility against compatible pathogens, indicating that OsPti1a functions as a negative regulator of plant immunity in rice.Citation2 The phosphorylation of threonine 233 (Thr-233) of OsPti1a, a conserved phosphorylation site of Pti family proteins in tomato,Citation3 arabidopsisCitation4 and rice, is required for the activation of basal resistance against Xanthomonas oryzae pv. oryzae (Xoo) infection.Citation5 This result suggests that the phosphorylation of OsPti1a has an important role in regulating basal resistance in rice.
In Arabidopsis, AtPTI1–2 directly interacts with, and is phosphorylated by, its upstream kinase OXI1 (oxidative stress-inducible 1)/AGC2–1, which is regulated by its upstream kinase AtPDK1 (phosphoinositide-dependent protein kinase 1).Citation4 In rice, we previously reported that OsPti1a also interacts with OsOxi1, the kinase that phosphorylates Thr-233 of OsPti1a. Further, OsOxi1 interacts with OsPdk1 that phosphorylates OsOxi1 in vitro and is rapidly phosphorylated after treatment with chitin, a major PAMP, in vivo. OsPdk1 also contributes to the activation of basal resistance against rice blast, suggesting that the Pdk1-Oxi1-Pti1 phosphorylation cascade plays an important role in activating PTI against pathogens in Arabidopsis and rice.Citation6 However, details of the regulatory mechanism underlying OsPti1a activation of disease resistance against pathogens are still unclear.
To investigate the subcellular localization of OsPti1a protein and its molecular function, we used reverse-genetic approaches that revealed that the 10 N-terminal amino acids of OsPti1a are important for complementing the ospti1a mutant phenotype.Citation7 Additionally, fractionation analysis and transient expression assays revealed that OsPti1a localizes to the plasma membrane through N-terminal palmitoylation, and OsPti1a forms a complex at the plasma membrane. Further, using liquid chromatography with tandem mass spectrometry (LC-MS/MS) analysis, OsPti1a-associated proteins were identified in the microsomal fraction, and several candidate proteins are associated with plant immunity. These findings suggest that the regulation of immune responses mediated by OsPti1a could depend on the interaction and regulation of OsPti1a-associated proteins at the plasma membrane.Citation7
We reported that upstream kinases, OsOxi1 and OsPdk1, are rapidly phosphorylated upon chitin treatment.Citation5,6 However, it remains unknown whether the phosphorylation of OsPti1a coincident with its membrane localization. Therefore, to analyze whether membrane-localized OsPti1a is phosphorylated, we purified OsPti1a protein and fractionated the preparation into soluble and microsomal fractions by ultracentrifugation. OsPti1a was previously shown to specifically localize at the plasma membrane, such that OsPti1a protein in the microsomal fraction came mostly from the plasma membrane.Citation7 After that, tagged OsPti1a protein (hemagglutinin-strepII-tag: HastrepII) was affinity-purified using Strep-Tactin beads. Phosphorylation was detected by Phos-tag molecules that have a high affinity to phosphate groups, thereby shifting the mobility of phosphorylated proteins compared to non-phosphorylated proteins in SDS-PAGE.Citation8 In this assay, we found a slow migrating band of OsPti1a protein in the microsomal fraction, compared to that in the soluble fraction (). This result indicates that a portion of OsPti1a protein in the plasma membrane is constitutively phosphorylated without stimuli. It is noted that only a fraction of the OsPti1a protein at the plasma membrane was phosphorylated (). This result raises the hypothesis that a small portion of OsPti1a is constantly phosphorylated at Thr-233 by OsOxi1, and that recognition of PAMPs or pathogen invasion triggers greatly enhanced OsPti1a phosphorylation. This hypothesis is supported by the fact that OsOxi1 as well as OsPdk1 is rapidly phosphorylated after chitin treatment.Citation5,6
Figure 1. Membrane-localized OsPti1a is phosphorylated in transgenic rice cultured cells. (A) Total protein derived from transgenic rice cultured cells expressing 35S:OsPti1a-HAstrepII in the OsPti1a mutant background was fractionated into soluble (S) and microsomal (M) fractions by ultracentrifugation. OsPti1a-HAstrepII protein was affinity-purified from fractionated samples using Strep-Tactin beads. Affinity-purified protein was separated on 10% acrylamide gels with or without Phos-tag included and subjected to immunoblot analysis with anti-HA antibody. The black arrowhead indicates phosphorylated OsPti1a. (B) The phosphorylation status of OsPti1a does not affect its cellular localization. Immunoblots of soluble (S) and microsomal (M) fractions derived from cultured cells expressing OsPti1aWT or OsPti1aT233A in the ospti1a mutant background. Cellular fractionation was confirmed by immunoblotting using anti-OsPIP2;1 antibody as a control for integral membrane proteins. Lower panels indicate CBB staining of membrane proteins after immunoblotting. (C) OsPti1aT233A-HAstrepII protein is not phosphorylated in the microsomal fraction. OsPti1a-HAstrepII and OsPti1aT233A-HAstrepII protein were affinity-purified from fractionated samples using Strep-Tactin beads. The black arrowhead indicates phosphorylated OsPti1a. (D) Gel filtration fractions of protein extracts from rice cultured cells expressing OsPti1aWT or OsPti1aT233A were subjected to immunoblot analyses (upper panel) using an anti-OsPti1a antibody. The fraction numbers and molecular masses (kDa) are indicated at the top of the figure. After immunoblotting, the membrane was stained with CBB (lower panel). On the right side of the figure, black arrowheads indicate non-specific bands, and the white arrowheads indicate OsPti1a protein.
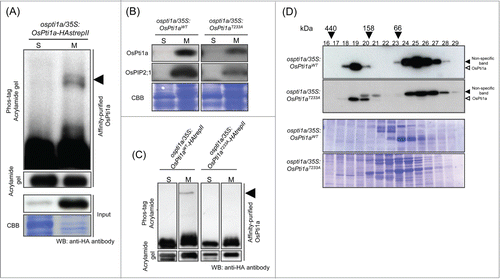
Next, to confirm whether the phosphorylation of OsPti1a affects its localization to the plasma membrane, we analyzed the localization of OsPti1aT233A, whose T233 was replaced with alanine by site-directed mutagenesis (). OsPti1aT233A was detected in the microsomal fraction in the same way as OsPti1aWT was, indicating that the phosphorylation of OsPti1a does not affect its cellular localization. Membrane localized-OsPti1aT233A was not phosphorylated (). This result agrees with findings that the molecular size of the OsPti1a T233A complex did not change () indicating that, at least, OsPti1a forms a stable complex that is attached to the membrane fraction whether OsPti1a is phosphorylated or not.
Despite this finding, T-233 phosphorylation of OsPti1a is necessary for the activation of basal resistance against Xanthomonas oryzae pv oryzae.Citation5 Our findings of OsPti1a phosphorylation at the plasma membrane probably have important implications for understanding the activation of basal resistance in rice. Identification of the phosphorylation status of OsPti1a protein at the plasma membrane during basal resistance responses sheds light on the signal transduction pathway for activating immunity and reveals the contribution of the OsPti1a complex in immunity. To elucidate the role of OsPti1a phosphorylation in basal resistance, a phosphoproteomics approach could help us characterize the phosphorylation status of OsPti1a during basal resistance responses. Indeed, phosphoproteomic techniques have been shown to be powerful tools for identifying signaling components in plants.Citation9 In the future, we will continue our efforts to reveal the regulatory mechanism of OsPti1a-mediated resistance by exploiting phosphoproteomic methods.
Methods
Protein extraction, affinity-purification and protein gel blot analysis
The subcellular fractionation methods were performed as described in Matsui et al.Citation7 To characterize the phosphorylation of OsPti1a protein in the microsomal fraction, rice suspension cultured cells were collected and homogenized in membrane isolation buffer (50 mM Tris-HCl (pH7.5), 0.25M sorbitol, 2 mM EDTA, 5 mM ascorbic acid, 50 mM β-glycerophosphate, 10 mM Na3VO4, 10 mM NaF, cOmplete Protease Inhibitor Cocktail Tablets [Roche, Mannheim, Germany]). For fractionation analysis, the resulting supernatants were used as the soluble fractions and the pellets were used as the microsomal fractions.
To determine the phosphorylation status of the OsPti1a-HAstrepII protein, microsomal fractions were mixed with 500 μl of membrane isolation buffer including 1% (w/v) Triton X-100. A total of 500 μl of soluble or microsomal fraction was incubated with 50 μl of Strep-Tactin beads (IBA GmbH, Gottingen, Germany) at 4°C for 2 hours with gentle rocking. The matrix was washed 5 times with the membrane isolation buffer, resuspended in 50 μl of 2 × SDS sample buffer, boiled for 5 minutes and 10 μl aliquots were loaded on SDS gels. To detect phosphorylated OsPti1a by SDS-PAGE, we used Phos-tag AAL-107 (NARD Institute Ltd., Amagasaki, Japan). Phosphorylated OsPti1a was electrophoretically separated on 8% (w/v) SDS-polyacrylamide gels containing 40 μM Phos-tag. Protein equivalents of the supernatant and pellet fractions were analyzed by immunoblotting using the anti-OsPti1a antibody,Citation2 anti-PIP2;1 (plasma membrane intrinsic protein 2;1) antibody (Cosmo Bio Co. Ltd., Tokyo, Japan) or anti-HA antibody (Covance). Prior to immunoblotting, the nitrocellulose membrane was analyzed by Coomassie brilliant blue (CBB) staining to confirm protein recovery and equal loading. Gel fractionation methods were described in Matsui et al.Citation7
Acknowledgments
We thank Mayuko Harada-Yamazaki for technical assistance and Pamela Gan at RIKEN CSRS for critically reading the manuscript.
Funding
This work was supported in part by a grant from the Ministry of Agriculture, Forestry, and Fisheries of Japan (Genomics for Agricultural Innovation, PMI0005) and by a Grant-in-Aid for Scientific Research (KAKENHI, No. 23780047 to A. T.; No. 23780048 and No. 25850034 to H. M.).
References
- Lacombe S, Rougon-Cardoso A, Sherwood E, Peeters N, Dahlbeck D, van Esse HP, Smoker M, Rallapalli G, Thomma BPHJ, Staskawicz B, et al. Interfamily transfer of a plant pattern-recognition receptor confers broad-spectrum bacterial resistance. Nature Biotechnol 2010; 28:365-369; PMID:20231819; http://dx.doi.org/10.1038/nbt.1613.
- Takahashi A, Agrawal GK, Yamazaki M, Onosato K, Miyao A, Kawasaki T, Shimamoto K, Hirochika H. Rice Pti1a negatively regulates RAR1-dependent defense responses. Plant Cell 2007; 19:2940-2951; PMID:17890377; http://dx.doi.org/10.1105/tpc.106.047142.
- Sessa G, D'Ascenzo M, Martin GB. The major site of the Pti1 kinase phosphorylated by the Pto kinase is located in the activation domain and is required for Pto-Pti1 physical interaction. Eur J Biochem 2000; 267, 171-178; PMID:10601864; http://dx.doi.org/10.1046/j.1432-1327.2000.00979.x.
- Anthony RG, Khan S, Costa J, Pais MS, Bӧgre L. The Arabidopsis protein kinase PTI1-2 is activated by convergent phosphatidic acid and oxidative stress signaling pathways downstream of PDK1 and OXI1. J Biol Chem 2006; 281, 37536-375346; PMID:17040918; http://dx.doi.org/10.1074/jbc.M607341200.
- Matsui H, Yamazaki M, Kishi-Kaboshi M, Takahashi A, Hirochika H. AGC kinase OsOxi1 positively regulates basal resistance through suppression of OsPti1a-mediated negative regulation. Plant Cell Physiol 2010; 51:1731-1744; PMID:20739304; http://dx.doi.org/10.1093/pcp/pcq132.
- Matsui H, Miyao A, Takahashi A, Hirochika H. Pdk1 Kinase regulates basal disease resistance through the OsOxi1-OsPti1a phosphorylation cascade in rice.Plant Cell Physiol 2010; 51:2082-2091; PMID:21051443.
- Matsui H, Fujiwara M, Hamada S, Shimamoto K, Nomura Y, Nakagami H, Takahashi A, Hirochika H. Plasma membrane localization is essential for Oryza sativa Pto-interacting protein 1a-mediated negative regulation of immune signaling in rice. Plant Physiol. 2014; 166(1):327-36; doi:10.1104/pp.114.243873.
- Kinoshita E, Kinoshita-Kikuta E, Takiyama K, Koike T. Phosphate-binding tag, a new tool to visualize phosphorylated proteins. Mol. Cell Proteomics 2006; 5:749-757; http://dx.doi.org/10.1074/mcp.T500024-MCP200.
- Nakagami H, Sugiyama N, Ishihama Y, Shirasu K. Shotguns in the front line: Phosphoproteomics in plants. Plant Cell Physiol 2012; 53(1):118-124; PMID:22039104; http://dx.doi.org/10.1093/pcp/pcr148.
