Abstract
The Eph receptor tyrosine kinases and their ephrin ligands direct axon pathfinding and neuronal cell migration, and mediate many other cell-cell communication events. The Ephs and ephrins both localize to the plasma membrane and, upon cell-cell contact, form extensive signaling assemblies at the contact sites. Recent structural, biochemical and cell-biological studies revealed that these assemblies are generated not only via Eph-ephrin interactions, but also via homotypic interactions between neighboring receptor molecules. In addition, Eph-Eph interactions mediate receptor pre-clustering, which ensures fast and efficient activation once ligands come into contact range. Here we summarize the current knowledge about the homotypic Eph-Eph interactions and discuss how they could modulate the initiation of Eph/ephrin signaling.
Introduction
Eph receptors, the largest family of receptor tyrosine kinases (RTKs), and their ephrin ligands, are membrane-anchored molecules that play important roles in axon pathfinding and neuronal cell migration, and in controlling other cell-cell interactions, including those of vascular endothelial cells.Citation1-3 The binding of ephrins to the extracellular region of Eph receptors results in activation of their cytoplasmic tyrosine kinase domainCitation4 and also leads to initiation of a reverse signal into the ephrin-bearing cell.Citation5,6 (see ). The Ephs and the ephrins are divided into A and B subclasses, based on their affinities for each other and on sequence conservation (https://eph-nomenclature.med.harvard.edu). With few exceptions, the 10 different EphA receptors promiscuously bind to and are activated by 6 A-ephrins while the EphB receptors (EphB1-B6) interact with 3 different B-ephrins (ephrin-B1-B3). The interactions of Ephs and ephrins lead to aggregation of both molecules in distinct clusters within their respective plasma membranes, resulting in the formation of signaling centers at the zones of cell-cell contact.Citation7
Figure 1. Schematic representation of an Eph/ephrin signaling assembly formed between 2 interacting cells. There are several types of protein-protein interactions stabilizing these assemblies: (i) hetero-dimerization and hetero-tetramerization interactions involving the Eph LBDs and the ephrin ectodomains; (ii) “clustering” Eph-Eph interactions involving the Eph Cys-rich regions; and iii) Eph-Eph LBD-FNIII interactions that are likely important for receptor pre-clustering (prior to contact with ligand) and for subsequent ligand-independent recruitment of unliganded Ephs to the signaling Eph/ephrin assemblies. Eph pre-clustering ensures a fast and efficient activation once ligands come within a contact distance. The ligand-independent recruitment of Ephs to Eph/ephrin assemblies modulates the Eph signal by allowing the size of the receptor clusters to exceed the size of the juxtaposed ephrin clusters, as well as by allowing recruitment of different receptor subtypes within the same signaling assemblies. The interacting FNIII and LBD regions are shown in cyan, other Eph domains are in blue; ephrins are shown in red. RBD, Receptor-Binding Domain; LBD, Ligand-Binding Domain; cys, Cysteine-Rich Domain (CDR); FN, Fibronectin type III Domain; TK, Tyrosine Kinase Domain; sam, Sterile Alpha Motive. Phosphorylated intracellular tyrosines are shown as small orange circles.
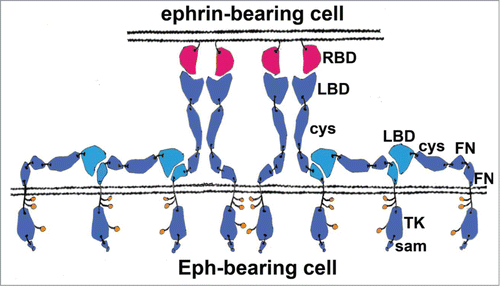
Ligand-induced and Ligand-Independent Eph Receptor Activation
A key feature of the activation of receptor tyrosine kinases is dimerization of the receptor upon ligand binding. Regardless of the precise details of receptor/ligand complex architecture and conformational changes caused by ligand binding, all RTKs share a common necessity for dimerization of their cytoplasmic kinase domains. The kinase domains exist in either an inactive or an active conformation, and dimerization is thought to drive activation by promoting trans phosphorylation of their activation loops and/or by directly stabilizing an active kinase conformation via kinase-kinase contacts.Citation8
Since the first crystal structures of Eph receptors were published, their activation mechanism has been described as ‘unique’.Citation9,10 This is based on the observation that Eph activation is the result of the association of at least 4 separate molecules, 2 ligands and 2 receptors, which is distinct from the canonical way of RTK activation where 2 receptors are brought together by a single ligand entity (either a monomeric ligand or a stable, receptor-independent, ligand dimer).Citation11 Although it was known that the Eph ligand-binding domain (LBD) is sufficient and necessary for ephrin binding, the precise stoichiometry and architecture of the activated Eph/ephrin complex were not known before crystal structures became available.Citation12-14 The structure of the complex between the minimal interacting domains of EphB2 and ephrin-B2, the first structure of an Eph/ephrin complex, revealed a ring-like assembly, where each receptor interacts with 2 ligands and each ligand with 2 receptors. One of the 2 distinct ligand-receptor interfaces is responsible for forming the initial high-affinity 1:1 dimer, the second one (a significantly smaller one) assembles the ligand/receptor dimers into hetero-tetramers.
Although this original concept has mostly stood the test of the many additional structural studies, a more complex and finessed picture of Eph receptor activation has appeared lately. Indeed, the structures of the full ectodomain of EphA2 revealed that the initial receptor/ligand recognition and binding steps are not enough for fully functional Eph activation.Citation13,15 Moreover, they suggested that ligand binding might not always be even needed for the activation of Eph receptors, and that homotypic Eph-Eph contacts are very important for the assembly of signaling clusters. Although requirement for additional contacts, via regions outside the minimal interacting domains, had been reported earlier using mutagenesis in combination with cell-based signaling assays,Citation16-18 the EphA2 full ectodomain structures provided the first direct evidence for this. The ligand-independent Eph-Eph interface observed in these structures, located C-terminally from the LBD, was named ‘clustering interface’. Thus, the combination of a ‘hetero-dimerization’ and a ‘clustering’ interface, was suggested to be required for the formation of actively signaling Eph assemblies, containing potentially hundreds of receptors.
The formation of Eph clusters on the cell surface following ligand binding has been known for a long time to be necessary for various, if not all, Eph-related downstream signaling events.Citation19 Interestingly, ligand-independent recruitment of Eph receptors into Eph/ephrin signaling clusters has also been observed. For example, it was shown that a mutated Eph receptor, unable to bind ligand, is recruited to and gets phosphorylated in receptor clusters containing wild-type Eph receptors.Citation20 The data indicated that direct Eph-Eph interactions, involving the LBD and CRD domains, are responsible for recruitment of different Eph receptor subtypes, with different ligand-binding preferences, within the same cell-surface signaling assemblies.Citation21 A schematic representation of an Eph/ephrin signaling cluster, including recruited unliganded Ephs, shown in , illustrates a molecular mechanism that could account for these observations.
While in some cases Eph signaling is tumor suppressive,Citation22,23 overexpression of RTKs in general, and Ephs in particular, is well known to occur in various cancers,Citation24 and it has been suggested that receptor dimerization may be driven by a direct mass action.Citation25 Consequently, enhanced basal (potentially tumorigenic) kinase activation would occur once the local receptor concentration is high enough to cause ligand-independent clustering. Recent structural data has clearly demonstrated that, in the case of the Eph RTKs, the same receptor clustering interfaces can be used for generating oligomeric assemblies both in the presence or absence of ligand. The role of the ephrin ligands, therefore, might be to simply increase the local concentration of receptors, facilitating the formation of higher-order Eph signaling assemblies.Citation13,26 This phenomenon presents a loophole that might be used by certain tumors to hijack the Eph/ephrin signaling pathway and induce a level of basal (ligand-independent) receptor activation enough to cause cellular havoc.Citation23
Other intriguing observations, relevant to Eph clustering, include the potential recruitment of cell-surface molecules outside of the Eph family into the higher-order Eph assemblies. Such interactions might be important for directing synapse formation (e.g. the Eph/NMDAR interactions)Citation27,28 or modulating other signaling pathways (e.g., the Eph/CXCR4 interactionsCitation29 or the Eph/FGFR interactionsCitation30).
In light of the now well-characterized ligand-independent Eph-Eph interactions, it is also notable that similar findings have been reported for the RTKs of the ErbB family.Citation11 Certain cancer cells overexpress one of the 4 members of the family, ErbB2, which has been shown to dimerize with the kinase-impaired ErbB3 in a ligand-independent manner, and to cause its phosphorylation. In fact, the ErbB2/ErbB3 heterodimer is possibly the most active of the ErbB dimers and efficiently activates several downstream pathways, including MAPK and Akt.
LBD-FNIII Interactions in Unliganded Eph Receptors
The structural and functional studies mentioned above raised questions about the biological relevance of receptor-receptor interactions not only in the context of the Eph-ephrin signaling clusters, but also in the ‘resting state’ of the Eph receptors prior to any contact with ligand. A structural study on EphA4 published recently,Citation31 for example, documented that the unliganded EphA4 uses its LBD to bind the second FNIII domain of a neighboring EphA4 molecule (see ). A closer inspection of the structure of the ligand-free EphA2 ectodomain published earlierCitation13 shows a similar LBD-FNIII interaction, although the interacting interface is considerably smaller. Interestingly B-class Ephs also form LBD-FNIII contacts as evident from the structure of the unliganded EphB2 ectodomain (unpublished data) (). This kind of head-to-tail interaction has not been reported for other RTKs, but might have been overlooked in other studies, just as the EphA2 LBD-FNIII interactions initially were. The implication that the LBD-FNIII interactions mediate functional ligand-independent receptor association (pre-clustering) was supported by mutagenesis experiments showing that mutations in EphA4 that strengthen this interface increase EphA4 activation, while mutations that weaken it, decrease the EphA4 signal.Citation31 Furthermore, a number of cancer-related Eph mutations have been reported to fall not only within the LBD and CRD, but also within the FN3 domains,Citation32 consistent with the biological importance of the LBD/FN head-to-tail interactions. Hence, the ‘head-to-head’ Eph-Eph interactions involving the LBD and the CRD domains and the ‘head-to-tail’ interactions probably represent 2 parallel receptor clustering mechanisms. Interestingly, previous studies have also suggested that the CRD domain might be directly involved in the recruitment of LBD-deleted EphA3 to wild-type EphA3 clusters.Citation18 These various types of receptor-receptor interactions could represent a new emerging paradigm for fine-tuning of membrane receptor activity.
Figure 2. LBD-FNIII Eph-Eph interactions. Top: The interacting LBD-FNIII regions of unliganded Ephs from the crystal structures of EphA2 (left), EphA4 (middle) and EphB2 (right). The LBDs are in green and the second FNIII repeats (FN2), in cyan. Bottom: The LBDs of the same Ephs bound to their ephrin ligands from the crystal structures of the Eph/ephrin complexes EphA2/ephrin-A1 (left), EphA4/ephrin-A5 (middle) and EphB2/ephrin-B2 (right). The LBDs are in green and the ephrins, in cyan.
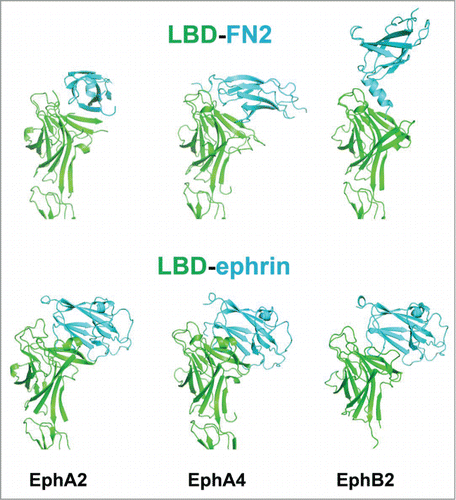
In this context, it is important to note that, as illustrated in , the surface area of the Eph LBD interacting with the FNIII region of a neighboring receptor overlaps with the LBD surface interacting with ephrin ligands, thus limiting the LBD-FNIII interaction to unliganded Ephs. The significantly larger LBD-FNIII interface in EphA4Citation31 might have important biological implications. Indeed, receptor pre-clustering would presumably ensure a fast and efficient activation once ligands come into contact range. Therefore, the fact that the EphA4 has a larger LBD-FNII interface than other Ephs, enabling a more efficient receptor pre-clustering, might account, at least partially, for the observed unique ligand promiscuity of the EphA4 receptor.Citation1-3
The schematic illustration in shows the LBD/FN interactions occurring in cis, on the same cell membrane. Nevertheless, we cannot fully exclude the possibility that these interactions occur in trans. In our opinion, this is a less likely scenario, since the maximum length of a fully-stretched Eph-ECD is around 150 Å and a head-to-tail in-trans LBD/FN Eph-Eph interaction might not allow for enough separation between interacting cells.Citation33
It is important to keep in mind that the strength of the downstream signal generated by Eph receptor activation depends on the size of the signaling clusters.Citation34,35 Furthermore, it has been shown that, upon cell-cell contact, the size of the receptor clusters on the Eph-expressing cell can significantly exceed the size of the opposing ephrin clusters on the ephrin-expressing cell.Citation20 The same LBD-FNIII Eph-Eph interactions discussed above and shown in might also (in addition to the ‘canonical’ LBD/LBD and CRD/CRD interactions) account for this phenomenon, as illustrated on the schematics representation of an Eph/ephrin cluster presented in .
While the head-to-tail LBD-FNIII Eph interactions were only recently reported, somewhat similar molecular interactions have been shown to play important roles in modulating other cellular events. Examples include Kinesin-1,Citation36 Myosin,Citation37 and ERM proteinsCitation38 where head-to-tail interactions mask biologically important binding sites.Citation39-41 Consequently, an activation step is required to expose the masked regions and confer biological activity.
Eph/Ephrin Interactions In cis
While the emerging concept of LBD-FNIII head-to-tail interactions as mediators of Eph signaling is novel and unexpected, it is somewhat related to a previously studied phenomenon, namely Eph/ephrin interactions in cis (by ligands and receptors expressed on the surface of the same cells). Unlike the prototypical Eph/ephrin interactions between molecules from opposing cells, the in cis Eph/ephrin interactions do not seem to result in receptor activation and initiation of downstream signaling.Citation6,42 There are conflicting reports regarding exactly which Eph and ephrin regions are involved in the in cis interactions,Citation43-45 with both the LBD and FNIII Eph domains being implicated. The in cis interactions were shown to have an inhibitory effect on the conventional in trans Eph/ephrin interactions and co-expressed ephrins appear to act primarily as signaling antagonists.Citation46 Importantly, the cis interaction between ephrin-A5 and EphA3 on retinal ganglion cells was shown to take place through the second FNIII domain (FN2) of the receptor, attenuating receptor phosphorylation and downstream signaling.Citation43 Since this interaction involves the same FNIII region as the FNIII-LBD Eph-Eph interactions discussed above, it's fascinating to speculate that these mechanisms have evolved as 2 competing ways to modulate the Eph/ephrin signaling.
The hypothesis that the in cis Eph/ephrin binding is structurally distinct form the canonical in trans Eph/ephrin binding is further supported by a recent study involving cells co-expressing Ephs and ephrins that do not interact or signal in trans.Citation47 Specifically, ephrin-B2 does not have measurable affinity for the EphA3 LBD, and thus cannot effect forward signaling in trans, but was shown to attenuate EphA3 signaling via in cis binding. Furthermore, this and other similar studiesCitation43 also show that mutations in the ligands and receptors can be identified that selectively affect only the in cis or in trans interactions. While the precise mechanism behind the antagonistic effects of co-expressed ephrins on Eph signaling is unknown, one could speculate that in cis interactions between ephrins and FNIII Eph regions might prevent the conformational rearrangements caused by in trans Eph/ephrin contacts that are required for the formation of ordered, signaling Eph/ephrin clusters between interacting cells (see ).
Figure 3. Schematic representation of the in cis vs in trans interactions between Eph receptors and ephrins. In many neurons the expression levels of A-class ephrins are high and they co-localize with Eph receptors to the same membrane patches. Within these patches, they are involved in cis interactions, which are generally inhibitory to the forward Eph signaling. Although the precise molecular mechanism of the inhibition is not well understood, it has been suggested that the in cis interactions prevent the conformational rearrangements normally effected by the in trans Eph/ephrin contacts that are required for the formation of the ordered Eph/ephrin signaling assemblies. The in cis interacting FNIII regions are shown in cyan, other Eph domains are in blue; ephrins are shown in red. RBD, Receptor-Binding Domain; LBD, Ligand-Binding Domain; cys, Cysteine-Rich Domain; FN, Fibronectin type III Domain; TK, Tyrosine Kinase Domain; sam, Sterile Alpha Motive.
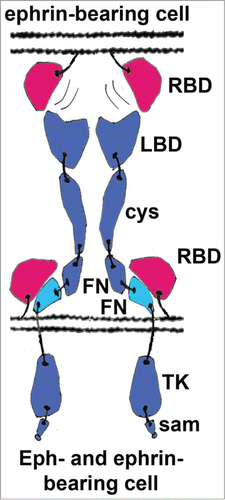
In some neuronal types, co-expressed Ephs and ephrins have been shown to segregate laterally into distinct membrane micro-domains, not only preventing the types of in cis Ephs/ephrin interactions discussed above, but also potentially leading to opposing signaling effects on the growth cones, depending on the precise co-expression levels of receptors and ligands.Citation48,49 Recent findings have added other pieces to the puzzle of the roles of co-expressed Eph's and ephrins: Kao and Kania reported that the relative contributions of trans vs cis interactions are influenced by the subcellular distribution of ligands in the receptor-containing membrane batches of spinal motor axons.Citation44 In aggregate, these studies suggest that the balance between in cis and in trans Eph-ephrin binding is more important than previously appreciated for the diversity of axon trajectories and other cellular/neuronal responses.
Conclusion
In this review we have outlined new structural observations on novel Eph-Eph interactions, which we suggest provide an efficient way to fine-tune both the strength of the downstream Eph signal and the Eph ligand specificity. Interestingly, while the LBD surface area involved in the homotypic LBD-FNIII interactions overlaps with the canonical high-affinity Eph-ephrin interface, the FNIII surface area involved in the LBD-FNIII interactions overlaps with an interface proposed to mediate Eph-ephrin interactions in cis. The intricate interplay of these competing interactions seems to fine-tune the precise Eph signaling response in a context-dependent manner.
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Acknowledgments
We thank Dorothea Robev and Momchil Kolev for technical support and Marina Himanen for preparation of the illustrations.
Funding
Our research on Eph receptors and ephrins is supported by a grant from the National Institutes of Health RO1NS038486.
References
- Flanagan JG, Vanderhaeghen P. The ephrins and Eph receptors in neural development. Annu Rev Neurosci 1998; 21:309-45; PMID:9530499; http://dx.doi.org/10.1146/annurev.neuro.21.1.309
- Frisen J, Holmberg J, Barbacid M. Ephrins and their Eph receptors: multitalented directors of embryonic development. EMBO J 1999; 18:5159-65; PMID:10508149; http://dx.doi.org/10.1093/emboj/18.19.5159
- Kullander K, Klein R. Mechanisms and functions of EPH and ephrin signalling. Nat Rev Mol Cell Bio 2002; 3:475-86; http://dx.doi.org/10.1038/nrm856
- Wilkinson DG. Multiple roles of EPH receptors and ephrins in neural development. Nat Rev Neurosci 2001; 2:155-64; PMID:11256076; http://dx.doi.org/10.1038/35058515
- Holland SJ, Gale NW, Mbamalu G, Yancopoulos GD, Henkemeyer M, Pawson T. Bidirectional signalling through the EPH-family receptor Nuk and its transmembrane ligands. Nature 1996; 383:722-5; PMID:8878483; http://dx.doi.org/10.1038/383722a0
- Egea J, Klein R. Bidirectional Eph-ephrin signaling during axon guidance. Trends Cell Biol 2007; 17:230-8; PMID:17420126; http://dx.doi.org/10.1016/j.tcb.2007.03.004
- Himanen JP, Saha N, Nikolov DB. Cell-cell signaling via Eph receptors and ephrins. Curr Opin Cell Biol 2007; 19:534-42; PMID:17928214; http://dx.doi.org/10.1016/j.ceb.2007.08.004
- Huse M, Kuriyan J. The conformational plasticity of protein kinases. Cell 2002; 109:275-82; PMID:12015977; http://dx.doi.org/10.1016/S0092-8674(02)00741-9
- Himanen JP, Henkemeyer M, Nikolov DB. Crystal structure of the ligand-binding domain of the receptor tyrosine kinase EphB2. Nature 1998; 396:486-91; PMID:9853759; http://dx.doi.org/10.1038/24904
- Himanen JP, Rajashankar KR, Lackmann M, Cowan CA, Henkemeyer M, Nikolov DB. Crystal structure of an Eph receptor-ephrin complex. Nature 2001; 414:933-8; PMID:11780069; http://dx.doi.org/10.1038/414933a
- Lemmon MA, Schlessinger J, Ferguson KM. The EGFR family: not so prototypical receptor tyrosine kinases. Cold Spring Harbor perspec Biol 2014; 6:a020768; PMID:24691965
- Himanen JP. Ectodomain structures of Eph receptors. Semin Cell Dev Biol 2012; 23:35-42; PMID:22044883; http://dx.doi.org/10.1016/j.semcdb.2011.10.025
- Himanen JP, Yermekbayeva L, Janes PW, Walker JR, Xu K, Atapattu L, Rajashankar KR, Mensinga A, Lackmann M, Nikolov DB, et al. Architecture of Eph receptor clusters. Proc Natl Acad Sci U S A 2010; 107:10860-5; PMID:20505120; http://dx.doi.org/10.1073/pnas.1004148107
- Himanen JP, Goldgur Y, Miao H, Myshkin E, Guo H, Buck M, Nguyen M, Rajashankar KR, Wang B, Nikolov DB. Ligand recognition by A-class Eph receptors: crystal structures of the EphA2 ligand-binding domain and the EphA2/ephrin-A1 complex. EMBO Rep 2009; 10:722-8; PMID:19525919; http://dx.doi.org/10.1038/embor.2009.91
- Seiradake E, Harlos K, Sutton G, Aricescu AR, Jones EY. An extracellular steric seeding mechanism for Eph-ephrin signaling platform assembly. Nat Struct Mol Biol 2010; 17:398-402; PMID:20228801; http://dx.doi.org/10.1038/nsmb.1782
- Smith FM, Vearing C, Lackmann M, Treutlein H, Himanen J, Chen K, Saul A, Nikolov D, Boyd AW. Dissecting the EphA3/Ephrin-A5 interactions using a novel functional mutagenesis screen. J Biol Chem 2004; 279:9522-31; PMID:14660665; http://dx.doi.org/10.1074/jbc.M309326200
- Day B, To C, Himanen JP, Smith FM, Nikolov DB, Boyd AW, Lackmann M. Three distinct molecular surfaces in ephrin-A5 are essential for a functional interaction with EphA3. J Biol Chem 2005; 280:26526-32; PMID:15901737; http://dx.doi.org/10.1074/jbc.M504972200
- Lackmann M, Oates AC, Dottori M, Smith FM, Do C, Power M, Kravets L, Boyd AW. Distinct subdomains of the EphA3 receptor mediate ligand binding and receptor dimerization. J Biol Chem 1998; 273:20228-37; PMID:9685371; http://dx.doi.org/10.1074/jbc.273.32.20228
- Davis S, Gale NW, Aldrich TH, Maisonpierre PC, Lhotak V, Pawson T, Goldfarb M, Yancopoulos GD. Ligands for EPH-related receptor tyrosine kinases that require membrane attachment or clustering for activity. Science 1994; 266:816-9; PMID:7973638; http://dx.doi.org/10.1126/science.7973638
- Wimmer-Kleikamp SH, Janes PW, Squire A, Bastiaens PI, Lackmann M. Recruitment of Eph receptors into signaling clusters does not require ephrin contact. J Cell Biol 2004; 164:661-6; PMID:14993233; http://dx.doi.org/10.1083/jcb.200312001
- Janes PW, Griesshaber B, Atapattu L, Nievergall E, Hii LL, Mensinga A, Chheang C, Day BW, Boyd AW, Bastiaens PI, et al. Eph receptor function is modulated by heterooligomerization of A and B type Eph receptors. J Cell Biol 2011; 195:1033-45; PMID:22144690; http://dx.doi.org/10.1083/jcb.201104037
- Day BW, Stringer BW, Boyd AW. Eph receptors as therapeutic targets in glioblastoma. British J Cancer 2014; 111:1255-61; PMCID:PMC4183860
- Pasquale EB. Eph receptors and ephrins in cancer: bidirectional signalling and beyond. Nat Rev Cancer 2010; 10:165-80; PMID:20179713; http://dx.doi.org/10.1038/nrc2806
- Blume-Jensen P, Hunter T. Oncogenic kinase signalling. Nature 2001; 411:355-65; PMID:11357143; http://dx.doi.org/10.1038/35077225
- Nagy P, Claus J, Jovin TM, Arndt-Jovin DJ. Distribution of resting and ligand-bound ErbB1 and ErbB2 receptor tyrosine kinases in living cells using number and brightness analysis. Proc Natl Acad Sci U S A 2010; 107:16524-9; PMID:20813958; http://dx.doi.org/10.1073/pnas.1002642107
- Nikolov DB, Xu K, Himanen JP. Eph/ephrin recognition and the role of Eph/ephrin clusters in signaling initiation. Biochim biophys Acta 2013; 1834:2160-5; PMID:23628727; http://dx.doi.org/10.1016/j.bbapap.2013.04.020
- Dalva MB, Takasu MA, Lin MZ, Shamah SM, Hu L, Gale NW, Greenberg ME. EphB receptors interact with NMDA receptors and regulate excitatory synapse formation. Cell 2000; 103:945-56; PMID:11136979; http://dx.doi.org/10.1016/S0092-8674(00)00197-5
- Drescher U. Excitation at the synapse: Eph receptors team up with NMDA receptors. Cell 2000; 103:1005-8; PMID:11163177; http://dx.doi.org/10.1016/S0092-8674(00)00204-X
- Salvucci O, de la Luz Sierra M, Martina JA, McCormick PJ, Tosato G. EphB2 and EphB4 receptors forward signaling promotes SDF-1-induced endothelial cell chemotaxis and branching remodeling. Blood 2006; 108:2914-22; PMID:16840724; http://dx.doi.org/10.1182/blood-2006-05-023341
- Fukai J, Yokote H, Yamanaka R, Arao T, Nishio K, Itakura T. EphA4 promotes cell proliferation and migration through a novel EphA4-FGFR1 signaling pathway in the human glioma U251 cell line. Mole Cancer Ther 2008; 7:2768-78; PMID:18790757; http://dx.doi.org/10.1158/1535-7163.MCT-07-2263
- Xu K, Tzvetkova-Robev D, Xu Y, Goldgur Y, Chan YP, Himanen JP, Nikolov DB. Insights into Eph receptor tyrosine kinase activation from crystal structures of the EphA4 ectodomain and its complex with ephrin-A5. Proc Natl Acad Sci U S A 2013; 110:14634-9; PMID:23959867; http://dx.doi.org/10.1073/pnas.1311000110
- Ding L, Getz G, Wheeler DA, Mardis ER, McLellan MD, Cibulskis K, Sougnez C, Greulich H, Muzny DM, Morgan MB, et al. Somatic mutations affect key pathways in lung adenocarcinoma. Nature 2008; 455:1069-75; PMID:18948947; http://dx.doi.org/10.1038/nature07423
- Jessell TM, Kandel ER. Synaptic transmission: a bidirectional and self-modifiable form of cell-cell communication. Cell 1993; 72 Suppl:1-30; PMID:8381334; http://dx.doi.org/10.1016/S0092-8674(05)80025-X
- Schaupp A, Sabet O, Dudanova I, Ponserre M, Bastiaens P, Klein R. The composition of EphB2 clusters determines the strength in the cellular repulsion response. J Cell Biol 2014; 204:409-22; PMID:24469634; http://dx.doi.org/10.1083/jcb.201305037
- Stein E, Lane AA, Cerretti DP, Schoecklmann HO, Schroff AD, Van Etten RL, Daniel TO. Eph receptors discriminate specific ligand oligomers to determine alternative signaling complexes, attachment, and assembly responses. Genes Dev 1998; 12:667-78; PMID:9499402; http://dx.doi.org/10.1101/gad.12.5.667
- Dietrich KA, Sindelar CV, Brewer PD, Downing KH, Cremo CR, Rice SE. The kinesin-1 motor protein is regulated by a direct interaction of its head and tail. Proc Natl Acad Sci U S A 2008; 105:8938-43; PMID:18579780; http://dx.doi.org/10.1073/pnas.0803575105
- Jung HS, Komatsu S, Ikebe M, Craig R. Head-head and head-tail interaction: a general mechanism for switching off myosin II activity in cells. Mole Biol Cell 2008; 19:3234-42; PMID:18495867; http://dx.doi.org/10.1091/mbc.E08-02-0206
- Fievet B, Louvard D, Arpin M. ERM proteins in epithelial cell organization and functions. Biochim Biophys Acta 2007; 1773:653-60; PMID:16904765; http://dx.doi.org/10.1016/j.bbamcr.2006.06.013
- Morales FC, Takahashi Y, Momin S, Adams H, Chen X, Georgescu MM. NHERF1/EBP50 head-to-tail intramolecular interaction masks association with PDZ domain ligands. Mole Cell Biol 2007; 27:2527-37; PMID:17242191; http://dx.doi.org/10.1128/MCB.01372-06
- Gary R, Bretscher A. Ezrin self-association involves binding of an N-terminal domain to a normally masked C-terminal domain that includes the F-actin binding site. Mole Biol Cell 1995; 6:1061-75; PMID:7579708; http://dx.doi.org/10.1091/mbc.6.8.1061
- Pearson MA, Reczek D, Bretscher A, Karplus PA. Structure of the ERM protein moesin reveals the FERM domain fold masked by an extended actin binding tail domain. Cell 2000; 101:259-70; PMID:10847681; http://dx.doi.org/10.1016/S0092-8674(00)80836-3
- Dudanova I, Klein R. The axon's balancing act: cis- and trans-interactions between Ephs and ephrins. Neuron 2011; 71:1-3; PMID:21745632; http://dx.doi.org/10.1016/j.neuron.2011.06.030
- Carvalho RF, Beutler M, Marler KJ, Knoll B, Becker-Barroso E, Heintzmann R, Ng T, Drescher U. Silencing of EphA3 through a cis interaction with ephrinA5. Nat Neurosci 2006; 9:322-30; PMID:16491080; http://dx.doi.org/10.1038/nn1655
- Kao TJ, Kania A. Ephrin-mediated cis-attenuation of Eph receptor signaling is essential for spinal motor axon guidance. Neuron 2011; 71:76-91; PMID:21745639; http://dx.doi.org/10.1016/j.neuron.2011.05.031
- Yin Y, Yamashita Y, Noda H, Okafuji T, Go MJ, Tanaka H. EphA receptor tyrosine kinases interact with co-expressed ephrin-A ligands in cis. Neurosci Res 2004; 48:285-96; PMID:15154674; http://dx.doi.org/10.1016/j.neures.2003.11.009
- Hornberger MR, Dutting D, Ciossek T, Yamada T, Handwerker C, Lang S, Weth F, Huf J, Wessel R, Logan C, et al. Modulation of EphA receptor function by coexpressed ephrinA ligands on retinal ganglion cell axons. Neuron 1999; 22:731-42; PMID:10230793; http://dx.doi.org/10.1016/S0896-6273(00)80732-1
- Falivelli G, Lisabeth EM, Rubio de la Torre E, Perez-Tenorio G, Tosato G, Salvucci O, Pasquale EB. Attenuation of eph receptor kinase activation in cancer cells by coexpressed ephrin ligands. PLoS One 2013; 8:e81445; http://dx.doi.org/10.1371/journal.pone.0081445
- Marquardt T, Shirasaki R, Ghosh S, Andrews SE, Carter N, Hunter T, Pfaff SL. Coexpressed EphA receptors and ephrin-A ligands mediate opposing actions on growth cone navigation from distinct membrane domains. Cell 2005; 121:127-39; PMID:15820684; http://dx.doi.org/10.1016/j.cell.2005.01.020
- Salaita K, Nair PM, Petit RS, Neve RM, Das D, Gray JW, Groves JT. Restriction of receptor movement alters cellular response: physical force sensing by EphA2. Science 2010; 327:1380-5; PMID:20223987; http://dx.doi.org/10.1126/science.1181729
