Abstract
We recently uncovered a novel genetic mechanism that generates the phenotypic uniformity, or canalization, of BMP signaling and cell fate specification during patterning of the dorsal-ventral (D/V) axis in D. melanogaster embryos. We went on to show that other wild-type Drosophila species lack this canalizing genetic circuitry and, consequently, have non-robust D/V patterning. In this review, we propose molecular mechanisms that may give rise to stereotyped BMP signaling, and we identify an additional species that could have decanalized D/V patterning. Extension of these analyses could in turn help explain why canalization is not a universal necessity for species survival.
Abbreviations
| BMP | = | Bone Morphogenetic Protein |
| Cv-2 | = | crossveinless-2 |
| D | = | Drosophila |
| D/V | = | dorsal-ventral |
| Dpp | = | Decapentaplegic |
| Egr | = | eiger |
| JNK | = | Jun N-terminal Kinase |
| Mad | = | Mothers against dpp |
| pMad | = | phosphorylated Mad |
| PWM | = | Position Weight Matrix |
| Sog | = | Short Gastrulation |
| Tkv | = | Thickveins |
| Tld | = | Tolloid |
| Tsg | = | Twisted Gastrulation |
| VRE | = | Ventral Repression Element |
| Zen | = | zerknüllt |
Early embryonic patterning is inherently critical to the viability of the developing organism. As such, the genetic circuits that specify the embryonic axes have long been considered to be both highly robust to perturbation and deeply evolutionarily conserved. Recently, our work has delineated a genetic network in D. melanogaster that ensures robustness of D/V axial patterning and identified Drosophila species that lack this network.1 These findings argue against the hypothesis that developmental robustness is universal. Here, we first speculate as to the molecular mechanisms underlying this phenomenon of phenotypic canalization. Then we consider the question of whether D. santomea is the only species with decanalized D/V patterning.
The role of Bone Morphogenetic Protein (BMP) signaling in directing cell fate specification of dorsal tissues in the Drosophila melanogaster embryo was first discovered by the Gelbart laboratory over 20 years ago.Citation2-5 Since then, many additional components of the BMP pathway, involved in either the extracellular movement of BMP ligands, or reception and transduction of the BMP signal, have been identified.Citation6-10 At the onset of zygotic transcription, in the early stage 5 blastoderm embryo, the BMP ligand Decapentaplegic (Dpp) is expressed throughout the dorsal half of the embryo, while a second BMP ligand, Screw, is expressed ubiquitously. The BMP type I receptors, Thickveins (Tkv) and Saxophone, and the type II receptor Punt, are maternally deposited and expressed throughout the embryo.Citation11 Despite this widespread expression of BMP ligands and the uniform distribution of the BMP receptors, BMP signaling - as visualized by an antibody specific to the phosphorylated form of the BMP signal transducer Mothers against dpp (Mad) - is initially observed only in a subset of the blastoderm nuclei as a low intensity, broad domain centered on the dorsal midline. By the onset of gastrulation, 30 minutes later, BMP signaling has intensified and refined to a sharp 6-8 cell stripe, which defines the dorsal midline. In the early gastrula embryo, dorsal cells posterior to the cephalic furrow with high levels of BMP signaling will be fated as the extra-embryonic amnioserosa tissue.Citation5,12,13 Once specified, amnioserosa cells no longer divide, rather they undergo endoreduplication cycles and form a polyploid squamous epithelium. The number of amnioserosa cells in later stage embryos therefore directly reflects the number of cells with high levels of BMP signaling at the onset of gastrulation.
Two processes cooperate to produce this pattern of BMP signaling. First, the extracellular BMP binding proteins Short Gastrulation (Sog) and Twisted Gastrulation (Tsg), and the metalloprotease Tolloid (Tld) facilitate the movement of BMP ligands to the dorsal midline. When bound to Sog and Tsg, BMPs are unavailable to the BMP receptors and thus diffuse freely in the extracellular perivitelline space. The Tld protease, which is expressed in the same pattern as Dpp, cleaves Sog only when it is bound to BMPs. This cleavage releases the BMP ligands from the inhibitory complex and allows them to bind to their receptors and signal.Citation7,14-16 Repeated cycles of complex formation, diffusion, proteolysis and receptor binding result in the concentration of BMP ligands and signaling in the dorsal regions of the embryo.
The second step in this process involves an intracellular positive feedback mechanism, which is necessary for the refinement and intensification of the signaling domain prior to gastrulation.Citation17 The binding of BMP ligands to their receptors causes the constitutively active kinase domain of Punt to phosphorylate the juxtamembrane domains of the type I receptors. The activated type I receptors then phosphorylate the C-terminal domain of Mad. Phosphorylated Mad (pMad) then complexes with its obligate transcriptional co-factor Medea and enters the nucleus to direct the expression of BMP target genes.Citation18,19 The transcriptional output of the BMP signaling pathway intensifies and refines BMP signaling by increasing the ability of cells with previous BMP signaling to bind and internalize BMP ligand.Citation17 Thus, the dorsally-directed extracellular movement of BMP ligands is coupled with an intracellular positive feedback circuit to produce a spatially-bistable pattern of receptor ligand interactions at the onset of gastrulation.
Recently, our laboratory and others quantified pMad staining in individual embryos and established that the spatial extent and signal intensity of BMP signaling is highly stereotyped in the D. melanogaster embryo.Citation1,10 We infer that this uniformity of the wild-type BMP signaling domain leads directly to low variability of the number of amnioserosa cells specified in D. melanogaster embryos. Thus, the phenotypic output of BMP signaling, which is dependent upon the long-range diffusion of a signaling ligand through the extracellular space, is strikingly uniform between individual wild-type D. melanogaster embryos. This phenotypic uniformity of the D. melanogaster embryonic axial patterning is an example of developmental canalization.
In our recent paper, we identified 2 additional genes that contribute to the establishment of the wild-type BMP signaling domain. We first examined the activity of a component of the positive-feedback circuit, eiger (egr). egr encodes of a Tumor Necrosis Factor - α ligandCitation20 that activates the Jun N-terminal Kinase (JNK) pathway. egr transcription is dependent on BMP signaling, and the injection of egr mRNA into the blastoderm embryo locally increases BMP-receptor interactions. However, while the absence of egr or maternally-supplied JNK activity reduces pMad intensity to half that of wild type, the average number of amnioserosa cells in egr mutants is nearly identical to that of wild-type embryos.
Next, we examined the activity of crossveinless-2 (cv-2) at the blastoderm stage. cv-2, which encodes an extracellular, membrane proximal BMP binding proteinCitation21, has both maternal and zygotic activities, and in the absence of both activities, the intensity of BMP signaling doubles, accompanied by a variable expansion in the width of the signaling domain. However, despite the quantitative change in BMP signaling, cv-2 null embryos have only a slightly higher number of amnioserosa cells. Thus, despite quantitative changes to BMP signaling, neither mutation disrupts organismal patterning, indicating that the early Drosophila embryo can tolerate a 2-fold variation in BMP signaling strength without gross phenotypic consequences.
The most compelling finding of our recent work, however, arose from our analysis of the phenotype of the egr cv-2 double mutant embryos. Superficially, the 2 mutations displayed incomplete epistasis; the average intensity of BMP signaling in the double mutant embryos was identical to that of wild-type embryos. However, the inter-embryonic variability in BMP signaling was extremely high in egr cv-2 embryos, as compared to wild type or either single mutant. Moreover, the increased variability in BMP signaling in the double mutant embryos was reflected in a significant increase in the variability in the number of amnioserosa cells. Thus, egr and cv-2 act together to ensure phenotypic canalization of BMP signaling and amnioserosa specification.
The incomplete epistasis between the 2 genes strongly suggests that both Egr and Cv-2 act on the level of BMP ligand availability and receptor-ligand interactions (). The dorsally directed concentration of BMP ligands through the extracellular action of Sog, Tsg and Tld results in an initial asymmetry in the intensity of BMP signaling, observed in Stage 5 embryos (). We propose that the BMP-dependent transcription of egr results in activation of the JNK homolog Basket, which then phosphorylates one or more proteins to increase the functional concentration of cell surface BMP receptors in regions of previous BMP signaling (). These regions thus become more able to compete for BMP ligands that have been concentrated dorsally by the extracellular ligand binding proteins. The increase in receptor concentration on the cell surface could be brought about through elevation of the rate of receptor secretion from either the recycling endosomes or a bulk exocytic process. Simultaneously, in the lateral region of the embryo the BMP binding protein Cv-2 acts as a non-signaling sink for BMP ligands. Thus, in the simplest form of our model, increased cell-surface receptor concentration in the dorsal-most regions, coupled with the presence of a sink for BMP ligands in the lateral regions, transforms the initial graded distribution of BMPs into a spatially bistable pattern of BMP-receptor interactions and signaling.
Figure 1. (A) Schematic depicting the activities of some of the known factors that shape the BMP signaling domain at the onset of gastrulation. Localization of Dpp to the dorsal midline is promoted by the binding of Dpp (green boxes) to an inhibitory complex of Sog (orange) and Tsg (red). Dpp is released from this complex by the cleavage of Sog by the metalloprotease Tld (blue scissors). Free Dpp can be bound by its receptor Tkv (green) or by Cv-2 (red dashes), which is under the control of early zen transcription and present throughout the dorsal half of the embryo. The level of pMad signaling in individual nuclei is represented by the intensity of green. (B) BMP signaling induces the expression of the positive feedback gene encoding Egr (blue) which can then activate the JNK cascade (teal arrows). JNK activity leads to an increase in BMP receptor-ligand interactions, possibly by increasing BMP receptor concentration on the cell surface, which in turn increases BMP signaling (compare left and right cells) (C) In regions of positive feedback (teal dashes), an increase in the BMP receptor concentration on the cell surface could convert the activity of Cv-2 from an antagonist to an agonist of BMP signaling (thick red dashes). The final signaling domain at the onset of gastrulation is constrained by the limiting of ligand to those cells with high concentrations of cell surface receptors.
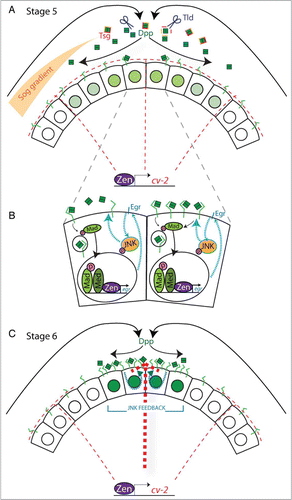
Additional genetic interactions suggested a refinement to the above model. We showed that in certain genetic backgrounds a lower level of Cv-2 can act as a BMP signaling agonist during D/V patterning. These data are consistent with previous experimental data and mathematical modeling that, in the developing wing vein, Cv-2 possesses dual functionality: while low levels of Cv-2 relative to the BMP receptors promote BMP signaling, high levels of Cv-2 antagonize BMP signaling.Citation22 Integrating these findings with ours, we suggest that the positive feedback circuit could, by significantly increasing the concentration of BMP receptors in the dorsal region, transform Cv-2 from an inhibitor that competes for limiting BMP ligands laterally to an agonist that presents BMP ligands to the receptors dorsally. Thus, the positive feedback circuit could additionally serve to convert Cv-2 from a non-signaling sink laterally to a local source of BMP ligands dorsally, thereby accentuating positive feedback ().
In summary, in the D. melanogaster embryo, we demonstrated that BMP signaling during D/V patterning has a topology that is very similar to computationally-derived minimal circuits necessary for switch like behavior, namely positive feedback enhancing activation coupled with a linear or non-linear negative regulation regime. These genetic interactions further support the hypothesis that BMP signaling generates spatial bistability during amnioserosa specification.
The second half of our paper provided insights into the more general question of the necessity of developmental canalization for species viability. In D. melanogaster, we showed that the transcription of both egr and cv-2 is dependent on the activity of the homeobox transcription factor zerknüllt (zen). zen is initially expressed in a broad domain in the dorsal 40% of the blastoderm stage D. melanogaster embryo. Later, at the onset of gastrulation, zen is expressed in a narrow domain centered on the dorsal midline.Citation23-25 While the later, BMP-dependent, zen expression has been shown to be required for amnioserosa specification,Citation26 the function, if any, of the early expression was not known. Previous work from our laboratory had shown that zen mutant embryos have an expanded pattern of BMP-receptor interactions at the later blastoderm stage,Citation17 suggesting that early zen expression plays a role in BMP signaling. When we examined zen mutant embryos, we discovered that the pregastrula expression of both egr and cv-2 was largely absent, and that BMP signaling was highly variable. Thus, the early expression of zen and its targets egr and cv-2 comprise a genetic network that canalizes embryonic D/V patterning.
The early broad dorsal expression of zen is dependent on activation by the Zinc-finger transcription factor Zelda, while zen is repressed ventrally by nuclear Dorsal protein. zelda mRNA is maternally deposited and Zelda protein accumulates in all blastoderm embryo nuclei throughout early embryogenesis.Citation27,28 While the exact molecular activity of Zelda is still unknown, it binds to DNA motifs, dubbed TAGteam sites, present in the regulatory regions of nearly all early zygotic genes.Citation27,29 Four footprinted TAGteam sites located in the highly conserved Ventral Repression Element (VRE), approximately 1.5 kb upstream of the zen transcriptional start site, were shown to be critical for early zen expression in D. melanogaster.Citation27
To determine whether the early expression of zen, and thus the canalization network, was conserved throughout the Drosophila lineage, we aligned the D. melanogaster VRE sequence to those found in sequenced genomes of related Drosophila species (). Two species, D. yakuba, and its sibling species D. santomea, had changes in 2 of the 4 TAGteam sites that are not in consensus with a Zelda Position Weight Matrix (PWM) defined motif; (G/TCAGGCAG/A).Citation30 Consistent with a functional relevance of these changes, early zen expression is absent in D. yakuba and D. santomea embryos. As a likely consequence of the absence of early zen expression, the early expression of egr was also greatly reduced in embryos of these species. The absence of expression of this canalization network results in an extremely variable pMad staining intensity and width in embryos of both species. Therefore, wild-type D. yakuba and D. santomea embryos have decanalized BMP signaling during D/V patterning.
Figure 2. Sequences of TAGteam motifs in the putative VRE of closely related Drosophila species. The region containing the TAGteam motif in the VRE was aligned using the highly conserved AT-rich binding site (gray) immediately proximal to a Dorsal binding site (gray with underlaid tildes). Zelda binding sites, as footprinted in D. melanogaster, are overlaid with an asterix. The low affinity (CAGGCAA) Zelda binding motifs are underlined. Changes from the consensus TAGteam motifs that are likely to render the site non-functional are overlaid with a diamond (◊), while the nucleotide changes that retain a Zelda binding site derived from the melanogaster PWM are denoted with an ≈. In the more distantly related pseudoobscura and persimilis, the A-T substitution that we believe maintains a valid Zelda site is denoted with a√.

When we examined the VRE sequences from additional species within the melanogaster subgroup, we found that all sequenced species have readily alignable and intact TAGteam sites (). The change in the second TAGteam site in D. erecta creates the reverse compliment of a known D. melanogaster site, CAGGTAA. These data suggest that, with the exceptions of D. santomea and D. yakuba, all members of the melanogaster subgroup have early expression of zen.
Analysis of Zelda binding sites in more distantly related Drosophila species, using the PWMs and affinities derived solely from D. melanogaster studies could be confounded by any significant change in Zelda binding preference, given its amino acid sequence divergence within the lineage. Recent work, however, has shown that the melanogaster Zelda protein is sufficient to activate the early zygotic genome of largely divergent Drosophila species to sufficient levels as to sustain embryogenesis.Citation31,32 We are therefore moderately confident that the Zelda binding sites, in fact, translate across considerable evolutionary time. Assuming that it is possible to extend the primary Zelda PWM outside the melanogaster subgroup, we find that D. ananassae has changes in the second and fourth TAGteam sites that, by analogy with the changes in D. yakuba, may reduce or abolish Zelda binding. While D. ananassae does have an alternate Zelda binding motif (CAGGCAA), this motif (underlined in ) is found in multiple species, including D. yakuba, and has the lowest binding affinity for melanogaster Zelda protein in vitro.Citation28 Unless there are additional compensatory Zelda binding sites outside the aligned VRE, it is likely that D. ananassae also lacks early zen expression and as such has non-canalized BMP signaling.
Two additional sequenced species, D. pseudoobscura and D. persimilis, have readily alignable VRE elements. The first 3 TAGteam sites in these species match D. melanogaster Zelda binding sites. The sequence of a fourth putative Zelda site in both species is not present within the melanogaster Zelda PWM. However, we suspect that this site, which differs from the melanogaster motif by a single A-T substitution, could be a respectable Zld binding site within these species. Four other sequenced Drosophila species, D. willistoni, D. mojavensis, D. virilis, and D. grimshawi, have small insertions into this region of the VRE (not shown). However, even in these species, 4 melanogaster TAGteam sites can be found by manual inspection. These data suggest that the early expression of zen, and likely its function in canalizing BMP signaling, is deeply conserved across Drosophila. Therefore, it is likely that only exceptional species have lost this early function. Detailed examination of these species, e.g., D. ananassae, may elucidate the evolutionary histories that have allowed for developmental decanalization.
We then explored whether the lack of developmental canalization during D/V patterning renders embryos sensitive to environmental perturbations and/or genetic variants. Strikingly, both egr cv-2 embryos and D. santomea embryos are highly susceptible to minor genetic perturbations: a very mild reduction in BMP signal transduction in the egr cv-2 background, or the presence of natural genetic variants in D. santomea, can lead to a high rate of developmental catastrophe, or failure to specify any amnioserosa cells. In contrast, we did not observe any increased variation in the number of amnioserosa cells, or of developmental catastrophe, when we raised egr cv-2 embryos or D. santomea embryos at extreme temperatures. Thus, in contrast to previous theoretical work that has proposed developmental canalization buffers the organism against both environmental perturbations and genetic variants,Citation33 our data suggest that this canalization network buffers against genetic variation, while its absence does not generate any increased susceptibility to extreme temperature regimes.
There could be multiple reasons why this canalization network buffers against genetic variation but not thermal perturbation. First, the molecular mechanisms underlying BMP signaling during D/V patterning may render this signaling cascade very susceptible to genetic variation in the BMP signaling pathway. In D. melanogaster, the dpp gene is haploinsufficient—lack of one copy of the gene causes most embryos to die because of failure to specify a sufficient number of amnioserosa cells. We propose that the genetic circuitry underlying bistability makes ligand very limiting in this system, and while one dose of dpp is sufficient to achieve the level of BMP signaling needed to specify the amnioserosa cell fate, its availability is severely constrained by the effects of positive feedback and non-signaling antagonists. Thus, the effects of mutations that decrease the amount of available ligand, or compromise the ability of cells to compete for ligand and transduce its signal, could have greater deleterious effects in this system than in many other signaling pathways. Second, we note that the BMP signaling pathway during D/V patterning may not be completely buffered against temperature even in the presence of the canalizing network, as dpp haploinsufficiency can be largely ameliorated by growth at low temperatures (unpublished results), indicating that the embryo's sensitivity to the amount of available Dpp is plastic with respect to temperature.
More generally, all experiments, both historical and contemporary, that have concluded that a specific mechanism confers robustness to both environmental and genetic perturbations were carried out in highly inbred laboratory strains.Citation34,35 Loss of heterozygosity and balancing epistatic alleles within the genomes of such inbred stocks over time could lead to a generalized lack of robustness to perturbation, which could have been revealed by ablation of one canalizing mechanism. That we have found a unimodal decanalized species, with an evident lack of robustness toward genetic variation, as opposed to temperature extremes, suggests that while a universal mode of canalization may exist, it may be evident primarily in highly inbred or otherwise genetically homogenous subpopulations. Regardless of the molecular cause of the divergence from the theoretical model of canalizing circuits, our demonstration of a new form of developmental canalization allows for future research to approach studies of canalization from a broader perspective.
Our discovery of a decanalized species is surprising in that it directly contradicts the theoretical speculation that canalization is, per se, adaptive and required for species survival.Citation33 While both D. yakuba and D. santomea are not canalized with respect to BMP signaling, only D. santomea lacks canalization with respect to an organismal phenotype of amnioserosa cell number, indicating that D. yakuba likely has at least one additional mechanism for phenotypic canalization not present in D. melanogaster. Possibly, the existence of this second mechanism rendered the necessity for the expression of the egr cv-2 canalization network less essential in D. yakuba. If so, it remains to be determined whether the presumptive loss of both canalization mechanisms in D. santomea, which recently speciated from D. yakuba, is exposing this species to selective pressure. Further research into the additional Drosophila species we have identified, D. ananassae, that may not express the zen -> egr cv-2 genetic circuit will help clarify this issue. The identification of additional non-canalized species would allow us to characterize factors shared between such species. Alternatively, if all other species are developmentally canalized, we would restrict our focus to the unique factors, such as its relatively recent speciation or its restricted range on an equatorial island, that have allowed D. santomea to become so very variable.
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
References
- Gavin-Smyth J, Wang YC, Butler I, Ferguson EL. A genetic network conferring canalization to a bistable patterning system in Drosophila. Curr Biol 2013; 23:2296-302; PMID:24184102; http://dx.doi.org/10.1016/j.cub.2013.09.055
- Gelbart WM. The decapentaplegic gene: a TGF-β homologue controlling pattern formation in Drosophila. Development 1989; 107 Suppl:65-74; PMID:2699859
- Irish VF, Gelbart WM. The decapentaplegic gene is required for dorsal-ventral patterning of the Drosophila embryo. Genes Dev 1987; 1:868-79; PMID:3123323; http://dx.doi.org/10.1101/gad.1.8.868
- Ray RP, Arora K, Nüsslein-Volhard C, Gelbart WM. The control of cell fate along the dorsal-ventral axis of the Drosophila embryo. Development 1991; 113:35-54; PMID:1765005
- Wharton KA, Ray RP, Gelbart WM. An activity gradient of decapentaplegic is necessary for the specification of dorsal pattern elements in the Drosophila embryo. Development 1993; 117:807-22; PMID:8330541
- Eldar A, Dorfman R, Weiss D, Ashe H, Shilo BZ, Barkai N. Robustness of the BMP morphogen gradient in Drosophila embryonic patterning. Nature 2002; 419:304-8; PMID:12239569; http://dx.doi.org/10.1038/nature01061
- Shimmi O, Umulis D, Othmer H, O’Connor MB. Facilitated transport of a Dpp/Scw heterodimer by Sog/Tsg leads to robust patterning of the Drosophila blastoderm embryo. Cell 2005; 120:873-86; PMID:15797386; http://dx.doi.org/10.1016/j.cell.2005.02.009
- Umulis D, O’Connor MB, Othmer HG. Robustness of embryonic spatial patterning in Drosophila melanogaster. Curr Top Dev Biol 2008; 81:65-111; PMID:18023724; http://dx.doi.org/10.1016/S0070-2153(07)81002-7
- Umulis DM, Serpe M, O’Connor MB, Othmer HG. Robust, bistable patterning of the dorsal surface of the Drosophila embryo. Proc Natl Acad Sci U S A 2006; 103:11613-8; PMID:16864795; http://dx.doi.org/10.1073/pnas.0510398103
- Umulis DM, Shimmi O, O’Connor MB, Othmer HG. Organism-scale modeling of early Drosophila patterning via bone morphogenetic proteins. Dev Cell 2010; 18:260-74; PMID:20159596; http://dx.doi.org/10.1016/j.devcel.2010.01.006
- Brummel TJ, Twombly V, Marques G, Wrana JL, Newfeld SJ, Attisano L, Massague J, Oconnor MB, Gelbart WM. Characterization and relationship of Dpp receptors encoded by the Saxophone and Thick veins genes in Drosophila. Cell 1994; 78:251-61; PMID:8044839; http://dx.doi.org/10.1016/0092-8674(94)90295-X
- Arora K, Nusslein-Volhard C. Altered mitotic domains reveal fate map changes in Drosophila embryos mutant for zygotic dorsoventral patterning genes. Development 1992; 114:1003-24; PMID:1618145
- Ferguson EL, Anderson KV. Decapentaplegic acts as a morphogen to organize dorsal-ventral pattern in the Drosophila embryo. Cell 1992; 71:451-61; PMID:1423606; http://dx.doi.org/10.1016/0092-8674(92)90514-D
- Decotto E, Ferguson EL. A positive role for Short gastrulation in modulating BMP signaling during dorsoventral patterning in the Drosophila embryo. Development 2001; 128:3831-41; PMID:11585808
- Holley SA, Jackson PD, Sasai Y, Lu B, De Robertis EM, Hoffmann FM, Ferguson EL. A conserved system for dorsal-ventral patterning in insects and vertebrates involving sog and chordin. Nature 1995; 376:249-53; PMID:7617035; http://dx.doi.org/10.1038/376249a0
- Oelgeschlager M, Reversade B, Larrain J, Little S, Mullins MC, De Robertis EM. The pro-BMP activity of Twisted gastrulation is independent of BMP binding. Development 2003; 130:4047-56; PMID:12874126; http://dx.doi.org/10.1242/dev.00633
- Wang YC, Ferguson EL. Spatial bistability of Dpp-receptor interactions during Drosophila dorsal-ventral patterning. Nature 2005; 434:229-34; PMID:15759004; http://dx.doi.org/10.1038/nature03318
- Hudson JB, Podos SD, Keith K, Simpson SL, Ferguson EL. The Drosophila Medea gene is required downstream of dpp and encodes a functional homolog of human Smad4. Development 1998; 125:1407-20; PMID:9502722
- Raftery LA, Wisotzkey RG. Characterization of Medea, a gene required for maximal function of the Drosophila BMP homolog Decapentaplegic. Ann N Y Acad Sci 1996; 785:318-20; PMID:8702167; http://dx.doi.org/10.1111/j.1749-6632.1996.tb56296.x
- Igaki T, Kanda H, Yamamoto-Goto Y, Kanuka H, Kuranaga E, Aigaki T, Miura M. Eiger, a TNF superfamily ligand that triggers the Drosophila JNK pathway. EMBO J 2002; 21:3009-18; PMID:12065414; http://dx.doi.org/10.1093/emboj/cdf306
- Conley CA, Silburn R, Singer MA, Ralston A, Rohwer-Nutter D, Olson DJ, Gelbart W, Blair SS. Crossveinless 2 contains cysteine-rich domains and is required for high levels of BMP-like activity during the formation of the cross veins in Drosophila. Development 2000; 127:3947-59; PMID:10952893
- Serpe M, Umulis D, Ralston A, Chen J, Olson DJ, Avanesov A, Othmer H, O’Connor MB, Blair SS. The BMP-binding protein Crossveinless 2 is a short-range, concentration-dependent, biphasic modulator of BMP signaling in Drosophila. Dev Cell 2008; 14:940-53; PMID:18539121; http://dx.doi.org/10.1016/j.devcel.2008.03.023
- Rushlow C, Doyle H, Hoey T, Levine M. Molecular characterization of the zerknullt region of the Antennapedia gene complex in Drosophila. Genes Dev 1987; 1:1268-79; PMID:2892759; http://dx.doi.org/10.1101/gad.1.10.1268
- Rushlow C, Levine M. Role of the zerknullt gene in dorsal-ventral pattern formation in Drosophila. Adv Genet 1990; 27:277-307; PMID:2112301; http://dx.doi.org/10.1016/S0065-2660(08)60028-0
- Ip YT, Kraut R, Levine M, Rushlow CA. The dorsal morphogen is a sequence-specific DNA-binding protein that interacts with a long-range repression element in Drosophila. Cell 1991; 64:439-46; PMID:1988156; http://dx.doi.org/10.1016/0092-8674(91)90651-E
- Frank LH, Rushlow C. A group of genes required for maintenance of the amnioserosa tissue in Drosophila. Development 1996; 122:1343-52; PMID:8625823
- Liang HL, Nien CY, Liu HY, Metzstein MM, Kirov N, Rushlow C. The zinc-finger protein Zelda is a key activator of the early zygotic genome in Drosophila. Nature 2008; 456:400-3; PMID:18931655; http://dx.doi.org/10.1038/nature07388
- Nien C, Liang H, Butcher S, Sun Y, Fu S, Gocha T, Manak JR, Rushlow C. Temporal coordination of gene networks by Zelda in the early Drosophila embryo. PLoS Genet 2011; 7:e1002339; PMID:22028675; http://dx.doi.org/10.1371/journal.pgen.1002339
- Ten Bosch JR, Benavides JA, Cline TW. The TAGteam DNA motif controls the timing of Drosophila pre-blastoderm transcription. Development 2006; 133:1967-77; PMID:16624855; http://dx.doi.org/10.1242/dev.02373
- Harrison MM, Li X-Y, Kaplan T, Botchan MR, Eisen MB. Zelda binding in the early Drosophila melanogaster embryo marks regions subsequently activated at the maternal-to-zygotic transition. PLoS Genet 2011; 7:e1002266; PMID:22028662; http://dx.doi.org/10.1371/journal.pgen.1002266
- Gavin-Smyth J, Matute DR. Embryonic lethality leads to hybrid male inviability in hybrids between Drosophila melanogaster and D. santomea. Ecol Evol 2013; 3:1580-9; PMID:23789069; http://dx.doi.org/10.1002/ece3.573
- Matute DR, Gavin-Smyth J. Fine mapping of dominant x-linked incompatibility alleles in Drosophila hybrids. PLoS Genet 2014; 10:e1004270; PMID:24743238; http://dx.doi.org/10.1371/journal.pgen.1004270
- Meiklejohn CD, Hartl DL. A single mode of canalization. Trends Ecol Evol 2002; 17:468-73; http://dx.doi.org/10.1016/S0169-5347(02)02596-X
- Rendel J, Sheldon B. Selection for canalization of the scute phenotype in Drosophila melanogaster. Aust J Biol Sci 1960; 13:36-47
- Frankel N, Davis GK, Vargas D, Wang S, Payre F, Stern DL. Phenotypic robustness conferred by apparently redundant transcriptional enhancers. Nature 2010; 466:490-3; PMID:20512118; http://dx.doi.org/10.1038/nature09158
