Abstract
The eukaryotic nucleus is structurally and functionally organized, as reflected in the distribution of its protein and DNA components. The genome itself is segregated into euchromatin and heterochromatin that replicate in a distinct spatio-temporal manner. We used a combination of fluorescence in situ hybridization (FISH) and DamID to investigate the localization of the early and late replicating components of the genome in a lymphoblastoid cell background. Our analyses revealed that the bulk of late replicating chromatin localizes to the nuclear peripheral heterochromatin (PH) in a chromosome size and gene density dependent manner. Late replicating DNA on small chromosomes exhibits a much lower tendency to localize to PH and tends to associate with alternate repressive subcompartments such as pericentromeric (PCH) and perinucleolar heterochromatin (PNH). Furthermore, multicolor FISH analysis revealed that late replicating loci, particularly on the smaller chromosomes, may associate with any of these 3 repressive subcompartments, including more than one at the same time. These results suggest a functional equivalence or redundancy among the 3 subcompartments. Consistent with this notion, disruption of nucleoli resulted in an increased association of late replicating loci with peripheral heterochromatin. Our analysis reveals that rather than considering the morphologically distinct PH, PCH and PNH as individual subcompartments, they should be considered in aggregate as a functional compartment for late replicating chromatin.
Abbreviations
| PCH | = | pericentromeric heterochromatin |
| PNH | = | perinucleolar heterochromatin |
| PH | = | peripheral heterochromatin |
| NOR | = | nucleolar organizing region |
| FISH | = | fluorescence in situ hybridization |
| LAD | = | lamina associated domain |
| DamID | = | Dam identification |
| Chr | = | chromosome |
| EU | = | 5-Ethynyl uridine |
Introduction
Over 150 years of microscopy studies have revealed that the eukaryotic nucleus is morphologically compartmentalized. The description of common intranuclear structures, like nucleoli, has suggested that this compartmentalization is functional. Current research now focuses on elucidating the interplay between structure and function within the nucleus. Proteins specific to particular nuclear processes (e.g. transcription, replication) cluster and form distinct subcompartments, and by extension the genome is equally segregated according to its chromatin state and thus its functional status.Citation1-4 At its most basic level, genomic DNA can be divided into euchromatin and heterochromatin, which form distinct compartments in the interphase nucleus that are easily visualized with a nucleic acid counterstain.Citation5,6 Heterochromatin consists of highly compacted chromatin that is marked by repressive histone modifications and largely devoid of genic transcription. It is principally concentrated in large clusters of pericentromeric heterochromatin (PCH), around the nuclear periphery and surrounding nucleoli (perinucleolar heterochromatin, PNH).Citation7 Euchromatin, the lighter staining component, contains decondensed DNA, is transcriptionally active and marked by activating histone modifications. Consistent with this, HiC analysis of the 3-dimensional organization of the human genome revealed that it is compartmentalized into 2 largely self-associating components.Citation8 Compartment ‘A’ was defined by gene rich open chromatin with high DNAse accessibility, mostly activating epigenetic marks and high transcriptional activity, while compartment ‘B’ was best described as consisting of closed chromatin. Compartment A thus bears all the hallmarks of euchromatin, while B encompasses the bulk of heterochromatin. A subsequent study revealed that the linear profile of these 2 compartments along all chromosomes closely follows that of replication timing.Citation9 Given that the genome does not replicate synchronously during S phase, but in coordinate clusters of early and late segments that replicate separately within the nucleus,Citation10-13 this correlation further supports the notion that the genome is physically and functionally compartmentalized.
Heterochromatin replicates late during S phase of the cell cycle and predominantly at the nuclear periphery, whereas euchromatin does so early and within the nucleoplasm. However, it is unclear whether a relatively small early replication domain can affect its localization when embedded in a larger late replicating domain (and vice versa). It is also unclear whether late replicating domains on small chromosomes contact the periphery, as small chromosomes localize more to the nuclear center. We therefore sought to analyze the localization of the late replicating component of the genome in more detail, in particular vis-à-vis the nuclear periphery, and to compare it to early replicating regions in a well characterized human lymphocyte cell line. Investigating the nuclear positioning of numerous loci on several chromosomes, both large and small, we sought to determine the prevalence of the peripheral localization of late replicating chromatin. We found that while late replicating loci have a strong tendency to localize to the periphery, especially on large chromosomes, this correlation breaks down with decreasing chromosomal size. A comprehensive analysis of the peripheral nuclear subcompartment by DamID revealed a strong correlation between replication timing and peripheral association on a genomic level, but again less so for the smaller chromosomes, some of which do not appear to associate with the periphery at all. Instead we found that late replicating regions that fail to localize to the periphery tend to associate with nucleoplasmic repressive subcompartments such as PNH and PCH. Moreover, late replicating loci can localize to more than one of these subcompartments simultaneously when they are adjacent to each other. Intriguingly, disruption of nucleolar integrity results in an increased association of late replicating chromatin with the nuclear periphery, suggesting the possibility of functional redundancy between these repressive subcompartments.
Results
Late replicating DNA on large chromosomes localizes to the periphery
We analyzed the localization of early and late replicating chromatin regions in GM06990 (GM) cells, as this human lymphoblastoid cell line is well characterized and comprehensive genome-wide data sets on replication timing, Hi-C and multiple chromatin marks are available. Initially we selected a set of Bacterial artificial chromosome probes (BACs) sampling 14 early and 15 late replicating loci on chromosomes of large (chr3, 4), intermediate (chr14, 17) and small (chr21, 22) size (for examples, see ; for exact locations and identities of all BACs used see supplemental materials). Using 3D fluorescence in situ hybridization (FISH) we determined the localization of these regions in interphase nuclei of GM cells relative to the nuclear periphery, which was marked fluorescently with an antibody against Lamin B1. illustrates examples of the localization of the 2 alleles of an early and a late replicating region as intranuclear and peripheral, respectively. Collectively the results for all analyzed loci demonstrate that early replicating parts of the genome do not tend to reside at the nuclear periphery (), as the association rates generally remained below 40% (3–37%), with only one outlier (discussed below). The exact replication time itself did not appear to be a significant determinant of localization, as long as it was classified as ‘early’ (for corresponding replication timing values, see Supplemental Table 1). By contrast, results for the late replicating regions showed a wider range for association with the periphery with a few loci remaining predominantly intranuclear (<20 % association) and others approaching an association close to 100%. When plotting the data as peripheral association versus the size of the chromosome harboring the test region, a clear trend between localization and chromosome size became apparent (). Late replicating chromatin on large chromosomes (e,g, chr3 and chr4 at close to 200Mb in size) was predominantly peripheral: however, with decreasing chromosome size the association declined. On intermediate sized chromosomes (chr14 and 17), late chromatin exhibited a mixed localization, while on small chromosomes (chr21, 22) it was increasingly found away from the periphery. Again the exact replication timing value of the test regions did not appear to correlate with the degree of peripheral association (as long as it was late), but it is of note that the loci on chr22 were significantly more intranuclear than those on chr21 (<20% vs 30–55%, see supplemental data for the identities). It is likely that chromosome 22 is generally located more centrally within the nucleus, as it harbors almost twice as many genes as chr21 (despite their comparable size) and also replicates much earlier - its mapped region has a considerably higher averaged replication timing value (0.722 vs −0.013). Even so, although the late replicating regions on chr22 did not exhibit a pronounced peripheral localization, the early replicating tested loci were still consistently less frequently associated with the periphery, in line with the general trend of early chromatin localizing more centrally within the nucleus. Overall, the trend of a lower association of late replicating regions with the nuclear periphery with decreasing chromosome size confirms results described for general chromosome positioning in human fibroblast nuclei.Citation14
Figure 1. Late replicating chromatin on large chromosomes resides at the nuclear periphery. (A) Replication timing profile of a large (Chr4), intermediate (Chr14) and small (Chr21) chromosome in GM06990 lymphoblastioid cells (adapted from replicationdomain.org, visualized on http://genome.ucsc.edu; E = early, L = late replicating regions). Gaps mark centromere locations or unmapped NOR repeat regions. Above the tracks blue (=early) and red (=late) bars indicate the positions of BAC FISH probes used in the analysis. (B) FISH images illustrating the localization of an early (left 2 panels) and late (right 2 panels) replicating region. Each pair of images represents a single z-section through the nucleus showing one of the alleles. (C) Localization of early and late regions sampled from 6 chromosomes vis-à-vis the nuclear periphery. Early replicating loci exhibit lower peripheral association than late replicating loci (percentage of alleles contacting the lamina; Tukey boxplot). (D) Peripheral localization of early and late replicating loci as a measure of the size of the chromosome the probes are located on. Data is the same as in C. The exponential best fit curve illustrates the correlation between chromosome size and peripheral localization of late replicating loci.
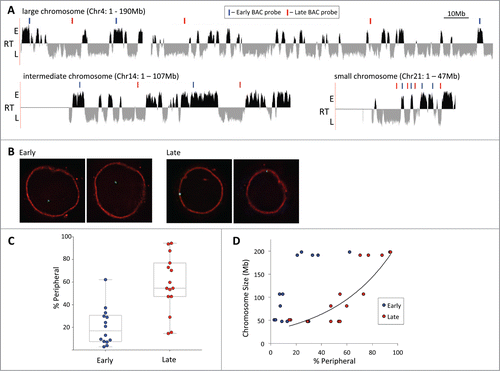
Finally, while the exact replication timing of both early and late test regions did not appear to exert a strong influence on positioning, the size of the replication domains may be quite significant. Two regions tested on chr21 were in early replication domains smaller than ∼1.2 Mb and surrounded by large late replicating regions. They exhibited peripheral localization for close to 30% of their alleles, whereas 2 other regions, located in considerably larger early domains, localized to the periphery at less than half that frequency (see supplemental Figure 1). Hence the larger the replication domain, the more consistent its localization. This appears to hold true also for large chromosomes, which show the greatest disparity between peripheral association of early and late replicating chromatin. While most early regions on chr3 and chr4 were peripheral fewer than 40% of the time, one test region on chr3 near the end of the q arm localized to the periphery at a frequency of 60%, significantly higher than all others and despite a high positive replication timing value. However, this early replicating domain encompasses roughly only 1.5 Mb, suggesting it may still be susceptible to the effects of adjacent late replicating chromatin. In summary, while late replicating loci tend to be peripheral, the sizes of their replication domain and of the chromosome they reside on also contribute to the strength of the correlation of their localization.
Late replicating DNA is not exclusively targeted to the nuclear periphery
Our microscopy results suggest that some late replicating regions do not consistently associate with the periphery, in particular on the smaller chromosomes. However, FISH analyses are inherently limited in scope. Thus to determine the correlation between peripheral localization and replication timing across the genome, we performed DamID chromatin profiling experiments via the expression of human LaminB1 fused to the bacterial Dam methylase.Citation15 Lamina associated chromatin was identified by hybridizing the isolated LaminB1-Dam methylated DNA to whole genome CGH microarrays (). We queried the DamID data for large contiguous domains marked by sharp LaminB1 binding boundariesCitation16 and established that GM cells have 1297 Lamina Associated Domains (LADs), averaging 870 kb in size (LAD track in ). Both the number and average size is comparable to that described for LADs in human fibroblast cells,Citation16 however, their identities do exhibit some significant differences (see tracks in Supplemental Fig. 4). About 1.5 Gb of the genome is not covered by LADs in either cell type, 684 Mb are covered in both and 448 Mb are unique LADs to GM cells, and another ∼456 Mb are unique to fibroblasts. Nonetheless, the total fraction of the genome covered by LADs thus remains constant at almost 37% between the 2 tissues, 39–40% of which is specific to each cell type (Supplemental Fig. 4).
Figure 2. Genome association with the nuclear lamina in human lymphoblastoid cells by DamID. (A) Map of LaminB1 interactions with chromosome 3 (orange = enrichment, blue = depletion), above the replication timing profile (RT) of chromosome 3 (E = early, L = late). The LADs track in the center shows contiguous segments of prominent lamina association. DamID maps of chromosome 21 (B) and 22 (C) associations with LaminB1 above their respective replication timing profiles.
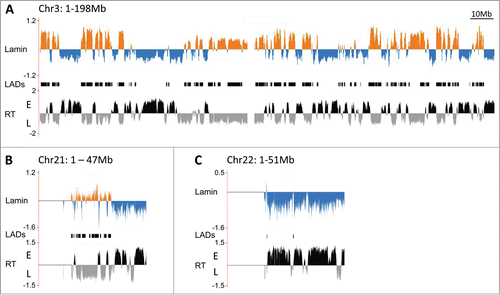
On a genomic level, our results also reveal a high correlation between the lamina association and replication timing in the GM lymphoblastoid cell line (global correlation coefficient of 0.66) and ∼84% of the LADs fall within late replicating chromatin. In particular for the large chromosomes, the DamID profile follows that of replication timing exceedingly well (, Supplemental Fig. 2), with only minor exceptions. Most differences arise within large genome segments contiguous for either LaminB1 depletion or enrichment that contain short late or early replicating regions, respectively (Supplemental Fig. 3). The localization of large multi-Mb replication domains may simply be dominant over any small intervening domain of opposite replication timing. Importantly, the DamID data is consistent with our microscopy assays in which a high colocalization frequency of genomic loci with the lamina observed by FISH translates to high LaminB1-Dam association, particularly on the large chromosomes.
On several of the smallest chromosomes, the Lamin-chromatin profile does not follow the replication timing data, and, to some extent, the FISH data. While on chr21 most tested regions exhibit LaminB1 association if late replicating and none if early, 2 late regions toward the 3′ end of the chromosome fail to show LaminB1 association (). In fact, the final 15.7 Mb of the chr21 q arm fail to exhibit any association with the periphery, despite the presence of at least 2 late replicating domains (to a total of 4.6 Mb) within that segment. However, given that these domains are comparatively small relative to the remainder of the chromosome arm, the long stretches of early replicating chromatin appear to be dominant for nuclear localization, as discussed above. Chr22 does not reveal any significant peripheral association by DamID, as this small chromosome likely locates primarily to the nuclear interior and is largely early replicating (). Chr19, on the other hand, does contain 17.3 Mb of late replicating chromatin in GM cells, almost 10 Mb of which in one large domain (Supplemental Fig. 2). However, the DamID analysis does not reveal significant association with the nuclear lamina over the entire length of the chromosome, suggesting chr19 also resides in the nuclear interior in lymphocytes, consistent with a previous report.Citation17 It has been proposed that its interior localization is a consequence of its high gene content (compared to chr18, which is peripheral and relatively gene poor), suggesting its late replicating regions may thus localize to alternate repressive subcompartments. However, not all small chromosomes localize to the nuclear center, as the DamID results reveal significant association of chr20 with the periphery, consistent with its replication timing profile, (Supplemental Fig. 2). Overall the genome wide lamina-chromatin profile confirms our microscopy analysis and demonstrates that the bulk of the late replicating lymphoid genome is in contact with the nuclear periphery. Furthermore, the data also reemphasizes that the positioning of small late replication domains may be context dependent, and that some entire small chromosomes remain predominantly in the nuclear interior, raising the question of to which subcompartment their late replicating chromatin localizes.
Late replicating chromatin localizes to multiple repressive subcompartments
The bulk of the late replicating DNA has been described to localize to the nuclear periphery forming the peripheral heterochromatin (PH), and our DamID data corroborates this for a majority of late replicating DNA. However, late replicating chromatin has also been observed in the interior nucleoplasm (our DamID data reveals that in lymphocytes 42% of late chromatin is not peripheral), where it presumably still resides in repressive subcompartments. We sought to identify and determine the localization of these intranuclear late repressive subcompartments. Late replicating pericentric heterochromatin (PCH) is not exclusively peripheral in many human tissue types, as is easily observed by the intensely DAPI staining regions within the nucleus. In addition, late replicating DNA has also been observed as part of the perinucleolar heterochromatin (PNH). Several of the late replicating loci we tested by FISH were found in the nuclear interior, in particular with increasing frequency on the smaller chromosomes, and our Lamin-chromatin profile similarly revealed large regions of late replicating DNA devoid of peripheral contact on the smaller chromosomes. We therefore investigated whether these late regions colocalize with the distinctly recognizable PCH and PNH subcompartments instead of PH. By multicolor ImmunoFISH we were able to visualize test loci simultaneously with the nuclear lamina (LaminB1 antibody), as well as PCH (a pan-centromeric FISH probe) and PNH (nucleolin antibody, marking nucleoli) (Supplemental Fig. 5). Using this approach, we determined the frequency with which late replicating loci localize to any of these 3 prevalent repressive subcompartments in any given cell. Test regions included late replicating regions on chr17, 21 and 22, as well as a pair of early replicating controls on chr17 and 21. This ImmunoFISH analysis revealed: (a) While all tested late loci exhibited peripheral localization for less than 50% of all alleles, inclusion of the other repressive subcompartments resulted in an increase in localization to a repressive subcompartment to 73%–97% (, Supplemental Table 2). (b) In contrast to loci on large chromosomes, late replicating chromatin on smaller chromosomes did not show a clear preference to localizing to any particular repressive subcompartment, but appeared to be distributed among them (Supplemental Table 2). (c) Late replicating loci can colocalize with more than one repressive subcompartment at the same time (examples shown in , Supplemental F. 5), a feature that is most prominent for PNH and PCH as they are often adjacent.
Figure 3. Late replicating loci associate with 3 repressive nuclear subcompartments. (A) The localization of several early and late replicating loci was determined simultaneously vis-à-vis the nuclear periphery (PH = peripheral heterochromatin), pericentromeric heterochromatin (PCH) and perinucleolar heterochromatin (PNH) by multicolor ImmunoFISH. Percent association of the loci to any of the 3 repressive subcompartments collectively (black) is significantly higher than association only with the PH compartment (blue). BAC probe identities: Chr17 A = RP11-142B17, B = RP11-42M14, Chr21 A = CTD-2059E17, B = RP11-315H16, C = CTD-2207L7, Chr22 = RP11-79G21. (B) Late loci in particular can localize to more than one repressive compartment at once. Venn diagrams show the association of two late and one early replicating region with the PH, PCH and PNH compartments. Early loci associate primarily with only one compartment. The size of the ellipses and their overlap is proportional to the percent association of a given locus with each compartment. BAC probe identities: Late Chr22 = RP11-79G21, Late Chr21 = RP11-315H16, Early Chr17 = RP11-142B17.
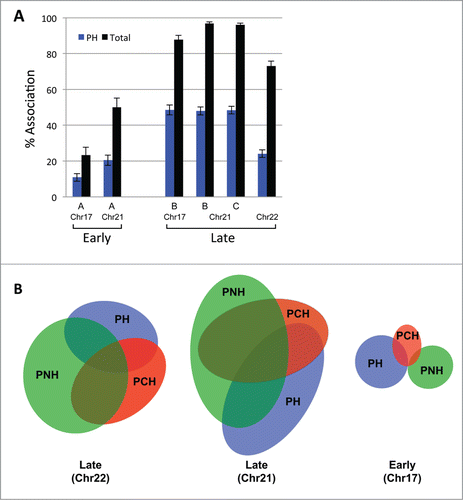
Early replicating loci also localize to PCH and PNH, albeit at a much lower frequency (reflected by the proportionally smaller ellipses in ), and their overall association with repressive subcompartments does not surpass 50% (Supplemental Table 2). Moreover, the early replicating loci usually do not contact more than one repressive subcompartment at a time. However, one BAC probe on chr21 showed an association with repressive subcompartments 50% of the time, and chr21 contains a nucleolar organizing region (NOR) on its p-arm. The highest association of this BAC is also with PNH, raising the question whether the presence of NORs skews the localization of chromatin on these chromosomes (chr13, 14, 15, 21, 22) toward PNH due to their linear proximity on the chromosome. Thus we tested whether NORs affect chromatin localization in cis by examining the positioning of both early and late replicating regions on non-NOR containing small chromosomes, chr19 and chr20. Our results show that the tested late replicating regions on these chromosomes associate with all 3 repressive subcompartments at a higher frequency than the early replicating ones (Supplemental Fig. 6). Consistent with the Lamin-chromatin profile, loci on chr19 exhibited low association with the periphery, but more so with PNH and PCH (Supplemental Table 3). While the late replicating regions localized to PNH at a lower frequency than on NOR containing chromosomes, it contributed significantly to the total association with repressive subcompartments (10–20% of late replicating alleles localized exclusively to PNH, while another fraction colocalized with PNH as well as the lamina or PCH; not shown). The data thus show that late replicating loci frequently reside at PNH even on non-NOR containing chromosomes, suggesting this is a common heterochromatin compartment. Importantly, since late replicating chromatin appears to associate interchangeably between at least 3 repressive nuclear subcompartments, we suggest that these subcompartments be considered collectively when analyzing the localization of the late replicating genome.
Redundancy between repressive subcompartments
As several of the analyzed late replicating loci localize with a high frequency to all 3 repressive subcompartments, we hypothesized that these subcompartments may be functionally redundant. To test this hypothesis, we determined whether the removal of one subcompartment alters the association of chromatin regions with the other subcompartments. We treated GM cells with actinomycin D, which at low concentrations selectively inhibits RNA polymerase I and leads to the functional disruption of nucleoli.Citation18,19 RNA polymerase II transcription remained active, as judged by nuclear EU incorporation in immunofluorescence assays (Supplemental Fig. 7). In addition, we performed qPCR controls on select annotated transcripts at test loci to ensure that the actinomycin D treatment did not alter expression at the analyzed regions by testing for changes in transcript levels in the nuclear RNA before and after actinomycin D treatment. Compared to GAPDH and actin controls, very few of the late replicating regions analyzed exhibited significant expression to begin with, and none of them were significantly upregulated after the treatment (Supplemental Fig. 8). The majority gave no reliable PCR product, indicating that the genes were (and remained) silent, consistent with their association with a repressive nuclear subcompartment. Thus exposing the cells to this concentration of actinomycin D did not change the activity state of the regions analyzed.
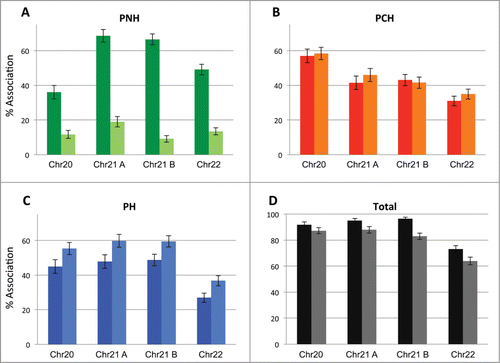
The localization of 4 previously analyzed loci (one each on chr20 and chr22, 2 on chr21) with respect to PH, PNH and PCH was simultaneously determined by ImmunoFISH in normal and actinomycin D treated GM cells. As expected, the association of all 4 loci with nucleoli was drastically reduced by up to 80% (, Supplemental Table 4A), and a large proportion of the nucleolin marker dispersed following actinomycin D treatment. While localization to PCH did not change significantly after treatment, the most striking and consistent finding is that for each tested region the association with the nuclear lamina increased by ∼10 percent (). Although the overall association with the repressive compartment decreased on average by a similar amount (), the loss of nucleolar association far exceeds that (60–80%). Even if many alleles colocalized with more than one repressive subcompartment in the control cells, between 15–26% of alleles colocalized exclusively with PNH (this decreased to less than 3% in the treated cells). Thus the majority of the late replicating loci are sequestered at the PH and PCH subcompartments following the functional disruption of the nucleolar subcompartment. Fitting our data to a mixed effects model confirms that the increase in peripheral localization is statistically highly significant (p < 0.001; see Methods). Importantly, early replicating regions did not show an alteration in PH association following nucleolar disruption, nor did a late replicating region that is predominantly associated with the lamina to begin with (>90 % frequency), supporting the specificity of the effect (Supplemental Table 4). Thus our results suggest a degree of functional equivalence or redundancy among different heterochromatic repressive subcompartments. In essence, many late replicating and silent chromatin regions localize seemingly interchangeably to multiple repressive nuclear subcompartments, and when one such subcompartment is disrupted, it may be replaced by association with an alternate repressive subcompartment.
Figure 4. Repressive nuclear subcompartments exhibit redundancy. The positioning of a total of 4 different late replicating loci located on 3 different chromosomes vis-à-vis the PNH (A), PCH (B) and PH (C) subcompartments as well as their association to all 3 compartments collectively (D) (percent association) was assessed prior to and after 2 hours of actinomycin D treatment. Localization to repressive subcompartments overall is largely maintained, even after the disruption of nucleoli. Full color bars = prior to treatment; lighter shade bars = post actinomycin D treatment.
Discussion
Our extensive FISH analysis of loci on multiple chromosomes in a human lymphoblastoid background revealed that while replication timing is a good predictor of nuclear localization (internal vs peripheral), chromosome size proved to be a second parameter significantly influencing localization. On large chromosomes, late replicating regions were predominantly located at the nuclear periphery, a correlation that became less prevalent with decreasing chromosome size (). This observation is consistent with the reported positioning of chromosomes in human fibroblasts,Citation14 with large chromosomes localizing to the nuclear periphery while small chromosomes cluster in the nuclear interior. Due to the closer proximity to the periphery, early replicating regions on large chromosomes colocalize with the lamina more frequently than those on small chromosomes as well, although the association still remains far below that of late replicating chromatin. Notably even on small chromosomes of similar size, the association of late replicating loci with the periphery is not equal. Unlike the localization of chromosomes in fibroblasts, in the more spherical lymphocyte cells gene density also influences chromosome positioning.Citation17,20-22 This behavior is reflected by the peripheral association of late replicating loci on the similarly sized chromosome pairs chr19 and chr20 (59, 64 Mb), and chr21 and chr22 (47, 51 Mb). In both cases the loci on the gene poorer chromosome (chr20 and chr21) associated 2–3 times more frequently with the lamina than on those on the gene denser chromosome (chr19 and chr22), suggesting a generally more peripheral localization of the whole chromosome.
At the genomic level, our LaminB1-DamID analysis confirms our FISH data, and nuclear lamina association correlates very well with replication timing (see , Supplemental Fig. 2). While just over half of the genome is in late replicating clusters in this lymphocyte background, about 36% resides in LADs at the nuclear periphery. The actual association with the lamina may be a few percentage points higher, because the algorithm that classifies LADs can miss some regions that contact LaminB1 but do not exhibit a sharp enough transition in the hybridization signal. A full 80% of the LADs overlap with late replicating regions (32% of the genome), the remainder represent largely transition regions where the LADs extend beyond late domains, rather than covering distinct early replicating regions. Again the DamID profile mirrors replication timing best on large chromosomes whereas some of the smaller chromosomes, in particular chr19 and 22, exhibit very little association with the lamina (as they did in the FISH analysis) despite the presence of several late replicating chromatin domains. This behavior likely reflects the predominantly intranuclear localization of the whole chromosomes 19 and 22. In fact, as a measure of correlation between peripheral localization and replication timing, by statistical analysis the overlap between the DamID data and replication timing is significantly greater than by chance (p < 0.05) for all chromosomes with the exception of those 2. Ultimately almost 17% of the lymphocyte genome is late replicating but not localized at the periphery; some of which results from relatively short late domains lying in between larger early replicating domains (Supplemental Fig. 3), as well as the interior localization of whole small chromosomes.
Despite the lack of association of some late replicating domains with the nuclear periphery, such domains likely still reside in a repressive subcompartment. Two prominent and often intranuclear subcompartments are obvious candidates – PCH and PNH, both of which have been shown previously to harbor late replicating chromatin, as measured by pulse labeling experiments.Citation11-13 PCH forms brightly staining constitutive heterochromatic foci (also known as chromocenters) in interphase nuclei and consists of centromeric and pericentromeric repetitive DNA that does not exhibit much genic transcription and is thus expected to be late replicating. BrdU pulse labeling experiments have shown significant overlap between PCH and late replicating intranuclear foci.Citation11-13,23 In addition, repressed transposable elements and genes have been shown to associate and cluster with PCH.Citation24-26 PNH is similarly easily visible by DAPI staining, as it forms a clear heterochromatic rim around nucleoli. In addition to containing the active and inactive rDNA repeats, nucleoli also associate with numerous specific sequences, including satellite repeats, zinc finger proteins, olfactory receptor genes, as well as members of the defensin and Ig gene families.Citation27,28 These identified nucleolar-associated regions encompassed about 4% of the genome and strongly correlate with low gene density and general transcriptional repression in the tissues examined. Moreover, due to the linear proximity of centromeres to NORs, PCH often colocalizes with PNH.Citation28-32
Our ImmunoFISH analysis of late replicating loci on small chromosomes (chr19, 20, 21 and 22) simultaneously with the 3 repressive subcompartments of PH, PCH and PNH clearly shows that even on chromosomes 19 and 22, which exhibit very little peripheral localization, the late loci largely remain associated with a repressive subcompartment (, Supplemental Tables 2, 3). Colocalization of these late regions with PCH and nucleoli therefore appears to account for the internal positioning of the DamID negative chromatin and highlights the importance of considering multiple repressive subcompartments simultaneously for visualizing late replicating chromatin. The assay also offers 2 additional important insights - that late replicating loci may associate with more than one repressive subcompartment simultaneously (most commonly PCH and PNH), and that with the exception of those on large chromosomes which tend to be peripheral, they often do not appear to exhibit preferential localization to any of the 3 repressive subcompartments. This raises the question of functional equivalence among the 3 subcompartments and whether they can compensate for each other. Disrupting nucleoli by inhibiting RNA pol I with actinomycin D suggests that this is indeed the case; for all analyzed late loci the association with the lamina increased, which is particularly notable given the short duration of the treatment. Given the limited mobility of chromatin in higher eukaryotes during interphase,Citation33 it is possible that relocalized loci were not far from the nuclear periphery to begin with, however, the net increase in peripheral localization is highly significant statistically and reproducible. Moreover, initially 15–27% of loci were exclusively localizing to PNH (Supplemental Table 4), a far greater proportion than post treatment or the loss in total repression as measured by association with any of the 3 subcompartments. On the other hand, an increase in PCH localization was not detected, which may in part be because many PNH associated loci were already contacting PCH beforehand. The possible equivalence of repressive subcompartments has also been suggested recently by the results of live cell assays. For example, labeled nucleolar chromatin has been shown to localize to either PH or PNH following mitosis, and localization of labeled LADs to either the periphery or nucleoli in the next cell cycle was shown to be stochastic and thus interchangeable.Citation27,34
The relationship between the 3 repressive subcompartments and gene loci associated with them is summarized in , illustrating that repressed loci can reside at either PNH or PH and apparently relocalize between them. This may happen to a limited extent during G1, for example upon the functional disruption of nucleoli, but also upon nucleolar reformation and chromosomal decondensation following mitosis. It remains to be determined whether repressed chromatin also shuttles between PCH and the other 2 repressive subcompartments, however, as several of the loci examined in this study localized extensively to all 3 subcompartments, this is highly likely. Ultimately the 3 repressive subcompartments, though morphologically distinct, may simply provide a uniform compartment for late replicating chromatin and should functionally be considered as a whole. Localization to any of the discernible subtypes may thus largely be dictated by chromosome placement within the nucleus (based on size and gene density, depending on cell type), as well as linear proximity of a chromatin domain to centromeres and NORs on a given chromosome.
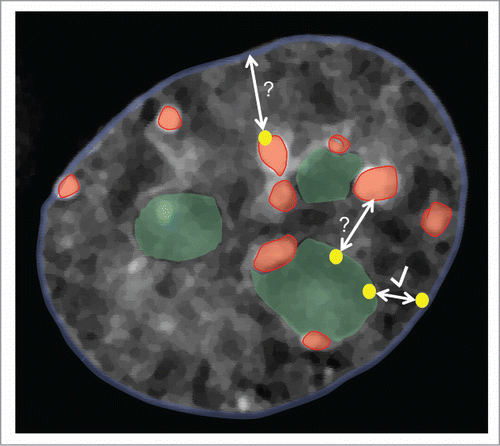
Figure 5. The late replicating genome localizes predominantly to 3 main repressive nuclear subcompartments: the nuclear periphery (PH, blue nuclear outline), pericentric heterochromatin (PCH, red) and perinucleolar heterochromatin (PNH, green). Our results suggest that a given late replicating region (yellow dots), particularly on small chromosomes, can associate with any (and multiple) of these subcompartments and relocalize from PNH to PH upon the loss of nucleoli (see arrow with check mark). While our results do not address the possibility of shuttling between the PCH and PH or PNH and PCH subcompartments (bidirectional arrows with question marks), work from other groups also suggest that localization between PH and PNH is interchangeable from one cell cycle to the next.
Materials and Methods
Cell lines and culture
GM06990 lymphoblastoid cells were mainained in RPMI 1640 (Life Technologies; 11875093) supplemented with 15% fetal bovine serum (Thermo Scientific; SH40007-13), 2mM L-Glutamine (Life Technologies; 25030-081) and Pen Strep (Life Technologies; 15140-122) at 37 C in 5% CO2. To avoid the onset of aneuploidy, cultures were generally exchanged for a fresh thaw every 4-6 weeks. For the actinomycin D treatment, cells were incubated for 2 hrs with 200 ng/ml actinomycin D (BioVision; 1036-50) and then harvested, washed in PBS and deposited on polylysine coated slides for fixation.
3D ImmunoFISH
3D ImmunoFISH was performed essentially as described.Citation20,35 Genomic regions were detected with nick translated BAC probes (for a complete list of BACs used and their genomic coordinates see Supplemental Table 5); about 100 ng of DIG- or biotin- or DNP-labeled probe (Roche; 11093088910, Roche; 11093070910, Perkin Elmer; NEL551001EA) and 10ug of human Cot-1 competitor DNA (Life Technologies; 15279-011) were used per hybridization. PCH was detected with directly labeled (FITC or Cy3) human pan-centromeric probe from Cambio (1695-F-02, 1695-C-02). BAC FISH spots were visualized with a FITC-conjugated monoclonal antibody to DIG (Sigma-Aldrich; F3523), fluorescently labeled streptavidin (Life Technologies; S-11223 or S-32356), or an Alexa-488-conjugated rabbit antibody to DNP (Life Technologies; A-11097). The lamina was detected using a goat anti-Lamin B1 antibody (Santa Cruz Biotechnology; sc6216) and a donkey Cy3 or Alexa-647 conjugated anti-goat antibody (Jackson ImmunoResearch; 705-166-147, Life Technologies; A-21447). Nucleoli were marked with a polyclonal rabbit antibody to nucleolin (Abcam; ab22758), which was detected either with a goat anti-rabbit secondary antibody conjugated with Alexa-405 (Life Technologies; A-31556) or a donkey anti-rabbit secondary antibody conjugated with Cy3 (Jackson ImmunoResearch; 711-166-152). While the majority of nucleolin became dispersed within the nucleoplasm following actinomycin D treatment, a sufficient amount remained within nucleoli to continue to demarcate them adequately using this marker.
DamID
DamID experiments with a LaminB1-Dam construct were performed essentially as described by Vogel et al.Citation36 Human LaminB1 (NM_005573.3) was cloned into the pLgw RFC1-V5-EcoDam vector (kindly provided by Bas van Steensel) via the Gateway system (Life Technolgies). Following verification of the correct localization of the transgene, the recovered and amplified Dam-methylated DNA was hybridized to a high density microarray covering the entire human genome with an average probe spacing of about 1100 bp (NimbleGen Human CGH 2.1M Whole-Genome Tiling v2.0D Array, Roche; 05541921001). In total, data from 6 replicate experiments was collected, including a dye swap. The data was normalized and LADs determined according to published protocols,Citation16 loaded into a genome browser and lifted onto the human genome build hg19.Citation37
Image acquisition and analysis
Image stacks of GM06990 cells were captured on an Olympus IX71 microscope (Olympus 100X/1.40, UPLS Apo objective) equipped with a cooled CCD camera and subsequently deconvolved using Deltavision SoftWorx software (Applied Precision). 3D colocalization analysis of the BAC signals with the lamina, PCH, and nucleoli was also performed using SoftWorx: FISH spots were judged to contact a subcompartment if at least several pixels in one plane overlapped with the stain for the respective subcompartment. Over 100 alleles and at least 2 independent hybridizations were scored for each BAC and condition.
Statistical analysis
We modeled the fraction of alleles that localized in the PNH, PCH, and PH subcompartment and was totally repressed (data in Supplemental Table 4) using a mixed effects model separately for each response (PH, PCH, PNH, total Repression). Actinomycin D treatment was analyzed as a fixed effect, and chromosome and experiment (nested within chromosome) were treated as random effects. Except for PCH, which was not significant, the other outcomes were all significantly associated with treatment with p < 10−4.
Genomic coverage statistics were analyzed using The Genomic HyperBrowser and Galaxy.Citation38-41
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Acknowledgments
We thank the members of the Groudine lab for their critical assessments of the data and manuscript.
Supplemental_File.pdf
Download PDF (3.2 MB)Funding
This work was funded by NIH grants DK44746 and HL065440.
Supplemental Material
Supplemental data for this article can be accessed on the publisher's website: http://www.tandfonline.com/kncl
References
- Pederson T. The spatial organization of the genome in mammalian cells. Curr Opin Genet Dev 2004; 14:203-9; PMID:15196468
- Misteli T. Concepts in nuclear architecture. Bioessays 2005; 27:477-87; PMID:15832379
- Rajapakse I, Groudine M. On emerging nuclear order. J Cell Biol 2011; 192:711-21; PMID:21383074; http://dx.doi.org/10.1083/jcb.201010129
- Cavalli G, Misteli T. Functional implications of genome topology. Nat Struct Mol Biol 2013; 20:290-9; PMID:23463314; http://dx.doi.org/10.1038/nsmb.2474
- Heitz E. Das heterochromatin der Moose. Jahrb Wiss Botanik 1928; 69:762-818.
- Dillon N. Heterochromatin structure and function. Biol Cell 2004; 96:631-7; PMID:15519697
- Padeken J, Heun P. Nucleolus and nuclear periphery: velcro for heterochromatin. Curr Opin Cell Biol 2014; 28:54-60; PMID:24690547; http://dx.doi.org/10.1016/j.ceb.2014.03.001
- Lieberman-Aiden E, van Berkum NL, Williams L, Imakaev M, Ragoczy T, Telling A, Amit I, Lajoie BR, Sabo PJ, Dorschner MO, et al. Comprehensive mapping of long-range interactions reveals folding principles of the human genome. Science 2009; 326:289-93; PMID:19815776; http://dx.doi.org/10.1126/science.1181369
- Ryba T, Hiratani I, Lu J, Itoh M, Kulik M, Zhang J, Schulz TC, Robins AJ, Dalton S, Gilbert DM. Evolutionarily conserved replication timing profiles predict long-range chromatin interactions and distinguish closely related cell types. Genome Res 2010; 20:761-70; PMID:20430782; http://dx.doi.org/10.1101/gr.099655.109
- Nakayasu H, Berezney R. Mapping replicational sites in the eucaryotic cell nucleus. J Cell Biol 1989; 108:1-11; PMID:2910875; http://dx.doi.org/10.1083/jcb.108.1.1
- O'Keefe RT, Henderson SC, Spector DL. Dynamic organization of DNA replication in mammalian cell nuclei: spatially and temporally defined replication of chromosome-specific alpha-satellite DNA sequences. J Cell Biol 1992; 116:1095-110; PMID:1740468; http://dx.doi.org/10.1083/jcb.116.5.1095
- Ferreira J, Paolella G, Ramos C, Lamond AI. Spatial organization of large-scale chromatin domains in the nucleus: a magnified view of single chromosome territories. J Cell Biol 1997; 139:1597-610; PMID:9412456; http://dx.doi.org/10.1083/jcb.139.7.1597
- Sadoni N, Langer S, Fauth C, Bernardi G, Cremer T, Turner BM, Zink D. Nuclear organization of mammalian genomes. Polar chromosome territories build up functionally distinct higher order compartments. J Cell Biol 1999; 146:1211-26; PMID:10491386; http://dx.doi.org/10.1083/jcb.146.6.1211
- Bolzer A, Kreth G, Solovei I, Koehler D, Saracoglu K, Fauth C, Müller S, Eils R, Cremer C, Speicher MR, et al. Three-dimensional maps of all chromosomes in human male fibroblast nuclei and prometaphase rosettes. PLoS Biol 2005; 3:e157; PMID:15839726
- Greil F, Moorman C, Van Steensel B. DamID: mapping of in vivo protein-genome interactions using tethered DNA adenine methyltransferase. Meth Enzymol 2006; 410:342-59; PMID:16938559; http://dx.doi.org/10.1016/S0076-6879(06)10016-6
- Guelen L, Pagie L, Brasset E, Meuleman W, Faza MB, Talhout W, Eussen BH, De Klein A, Wessels L, De Laat W, et al. Domain organization of human chromosomes revealed by mapping of nuclear lamina interactions. Nature 2008; 453:948-51; PMID:18463634; http://dx.doi.org/10.1038/nature06947
- Boyle S, Gilchrist S, Bridger JM, Mahy NL, Ellis JA, Bickmore WA. The spatial organization of human chromosomes within the nuclei of normal and emerin-mutant cells. Human Molecular Genetics 2001; 10:211-9; PMID:11159939; http://dx.doi.org/10.1093/hmg/10.3.211
- Penman S, Vesco C, Penman M. Localization and kinetics of formation of nuclear heterodisperse RNA, cytoplasmic heterodisperse RNA and polyribosome-associated messenger RNA in HeLa cells. J Mol Biol 1968; 34:49-69; PMID:4106524; http://dx.doi.org/10.1016/0022-2836(68)90234-9
- Perry RP, Kelley DE. Inhibition of RNA synthesis by actinomycin D: characteristic dose-response of different RNA species. J Cell Physiol 1970; 76:127-39; PMID:5500970; http://dx.doi.org/10.1002/jcp.1040760202
- Croft JA, Bridger JM, Boyle S, Perry P, Teague P, Bickmore WA. Differences in the localization and morphology of chromosomes in the human nucleus. J Cell Biol 1999; 145:1119-31; PMID:10366586; http://dx.doi.org/10.1083/jcb.145.6.1119
- Cremer M, Hase von J, Volm T, Brero A, Kreth G, Walter J, Fischer C, Solovei I, Cremer C, Cremer T. Non-random radial higher-order chromatin arrangements in nuclei of diploid human cells. Chromosome Res 2001; 9:541-67; PMID:11721953; http://dx.doi.org/10.1023/A:1012495201697
- Cremer M. Inheritance of gene density-related higher order chromatin arrangements in normal and tumor cell nuclei. J Cell Biol 2003; 162:809-20; PMID:12952935
- Wu R, Terry AV, Singh PB, Gilbert DM. Differential subnuclear localization and replication timing of histone H3 lysine 9 methylation states. Mol Biol Cell 2005; 16:2872-81; PMID:15788566; http://dx.doi.org/10.1091/mbc.E04-11-0997
- Brown KE, Guest SS, Smale ST, Hahm K, Merkenschlager M, Fisher AG. Association of transcriptionally silent genes with Ikaros complexes at centromeric heterochromatin. Cell 1997; 91:845-54; PMID:9413993; http://dx.doi.org/10.1016/S0092-8674(00)80472-9
- Clowney EJ, LeGros MA, Mosley CP, Clowney FG, Markenskoff-Papadimitriou EC, Myllys M, Barnea G, Larabell CA, Lomvardas S. Nuclear aggregation of olfactory receptor genes governs their monogenic expression. Cell 2012; 151:724-37; PMID:23141535; http://dx.doi.org/10.1016/j.cell.2012.09.043
- Tanaka A, Tanizawa H, Sriswasdi S, Iwasaki O, Chatterjee AG, Speicher DW, Levin HL, Noguchi E, Noma K-I. Epigenetic regulation of condensin-mediated genome organization during the cell cycle and upon DNA damage through histone H3 lysine 56 acetylation. Mol Cell 2012; 48:532-46; PMID:23084836; http://dx.doi.org/10.1016/j.molcel.2012.09.011
- van Koningsbruggen S, Gierlinski M, Schofield P, Martin D, Barton GJ, Ariyurek Y, Dunnen den JT, Lamond AI. High-resolution whole-genome sequencing reveals that specific chromatin domains from most human chromosomes associate with nucleoli. Mol Biol Cell 2010; 21:3735-48; PMID:20826608; http://dx.doi.org/10.1091/mbc.E10-06-0508
- Németh A, Conesa A, Santoyo-Lopez J, Medina I, Montaner D, Péterfia B, Solovei I, Cremer T, Dopazo J, Längst G. Initial genomics of the human nucleolus. PLoS Genet 2010; 6:e1000889; PMID:20361057
- Manuelidis L. Different central nervous system cell types display distinct and nonrandom arrangements of satellite DNA sequences. Proc Natl Acad Sci USA 1984; 81:3123-7; PMID:6587343
- Carvalho C, Pereira HM, Ferreira J, Pina C, Mendonça D, Rosa AC, Carmo-Fonseca M. Chromosomal G-dark bands determine the spatial organization of centromeric heterochromatin in the nucleus. Mol Biol Cell 2001; 12:3563-72; PMID:11694589; http://dx.doi.org/10.1091/mbc.12.11.3563
- Solovei I, Schermelleh L, Düring K, Engelhardt A, Stein S, Cremer C, Cremer T. Differences in centromere positioning of cycling and postmitotic human cell types. Chromosoma 2004; 112:410-23; PMID:15197559; http://dx.doi.org/10.1007/s00412-004-0287-3
- Wiblin AE, Cui W, Clark AJ, Bickmore WA. Distinctive nuclear organisation of centromeres and regions involved in pluripotency in human embryonic stem cells. J Cell Sci 2005; 118:3861-8; PMID:16105879; http://dx.doi.org/10.1242/jcs.02500
- Chubb JR, Boyle S, Perry P, Bickmore WA. Chromatin motion is constrained by association with nuclear compartments in human cells. Curr Biol 2002; 12:439-45; PMID:11909528; http://dx.doi.org/10.1016/S0960-9822(02)00695-4
- Kind J, Van Steensel B. Stochastic genome-nuclear lamina interactions: modulating roles of Lamin A and BAF. Nucleus 2014; 5:124-30; PMID:24717229; http://dx.doi.org/10.4161/nucl.28825
- Ragoczy T, Bender MA, Telling A, Byron R, Groudine M. The locus control region is required for association of the murine beta-globin locus with engaged transcription factories during erythroid maturation. Genes Dev 2006; 20:1447-57; PMID:16705039; http://dx.doi.org/10.1101/gad.1419506
- Vogel MJ, Peric-Hupkes D, Van Steensel B. Detection of in vivo protein-DNA interactions using DamID in mammalian cells. Nat Protoc 2007; 2:1467-78; PMID:17545983; http://dx.doi.org/10.1038/nprot.2007.148
- Kent WJ, Sugnet CW, Furey TS, Roskin KM, Pringle TH, Zahler AM, Haussler D. The human genome browser at UCSC. Genome Res 2002; 12:996-1006; PMID:12045153; http://dx.doi.org/10.1101/gr.229102.
- Giardine B, Riemer C, Hardison RC, Burhans R, Elnitski L, Shah P, Zhang Y, Blankenberg D, Albert I, Taylor J, et al. Galaxy: a platform for interactive large-scale genome analysis. Genome Res 2005; 15:1451-5; PMID:16169926; http://dx.doi.org/10.1101/gr.4086505
- Blankenberg D, Kuster Von G, Coraor N, Ananda G, Lazarus R, Mangan M, Nekrutenko A, Taylor J. Galaxy: a web-based genome analysis tool for experimentalists. Curr Protoc Mol Biol 2010; Chapter 19:Unit19.10.1-21; PMID:20373502
- Goecks J, Nekrutenko A, Taylor J, Galaxy Team. Galaxy: a comprehensive approach for supporting accessible, reproducible, and transparent computational research in the life sciences. Genome Biol 2010; 11:R86; PMID:20738864
- Sandve GK, Gundersen S, Rydbeck H, Glad IK, Holden L, Holden M, Liestøl K, Clancy T, Ferkingstad E, Johansen M, et al. The Genomic HyperBrowser: inferential genomics at the sequence level. Genome Biol 2010; 11:R121; PMID:21182759
