Abstract
Immunotherapy is at the forefront of cancer treatment, and biomarkers are urgently needed to predict patient response to therapy. Recently, we discovered a 4-gene peripheral blood mRNA signature for prolonged survival in patients treated with tremelimumab. Peripheral blood mRNA is a readily accessible and under-utilized source of clinically relevant biomarkers.
The tumor environment can suppress T cells and other immune cells through release of humoral factors and direct cell-cell contact. Malignant cells may also induce myeloid cells to secrete pro-angiogenic factors. These signals are strongest within the local tumor microenvironment.Citation1 However, particularly in advanced tumors, the systemic immune system including peripheral blood leukocytes may also be affected.Citation2 Peripheral blood has the advantage of being easily accessibile in the clinical setting and is a practical source of biomarkers. Immune cells in the peripheral blood are migratory cells whose transcriptional profile may be influenced by the presence of tumor cells either residing in solid tissue as well as those present in blood. The transcriptional profile of peripheral blood may, in turn, reflect the degree of tumor-mediated immunosuppression and impact potential response to immunotherapy.
In our recent publication, “Blood mRNA Expression Profiling Predicts Survival in Patients Treated with Tremelimumab,” we identified a panel of 4 gene transcripts predictive of survival in patients treated with tremelimumab.Citation3 Tremelimumab (ticilimumab, Pfizer), an IgG2 antibody that targets cytotoxic T-lymphocyte associated protein (CTLA-4), an inhibitory marker expressed on T cells. Tremelimumab has established activity in melanoma and is comparable in terms of toxicity and proposed mechanism of action to the FDA approved CTLA-4 blocking antibody, Yervoy® (ipilimumab, Bristol Myers Squibb). Tremelimumab is currently being administered in experimental protocols as part of combination regimens for melanoma but it has not yet been submitted for FDA approval since it has yet to show superiority to dacarbazine.Citation4 However, Yervoy® was never directly tested against either dacarbazine or tremelimumab so the relative efficacies of the 2 antibodies remain unknown.
In order to identify a genomic signature of prolonged survival in patients receiving tremelimumab therapy, peripheral blood samples were obtained from 218 patients enrolled in a multi-center Phase II clinical trial of tremelimumab between 2005 and 2006.Citation5 Reverse transcription polymerase chain reaction (RT-PCR) was performed on whole peripheral blood samples to quantify mRNA levels of 169 candidate genes selected on the basis of prior studies of gene expression in cancer patients, and on the known mechanisms of action of CTLA-4. We found that 4 genes, cathepsin D (CTSD), phospholipase A2 group VII (PLA2G7), thioredoxin reductase 1 (TXNRD1), and interleukin 1 receptor associated kinase 3 (IRAK3) predicted overall survival in this population. These results were then confirmed in a validation set of 260 patients enrolled in the treatment arm of a multicenter Phase III study comparing tremelimumab and chemotherapy between 2006 and 2007 (P = 0.001).Citation4 Gene signature scores added significantly to standard clinical predictors in a multivariable model (P < 0.001). A limitation to this study is that unfortunately blood was not available from the control arm of the Phase III study so that the determination could not be made as to whether the 4-gene panel is a predictive or a prognostic marker (or both).
Importantly, this work (refer to ) provides evidence to the feasibility of using genomic blood biomarkers to predict outcomes in metastatic melanoma. Expression levels of the 169 genes measured correlated very closely between the 2 populations at baseline. Further, relative expression levels of the genes between patients who survived more than 1 y and patients who survived less than 1 y were also closely matched.Citation3 Thus, gene expression correlated reproducibly with outcome in 2 independent populations. Blood is a readily accessibly source of biomarkers, and relatively small amounts are sufficient for RNA extraction. Thus, peripheral blood seems a very promising resource for future research seeking to define novel, clinically applicable biomarkers.
Figure 1. Schema of the workflow for mRNA expression profiling in peripheral blood of patients treated with Tremelimumab.
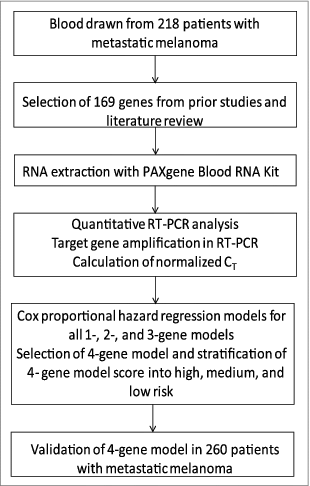
Biomarkers are urgently needed to predict responsiveness to immunotherapy. Although immunotherapy does not have the toxicities of conventional chemotherapy, the potential for severe life threatening auto-immunity makes CTLA-4 blockade difficult to tolerate. Further, most patients do not respond or respond only transiently to CTLA-4 blockade, so that targeted patient selection would provide greatly improved clinical benefit.Citation6 Antibodies targeting programd cell death 1 (PD-1) have a more favorable risk benefit ratio but many patients progress or respond only transiently and treatment can induce severe auto-immune toxicity, particularly if given in combination with ipilimumab. Prior studies have identified many immune correlates in the blood of patients responding to Yervoy®, including alterations in the numbers, phenotype, and clonal distributions of CD4+ and CD8+ T cells.Citation7 There is likewise evidence that characteristics of the immune microenvironment within the tumor context may predispose particular patients to beneficial responses to immunotherapy.Citation8 A biomarker defined based on sampling the immune milieu in peripheral blood based on gene expression studies prior to initiation of therapy is conceivable and would be ideal for clinical applications.
RNA in whole blood is likely primarily reflective of the transcriptional activity of peripheral blood leukocytes. Our research is thus in line with reported findings that alterations in peripheral blood suppressor myeloid cell populations correlate with, and are predictive of, response to CTLA-4 blockade.Citation9 Furthermore, the genes identified in our panel to correlate inversely with survival, CTSD and IRAK3, have established roles in cancer growth and immunosuppression. IRAK3, is primarily expressed by myeloid cells and suppresses toll-like receptor signaling, diminishing the perception of “danger signals” by the immune system. CTSD is expressed by most mammalian cells and is overexpressed in melanoma. Similar to vascular endothelial growth factor (VEGF), another proposed marker for poor response to both Yervoy® and IL-2, CTSD favors angiogenesis and metastasis.Citation10
As the paradigm in cancer treatment shifts toward immunotherapy, understanding the complex interplay between cancer cells and the host immune system becomes increasingly critical. We know that this interaction occurs and is reflected by changes in phenotype and activation status of peripheral blood leukocytes. Assessing gene expression is a straightforward way to assess the systemic immune milieu and may be relevant to the development of clinically useful predictive biomarkers, both those specifically for CTLA-4 blockade as well as markers more broadly relevant to immunotherapeutic regimens in general.
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
References
- Gajewski TF, Fuertes M, Spaapen R, Zheng Y, Kline J. Molecular profiling to identify relevant immune resistance mechanisms in the tumor microenvironment. Curr Opin Immunol 2011; 23:286-92; PMID: 21185705; http://dx.doi.org/10.1016/j.coi.2010.11.013
- Ross RW, Galsky MD, Scher HI, Magidson J, Wassmann K, Lee GS, Katz L, Subudhi SK, Anand A, Fleisher M, et al. A whole-blood RNA transcript-based prognostic model in men with castration-resistant prostate cancer: a prospective study. Lancet Oncol 2012; 13:1105-13; PMID: 23059047; http://dx.doi.org/10.1016/S1470-2045(12)70263-2
- Saenger Y, Magidson J, Liaw B, de Moll E, Harcharik S, Fu Y, et al. Blood mRNA Expression Profiling Predicts Survival in Patients Treated with Tremelimumab. Clin Cancer Res Off J Am Asso Cancer Res 2014; 20(12):3310-18
- Ribas A, Kefford R, Marshall MA, Punt CJ, Haanen JB, Marmol M, Garbe C, Gogas H, Schachter J, Linette G, et al. Phase III randomized clinical trial comparing tremelimumab with standard-of-care chemotherapy in patients with advanced melanoma. J Clin Oncol Off J Am Soc Clin Oncol 2013; 31:616-22; PMID: 23295794; http://dx.doi.org/10.1200/JCO.2012.44.6112
- Kirkwood JM, Lorigan P, Hersey P, Hauschild A, Robert C, McDermott D, Marshall MA, Gomez-Navarro J, Liang JQ, et al. Phase II trial of tremelimumab (CP-675,206) in patients with advanced refractory or relapsed melanoma. Clin Cancer Res Official J Am Asso Cancer Res 2010; 16:1042-8; PMID: 20086001; http://dx.doi.org/10.1158/1078-0432.CCR-09-2033
- Ascierto PA, Kalos M, Schaer DA, Callahan MK, Wolchok JD. Biomarkers for immunostimulatory monoclonal antibodies in combination strategies for melanoma and other tumor types. Clin Cancer Res official J Am Assoc Cancer Res 2013; 19:1009-20; PMID: 23460532; http://dx.doi.org/10.1158/1078-0432.CCR-12-2982
- Cha E, Klinger M, Hou Y, Cummings C, Ribas A, Faham M, Fong L. Improved Survival with T Cell Clonotype Stability After Anti-CTLA-4 Treatment in Cancer Patients. Sci Trans Med 2014; 6:238ra70; http://dx.doi.org/10.1126/scitranslmed.3008211
- Ji RR, Chasalow SD, Wang L, Hamid O, Schmidt H, Cogswell J, Alaparthy S, Berman D, Jure-Kunkel M, Siemers NO, et al. An immune-active tumor microenvironment favors clinical response to ipilimumab. Cancer Immunol Immun: CII 2012; 61:1019-31; PMID: 22146893; http://dx.doi.org/10.1007/s00262-011-1172-6
- Kitano S, Postow MA, Ziegler CG, Kuk D, Panageas K, Cortez C, Rasalan T, Adamow M, Yuan J, Wong P, et al. Computational Algorithm Driven Evaluation of Monocytic Myeloid Derived Suppressor Cell Frequency For Prediction of Clinical Outcomes. Cancer Immunol Res 2014; 2(8):1-10.
- Yuan J, Zhou J, Dong Z, Tandon S, Kuk D, Panageas KS, Wong P, Wu X, Naidoo J, Page DB, et al. Pretreatment serum VEGF is associated with clinical response and overall survival in advanced melanoma patients treated with ipilimumab. Cancer Immunol Res 2014; 2:127-32; PMID: 24778276; http://dx.doi.org/10.1158/2326-6066.CIR-13-0163
