Abstract
To assess immunogenicity and development of antibodies in the context of vaccination, it is critical to quantify titers of neutralizing antibodies. We have been employing the 293TT cell-based neutralization assay system to quantify anti-HPV neutralizing antibodies. In this system, human papillomavirus (HPV) pseudovirion (PsV) particles encapsidating secreted alkaline phosphatase (SEAP) gene are used to measure infection of 293TT cells in 72-hr cell-culture supernatants. SEAP has traditionally been measured by Great EscAPe™ SEAP Chemiluminescence Kit 2.0 (GE). To reduce the cost, and to potentially increase efficiency, we sought a cheaper kit with better detection capability. Performance characteristics of the newer chemiluminescence kit, ZiVa® Ultra SEAP Plus Assay (Ziva) and GE were compared using the 293TT system. Dose titration of HPV PsV 16 or 18 showed that signal-to-noise ratios at 48 and 72 hr post-infection were higher for ZiVa at nearly all doses. ZiVa was superior to GE as it was able to detect SEAP at 48 hr, as well as when lower numbers of 293TT cells were used. The ability of ZiVa to quantitate HPV-16 and -18 neutralizing antibody titers was tested using sera from Cervarix® immunized individuals. Spearman rank correlational analyses showed excellent correlations between the titers obtained with ZiVa and GE for anti-HPV16 (r = 0.9822, p < 0.0001) and anti-HPV18 (r = 0.9832, p < 0.0001) antibodies. We concluded that ZiVa is superior to GE in detecting SEAP, and the antibody titers in sera of vaccinated individuals were similar to those obtained with GE. Thus, Ziva is a suitable alternative to GE.
Introduction
The most common sexually transmitted viral infections are caused by human papillomaviruses (HPV).Citation1,2 HPV are also the major etiological agents responsible for development of benign genital warts (types 6 and 11), precancerous and cancerous lesions of the cervix and for a portion of anogential and oral malignancies (types 16 and 18).Citation3-7 HPV types 16 and 18 are responsible for approximately 70% of cervical cancers.Citation8 In 2006 and in 2009, the United States Food and Drug Administration (FDA) approved 2 different prophylactic HPV vaccines: one of them is for females and males,Citation9 and the other for females only.Citation10 These 2 HPV vaccines, Gardasil® and Cervarix®, are comprised of late 1 (L1) major capsid protein-based virus-like-particles (VLPs) of HPV6, 11, 16 and 18 (Gardasil®) or HPV16 and 18 (Cervarix®).Citation11 These vaccines have demonstrated excellent efficacy in clinical trials. In response to these L1 VLP vaccines, virus-neutralizing L1-specific antibodies are generated, whose levels and neutralization activity can be quantitated by enzyme-linked immunosorbent assay (ELISA) and in vitro neutralization assay, respectively.
In addition to these licensed vaccines, second-generation HPV vaccines are in development. For example, the nonavalent L1 VLP vaccine that contains VLPs for HPV types 6, 11, 16, 18, 31, 33, 45, 52, and 58 has been developed to widen the range of protection against other carcinogenic HPV types,Citation11 and it is under review for licensure by the FDA. Another second-generation vaccine that is based on the minor capsid protein L2 has also been in the development because of L2's potential to be broadly reactive;Citation12 thereby providing a wider range of protection against different HPV types.Citation11,13 A recombinant fusion protein composed of amino acid sequence positions 11 to 88 of L2 proteins has shown promising preclinical results.Citation14,15 However, L2-based vaccines have yet to be evaluated in clinical trials.
A measure of the success of a given vaccine is dependent on its ability to provide protection from the targeted infectious agent and the disease that it causes. Although no correlates of protection have formally been identified for the HPV vaccines, neutralizing antibodies are believed to play a key role in providing protection against HPV infection. Therefore, quantitation of titers of biologically active antigen-specific antibodies becomes critical in understanding the development of humoral immunity in response to a vaccine. As such, cell-based in vitro neutralization assay is an ideal system to measure the levels of biologically active antibodies in clinical samples. Although different neutralization assays have been reported for HPV,Citation16-25 our laboratory has adopted, optimized, and validatedCitation26-29 a pseudovirion (PsV)-based HPV neutralization assay that was originally reported by Pastrana et al.Citation30 In this assay system, PsV particles that are composed of L1 and L2 capsid proteins from a particular HPV type are used to infect 293TT cells for 72 hr. Because a PsV particle encapsidates a reporter gene during its production,Citation31,32 magnitude of PsV entry and infection can be measured through the detection of the reporter gene product. Using this 293TT cell-based system and secreted alkaline phosphatase (SEAP) as a reporter, we have measured titers of biologically active anti-L1 specific antibodies in HPV L1VLP vaccine recipients as well as in those who were naturally infected with HPVs.Citation27,28,30
As the number of samples from clinical trials can be large, conducting the cell-based neutralization assay and SEAP assay can be labor-intensive and expensive. This 293TT cell-based neutralization assay takes 72 hr before collecting and storing cell-culture supernatants for the subsequent SEAP assay. Furthermore, production of and the use of PsVs in the neutralization assay can also be costly when testing hundreds to thousands of clinical samples. Hence, it is desirable to use the lowest amount possible of virus particles that would still lead to a reasonable induction of signal over background. For these reasons, we sought a cheaper SEAP detection kit with a better detectability to reduce the expenditures of resources and to increase efficiency. However, we wanted to ultimately examine whether such kit can still be used in measuring HPV neutralizing anti-L1 antibodies in clinical serum samples.
We and others have previously used Great EscAPe™ SEAP Chemiluminescence Kit 2.0 from Clontech (GE) for the detection of SEAP,Citation28-30 while a different group of investigators has used the SEAP Reporter Gene Assay for Chemiluminescence from Roche.Citation33 In addition to these 2 well-known brand name assay kits, we found 2 additional products after performing on-line searches. One of them, ZiVa® Ultra-Sensitive SEAP Detection Kit (ZiVa) from Jaden Bioscience, Inc., appeared to fit our need of a kit that is less expensive and highly sensitive. In this study, we tested whether ZiVa can be an alternative product to GE. We conducted a side-by-side comparison of ZiVa and GE using the 293TT cell-based assay system. This comparison included evaluation of kits’ ability to detect SEAP through performing infectivity assays using different concentrations of PsV particles of HPV16 and 18, and by using different numbers of target 293TT cells. In addition, SEAP activity was examined at 48 and 72 hr post-infection to evaluate whether the duration of 293TT cell-based assay could be shortened. Most important, we examined the ability of ZiVa to quantitate HPV neutralizing titers in sera from a subset of Cervarix® immunized individuals who received 1, 2, or 3 doses of the vaccine as part of a Uganda-government sponsored vaccine demonstration program,Citation34 and compared how well these titers correlated with those obtained with the GE kit.
Results
Comparison of different SEAP assay kits
To find an alternative to Clontech's Great EscAPe™ SEAP Kit 2.0 (hereon, referred to as GE), we searched for SEAP chemiluminescence kits from different vendors and compared their characteristics (). In addition to GE, we found kits from Roche, Life Technologies, and Jaden Bioscience, Inc. All four vendors offered product size of approximately 1,000 tests, except for Roche, which had 500 tests as its biggest size. Based on the available information from the manufacturers, we found that the majority of the characteristics were similar between the kits (). However, the product from Jaden Bioscience had other unique, attractive features. These included lower cost and better detectability of SEAP (). These characteristics led us to compare the ability of ZiVa® Ultra SEAP Plus Detection Kit (hereon, referred to as ZiVa) and GE to detect SEAP in supernatants from cell-based bioassays.
Table 1. Comparison of different chemiluminescence-based SEAP assay kits
ZiVa is superior to GE in its ability to detect SEAP in cell-culture supernatants
To compare the ability of ZiVa and GE to detect SEAP levels, 293TT cells were infected with SEAP gene encapsidated HPV pseudovirion (PsV)16 or PsV18 particles that were 2-fold serially diluted (range: 1,000 – 1,024,000). To express the magnitude of infections, signal-to-noise ratio (fold-increase) was calculated by dividing the relative-light-unit (RLU) of an infected sample by the mean RLU of the no-virus controls, in which 293TT cells were incubated without any viral particles. The mean fold-increase was calculated using individual values of the duplicate wells, and the results of 3 independent experiments for both PsV16 and PsV18 are shown in .
Figure 1. ZiVa can better detect SEAP than GE. Pseudovirion particles of HPV types 16 and 18 were serially diluted, and used to infect 0.03 × 106 293TT cells. Two-fold serial dilutions were performed starting from 1:1000 to 1:1024000. Supernatants were harvested at 48 and 72 hr post-infection. Each PsV dilution was setup in duplicate wells. Mean ± standard deviation (SD) of 3 independent experiments are shown. (A) Infectivity levels (fold-increase) by PsV16 particles after 48 or 72 hr of incubation are shown. (B) Infectivity levels by PsV18 particles after 48 or 72 hr of incubation are shown. In both (A and B), the same supernatant samples were tested using ZiVa or GE SEAP detection kit. The plots show fold-increase in log10 scale as a function of viral concentration (log10). Some of the error bars are too small to be seen in the graphs.
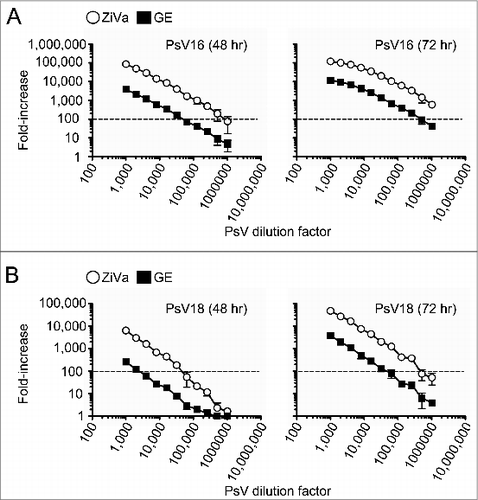
For both PsV16- () and PsV18-infected () samples, ZiVa out performed GE at both 48- and 72-hr time points, despite using only 5 μL of the supernatants vs 25 μL of GE. With ZiVa, higher fold-increases were achieved for all dilutions tested for PsV16 at both time points with the mean ZiVa/GE ratios that ranged from 16–24 at 48 hr, and 10–16 at 72 hr (). For PsV18, the mean ratios ranged from 1–26 and 12–16 at 48 and 72 hr, respectively (). The last 2 PsV18 viral dilution factors (512,000 and 1,024,000) showed a minimal to no difference at the 48-hr time point for PsV18. A further indication that ZiVa can better detect SEAP than GE was that when we arbitrarily chose to examine the amount of viral particles required to induce 100-fold increase in signal, we found that ZiVa required at least 10-fold less particles than those required for GE (, dotted lines). At 72 hr post-infection, the values of fold-increase for PsV16 remained above 100-fold for ZiVa even at the lowest dilution of 1:1,024,000 (, right panel). Furthermore, mean RLUs of the no-virus controls (n = 8 wells per experiment) from the 3 experiments at 48 hr were 57, 67, and 73 (mean = 66), and were 118, 143, and 168 (mean = 143) at 72 hr for ZiVa. For GE, they were 170, 190, and 228 (mean = 196) at 48 hr, and 241, 278, and 247 (mean = 255) at 72 hr. The values were higher for GE by factors of 3.0 (196/66) and 1.8 (255/143) for 48- and 72-hr time points, respectively. Therefore, the higher background RLU for GE does not explain better fold-increases obtained by using ZiVa.
Table 2. Ratios of ZiVa to GE in detecting SEAP
To examine whether there was an added benefit to culturing 72 hr when compared to 48 hr, we divided the values of mean fold-increase of the 72-hr time point by those of the 48-hr time point within each assay for both PsV16 and PsV18 (Tables S1 and 2). For ZiVa, the overall ratios of 72-hr to 48-hr time points ranged from 1 to 19 over 3 independent experiments for PsV16, while the range was 3 to 18 for GE (Table S1). For PsV18, they were 6 to 106 for ZiVa, and 3 to 30 for GE (Table S2). As expected, with higher dilutions, the ratios were higher for both PsV types. However, because the last 5 dilutions were border line limiting factors for PsV18, the ratios were skewed toward higher values. Overall, there was an added benefit to culturing for 72 hr for both ZiVa and GE.
Lower numbers of 293TT cells can be used to detect SEAP with ZiVa
We further tested the ability of the 2 kits to detect SEAP by using different numbers of 293TT cells per well (range: 0.002 × 106 to 0.3 × 106). For this study, we used 2 different viral concentrations for PsV16 (1:100,000 and 1:600,000), and for PsV18 (1:10,000 and 1:60,000) to infect different numbers of 293TT cells. These viral dilutions factors were expected to provide approximately 100,000 to 300,000 RLUs using either ZiVa or GE based on the results from the infectivity assay (). The lower dilution factors were to provide the expected RLU values when using GE, and the higher dilutions factors were for ZiVa. Cell-culture supernatants were collected as in , and the results from 3 independent experiments are shown in and .
Figure 2. ZiVa can detect SEAP even using limiting numbers of 293TT cells. Different numbers of 293TT cells were plated at the start of the infections by PsV16 or PsV18. The number of cells ranged from 0.002 - 0.3 × 106 cells per well, and 2-fold serial dilutions were performed. Each cell number was tested in triplicates. A fixed dilution factor of 1:100000 for PsV16 (A) and 1:10000 for PsV18 (B) were used to infect 293TT cells. At 48 or 72 hr post-infection, the supernatants were harvested. Cell numbers were plotted in log10 scale. Mean ± SD of 3 independent experiments are shown. Some of the error bars are too small to be seen in the graphs.
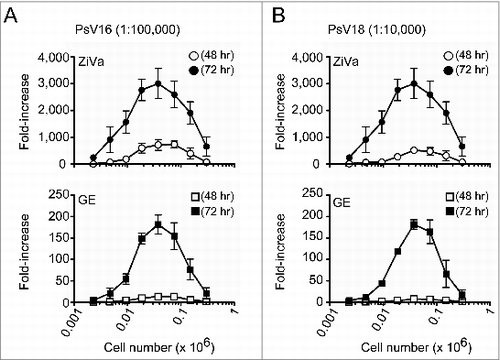
Figure 3. ZiVa can detect SEAP at lower cell numbers using lower viral dilution concentrations. The experiments were conducted as described in , except higher viral dilution factors were used: 1:600000 for PsV16 and 1:60000 for PsV18. Mean ± SD of 3 independent experiments are shown. Some of the error bars are too small to be seen in the graphs.
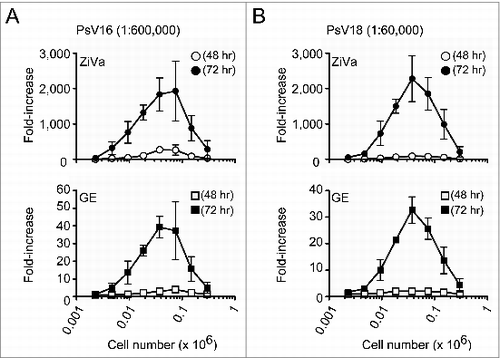
In general, the magnitude of signal increased until the cell number reached 0.038 × 106 cells per well, and then, it began to decline ( and ). The maximal fold-increases were detected at 0.038 × 106 cells per well for both HPV16 ( and ) and HPV18 ( and ), regardless of the viral concentrations. However, the ability of GE to detect SEAP was inferior to ZiVa at both 48- and 72-hr time points; for the majority of cell numbers tested, the magnitude of fold-increase was much less than those of ZiVa.
Overall, the data showed that ZiVa is capable of detecting the amount of SEAP that is limiting for GE in cell-culture supernatants, and that ZiVa can provide higher level of signal than GE. As a result, lower amount of PsV can be used and still obtain a high signal-to-noise ratio. Furthermore, although we observed an optimal signal induction at 0.038 × 106 cells per well, the data showed that ZiVa can detect substantial amount of the enzyme even when using lower numbers of cells.
HPV-specific antibody titers measured by ZiVa correlate with those of GE
HPV immunogenicity studies often involve testing of large numbers of clinical serum samples from HPV vaccinated individuals to determine neutralizing antibody titers. In our previous studies, we used GE to conduct SEAP assay to measure HPV neutralizing antibody titers.Citation26,29,35 To determine if ZiVa could be an alternative to GE in clinical studies, we evaluated whether the neutralization titers measured by ZiVa correlate to those measured by GE. For this, we used culture supernatants from 293TT cell-based neutralization assays that were performed for the Uganda Immunogenicity Study.Citation36 Anti-HPV16 and -HPV18 antibody titers were determined using randomly selected sera from those who received 1 (n = 25), 2 (n = 24), or 3 doses (n = 23) of the vaccine.
The results showed excellent overall correlations between the values obtained by GE and ZiVa for both anti-HPV16 and -HPV18 antibody titers (p < 0.0001 for both), with r = 0.9822 for anti-HPV16, and r = 0.9832 for anti-HPV18 (). We further tested for the correlations within the 3 different dosage groups. Regardless of the number of doses received, we found significant correlations (p < 0.0001 for all) between the values of titers measured by ZiVa and GE for anti-HPV16 (r = 0.9592, 0.9730, and 0.9427 for 1, 2, and 3 doses, respectively), and for anti-HPV18 antibodies (r = 0.9862, 0.9760, and 0.9753 for 1, 2, and 3 doses, respectively). The anti-HPV16 and -HPV18 antibody titers in these serum samples covered wide ranges; they were from 1:27 to 1:109,285 for anti-HPV16 for ZiVa, and from 1:16 to 1:17,828 for GE. For anti-HPV18, they were 1:9 to 1:8,745 for ZiVa, and 1:8 to 1:7667 for GE. These data showed that ZiVa can be used to quantitate the titers of samples from vaccinated individuals, demonstrating the feasibility of its use in clinical studies. Moreover, when we compared the values of 50% neutralization titers measured by GE or ZiVa, we found that the titer values were very similar, except for sample ID 64 for anti-HPV16 (). The data in provide further support that ZiVa can be used to measure the antibody titers in clinical serum samples that are comparable to the ones obtained with GE.
Table 3. Comparison of 50% neutralization titer values measured by GE or ZiVa
Figure 4. HPV neutralizing antibody titers measured by ZiVa correlate with those of GE. Cell-culture supernatants from 293TT cell-based neutralization assay from the Uganda Immunogenicity Study were used to quantitate 50% neutralization titers in sera of Cervarix®-immunized individuals using ZiVa or GE. The supernatant samples collected after 72 hr of incubation were used to measure the SEAP activity. Spearman rank-correlational analyses were performed between the titers obtained by ZiVa or GE for both anti-HPV16 and -HPV18 antibodies.
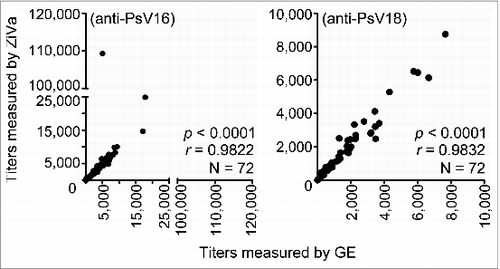
Together, the data showed that HPV neutralizing titers in serum samples from Cervarix® immunized individuals determined by ZiVa significantly correlated to the titers obtained by GE, regardless of the number of doses received. In addition, amounts of PsV particles required to obtain similar levels of fold-induction were at least 10-fold less for ZiVa compared to GE. Therefore, the ability of ZiVa to detect SEAP is superior to that of GE, and that ZiVa can serve as an alternative SEAP detection kit with the potential to substantially lower the cost of immunogenicity studies of HPV vaccines.
Discussion
In this study, we compared the ability of Ziva® Ultra SEAP Plus Assay and Great EscAPe™ SEAP Chemiluminescence Kit 2.0 to detect SEAP. Because the product information for ZiVa indicated that it was more sensitive (at polymerase chain reaction [PCR]-level, per Jaden BioScience, Inc.) and cheaper than GE, we wanted to compare the ability of the 2 kits to detect SEAP in cell-culture supernatants. However, our major aim was to examine whether ZiVa could measure HPV neutralizing antibody titers in sera of vaccinated individuals at comparable levels to those measured by GE. We found that ZiVa was able to detect SEAP better than GE, and that it quantitated the titers of anti-HPV antibodies that highly correlated to those measured by GE.
Although we used SEAP as a reporter gene, green fluorescent protein (GFP),Citation30,37 Gaussia luciferase,Citation33 or firefly luciferaseCitation38 has been used in the 293TT cell-based system as an alternative reporter gene. Our laboratory has chosen SEAP as the reporter gene of choice because of its high sensitivity as well as its potential application in a high-throughput system. GFP is useful in quantitating the number of cells that were infected by PsV particles, while the luciferase system can also be used to measure in vivo infection in animal models. However, GFP and firefly luciferase require collection or lysis of cells for subsequent measurement by a flow cytometer or by a luminometer with an injector, respectively. In contrast, the activity of SEAP can directly be measured using only a few microliters of cell-culture supernatants, and does not require an injector. Although Gaussia luciferase can also be detected using cell-culture supernatants, it still requires a luminometer with an injector. Furthermore, an entire 96-well plate can be read in seconds to minutes for SEAP, and once a maximal level of signal is reached, it is stably maintained for hours. In contrast, signals generated by luciferase starts to decay within seconds to minutes, and collection of cells by a flow cytometer can take several minutes to hours even if the samples were measured in a plate-format.
Our data showed that less amounts of PsV particles can be used with ZiVa, which could potentially enable the use of the same lot of viral particles for multiple studies. Our data from infection assays demonstrated that ZiVa can detect SEAP even at the PsV doses that were limiting for GE. However, when we compared the ratios of signals by dividing those from the 72-hr time point by those from the 48-hr time point, we found similar magnitude of increases for both kits. We concluded that there was an added benefit to culturing 72 hr. However, because of ZiVa's superior detectability, the assay could potentially be terminated after 48 hr of culture. In addition, although we observed that the optimal fold-increase was obtained with 0.038 × 106 cells per well, we did not observe much of a difference when half the number of cells (0.019 × 106 cells per well) was used, indicating that the cell number could be lowered, which could reduce the cost associated with cell culture.
Although Sehr et al.Citation33 have reported that Gaussia luciferase in their high-throughput system was more sensitive than SEAP, they used the reporter assay kit from Roche. In addition, ZiVa is supposed to provide a PCR-level of sensitivity. Nevertheless, a future, formal comparison of SEAP assay detection kits from Roche and Jaden BioScience Inc. may be needed, along with Gaussia luciferase in the context of vaccination and/or natural infection.
We found significant correlations between the titers obtained by both kits. The correlations were equally high regardless of whether the participants had received 1, 2, or 3 doses. Although we did not test sera from naturally infected individuals, whose titers are expected to be lower than those of vaccinated individuals, the samples tested in this study covered a wide range of titers, including some that were near the cut-off titers (1:10). Hence, we expect that ZiVa will also be applicable to epidemiological studies of HPV natural infection.
Correlational analyses showed that there was a high agreement (r > 0.98 for both HPV16 and 18) between titers of antibodies spanning a wide range of levels induced by 1, 2, or 3 doses; indicating that the performance of the assay is independent of the levels of HPV neutralizing antibodies. Although one of the samples (ID64) showed a higher titer value for ZiVa, we believe this is an outlier. As the titers for anti-HPV18 were similar in this sample, we will need to increase the study samples size to determine the frequency of such event. Of note, because ZiVa could allow the use of lower concentrations of PsV particles, it is possible that the usage of lower numbers of PsV particles would change the values of 50% neutralization titers. However, this should not change the overall results and interpretation of the data, such that samples with high or low titers will be expected to be ranked as such. Nevertheless, formal future studies may be warranted using a lower PsV concentration with vaccinated and/or naturally infected serum samples.
In conclusion, we believe that ZiVa can be an alternative to GE, because of its superior detectability of SEAP and its lower price. In addition, our data showed that the length of the experiments could potentially be shortened, and a lower number of cells could be used. Both of these parameters need to be further evaluated. They may further increase the efficiency of the experiments, and add to the system that is already high-throughput in the generation of data.
Materials and Methods
Cell culture and culture media
293TT cells have previously been described.Citation32 The cells were maintained in Dulbecco's Modified Eagle Medium (DMEM) (11965; Gibco/Life Technologies) supplemented with 10% heat-inactivated fetal bovine serum (FBS) (SH30070/03; Hyclone), 1% MEM non-essential amino acids (H11140–050) and 1% Glutamax (35050–061) (both from Gibco/Life Technologies). To maintain the expression of large-T antigen, the cells were cultured in the presence of 400 μg/mL hygromycin B (10687–010; Gibco/Life Technologies). For 293TT cell infectivity assay and 293TT cell-based neutralization assay, phenol red-free DMEM (21063–029; Gibco/Life Technologies) supplemented with above reagents were used along with 1% antibiotic/antimycotic (15240–062) and 1% HEPES (15630–080) (both from Gibco/Life Technologies); however, hygromycin B was not included in the media.
Production of PsV particles with SEAP reporter gene
The production of PsV particles has previously been reported.Citation32 Briefly, for the production of PsV particles with encapsidated SEAP gene, 293TT cells were transfected with a pShell plasmid that contains codon-modified L1 and L2 genesCitation32 of HPV16 or HPV18 with pYSEAP plasmid Citation30 using Lipofectamine 2000 (11668–019; Invitrogen/Life Technologies). The cells were cultured for 2 d in a 37°C CO2 incubator after the transfection. To collect the PsV particles produced inside 293TT cells, the cells were harvested, lysed in Lysis Buffer (PBS with 10 mM MgCl2 [14040–141; Gibco/Life Technologies], 0.5% Brij58 [P-5884; Sigma], and RNase A/T1 cocktail [AM2286; Ambion]) without Benzonase. Released PsV particles in Lysis Buffer were maturedCitation39 by incubating them in a 37°C water bath for 24 hr. The matured PsV particles were purified through Optiprep (D1556; Sigma) gradient (27%, 33%, and 39%) by ultracentrifugation at 50000rpm (Beckman Coulter Optima™ L-80 XP) for 3.5 hr. Purified PsV particles were collected, aliquoted into siliconized tubes (4203SLS, Bio Plas, Inc.), and stored at −80°C until use.
293TT infectivity assay
To perform infection of 293TT cells with PsV16 or PsV18 particles encapsidating the SEAP gene, 0.03 × 106 of 293TT cells per well were plated in 100 μL of phenol red-free culture media in 96-well flat-bottom plates (3596; Costar). The cells were incubated in a 37°C tissue culture incubator for a minimum of 2 hr prior to the addition of serially diluted PsV particles. Two-fold serial dilutions of PsV particles were performed in a 2 mL round-bottom 96-well plate (40002–014; VWR) using phenol-red free culture media. From this 2 mL 96-well round-bottom plate, 100 μL of the serially diluted PsV particles was transferred to a regular round-bottom 96-well plate (3788; Costar), and 25 μL of phenol red-free culture media was added. The serially diluted PsV particles were incubated at 4°C for 1 hr to parallel the procedure for the 293TT cell-based neutralization assay. After the 1-hr incubation, 100 μL of the diluted PsV particle solutions were transferred to the pre-plated 293TT cells. At 48 hr post-infection, 50 μL of supernatants were collected from each well, and the plates were cultured for an additional 24 hr. At 72 hr, an additional 100 μL of the supernatants were harvested. The collected supernatants were stored at −80°C for a minimum of 1 day until SEAP assays were performed.
To perform infectivity assay using different numbers of 293TT cells, the assays were carried out as described above using 2 dilution factors of PsV16 (1:100,000 or 1:600,000) or PsV18 (1:10,000 or 1:60,000) particles to infect different numbers of 293TT cells. These PsV dilutions factors were chosen based on the results from the PsV titration study, and these selected dilution factors were projected to provide 100,000 to 300,000 RLUs after a 72-hr incubation with 0.03 × 106 cells/well. Two-fold serial dilutions of 293TT cells were performed starting from 0.3 × 106 cells to 0.002 × 106 cells per well. The supernatants were collected after 48 and 72 hr of incubation at 37°C, and stored at −80°C until SEAP assays were performed.
Information on the study population and the assays
The neutralization/SEAP assays were conducted as part of the Uganda Immunogenicity Study,Citation36 which was a follow-up vaccination study of Cervarix® among 10–11 y old girls who were vaccinated as part of a government-run HPV vaccination demonstration program in Uganda. The program was designed to provide the recommended 3 doses of the vaccine; however, some of the participants received less than 3 doses because of absenteeism from school by these girls.34 These serum samples were collected more than 24 mo after the receipt of the last dose of the vaccine.
In the Uganda Immunogenicity Study,Citation36 our laboratory performed the anti-HPV16 and HPV18 ELISAs, and 293TT cell-based neutralization/SEAP assays on randomly selected subset of the samples from each vaccine dose group. This enabled us to examine the titers obtained from the ELISAs correlated with those from the SEAP assays, as we have done in our previous study.Citation28 In the current study, these results were used to evaluate 72 samples, and the study protocol was approved by the National Cancer Institute Special Studies Institutional Review Board and by PATH Research Ethics Committee, Seattle, Washington, USA.
Neutralization assay and SEAP assay
For the neutralization assay, 0.03 × 106 293TT cells were plated and incubated for a minimum of 2 hr at 37˚C. Four-fold serial dilutions of serum samples were performed starting from 1:10 to 1:2,621,440, and incubated with PsV16 (1:350,000 dilution) or PsV18 (1:40,000 dilution) for 1 hr at 4°C in a final volume of 125 μL. After the incubation, 100 μL of the serum sample/PsV mixture was transferred to the pre-plated 293TT cells, and incubated for 72 hr at 37°C. The cell-culture supernatants were harvested and stored at −80°C until SEAP assays were performed.
The level of SEAP in 293TT cell-culture supernatants was measured using Great EscAPe™ SEAP Chemiluminescence Kit 2.0 (GE) from Clontech (631738) and ZiVa® Ultra-Sensitive SEAP Detection Kit (ZiVa) from Jaden Bioscience, Inc. (CM025). The two assays were conducted simultaneously to test the same samples using white OptiPlate-96 plates (6005290; Perkin-Elmer). For GE, 25 μL of supernatant was incubated with 75 μL of 1X Dilution Buffer, and incubated at 65°C in an oven (Heratherm OMH100, Thermo Scientific) for 45 min.Citation27,28 After equilibrating the plates to room temperature, 100 μL of substrate was added and incubated in dark at room temperature for 20 min before measuring SEAP activity. For ZiVa, 5 μL of supernatant samples were incubated with 75 μL of SEAP Sample Preparation Solution at 65°C in the oven for 45 min. After equilibrating the plates to room temperature, 25 μL of substrate was added and incubated in the dark at room temperature for 30 min. For both kits, the activity of SEAP was measured using a plate reader (SpectraMax M5; Molecular Devices) using 0.2 s integration time. The neutralization titers were calculated by linear interpolation, and defined as the reciprocal of the dilution that showed 50% reduction in secreted alkaline phosphatase activity compared to control wells (virus-only without serum sample). Neutralization titers below the starting dilution of 1:10 were arbitrarily assigned a value of 5.
Statistical analysis
Spearman rank correlation was performed to compare anti-HPV titers in serum samples of Cervarix®-immunized individuals. GraphPad Prism 4 (GraphPad Software) was used for both analyses. Coefficient of variation (CV) for duplicate wells were calculated as follows. The replicate titer values were log transformed, and then Proc varcomp (SAS Institute, Inc.) was used to estimate the variance component (repeat measures). CVs for the duplicate wells for anti-HPV16 and -HPV18 were 24.9% and 21.9%, respectively, for GE, and were 33.0% and 29.2% for ZiVa, which were comparable to previous findings using GE.Citation29
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Supplemental Material
Supplemental data for this article can be accessed on the publisher's website.
Author Contributions
All authors have reviewed the data and read the manuscript. LAP was responsible for the oversight of the study. TJK and GS designed and performed the experiments. MS is a co-PI of the Uganda Immunogenicity Study and have provided us with access to vaccinated samples. KM and LAP formulated the manuscript. TJK, KM, and LAP wrote the manuscript. TJK and KM contributed equally to this work.
Supplementary_Tables.docx
Download MS Word (28.6 KB)Acknowledgments
We would like to extend our thanks and appreciations to PATH for making it possible for us to test the supernatant samples from the Uganda Immunogenicity Study. We would also like to thank Dr. D. Scott LaMontagne, principal investigator for the Uganda Immunogenicity Study, for his suggestions and comments on the manuscript. We would also like to thank Yuanji Pan for his technical advice on pseudovirion production.
Funding
This project has been funded in whole or in part with federal funds from the National Cancer Institute, National Institutes of Health, under Contract No. HHSN261200800001E. The content of this publication does not necessarily reflect the views or policies of the Department of Health and Human Services, nor does mention of trade names, commercial products, or organizations imply endorsement by the US. Government.
References
- Cates W, Jr. Estimates of the incidence and prevalence of sexually transmitted diseases in the United States. american social health association panel. Sex Transm Dis 1999; 26:S2-7; PMID:10227693; http://dx.doi.org/10.1097/00007435-199904001-00002
- Bernard HU, Burk RD, Chen Z, van Doorslaer K, zur Hausen H, de Villiers EM. Classification of papillomaviruses (PVs) based on 189 PV types and proposal of taxonomic amendments. Virology 2010; 401:70-9; PMID:20206957; http://dx.doi.org/10.1016/j.virol.2010.02.002
- Einstein MH, Schiller JT, Viscidi RP, Strickler HD, Coursaget P, Tan T, Halsey N, Jenkins D. Clinician's guide to human papillomavirus immunology: knowns and unknowns. Lancet Infect Dis 2009; 9:347-56; PMID:19467474; http://dx.doi.org/10.1016/S1473-3099(09)70108-2
- zur Hausen H. Papillomaviruses in the causation of human cancers - a brief historical account. Virology 2009; 384:260-5; PMID:19135222; http://dx.doi.org/10.1016/j.virol.2008.11.046
- Bosch FX, de Sanjose S. Chapter 1: human papillomavirus and cervical cancer–burden and assessment of causality. J Natl Cancer Inst Monogr 2003:3-13; PMID:12807939; http://dx.doi.org/10.1093/oxfordjournals.jncimonographs.a003479
- Castellsague X, Diaz M, de Sanjose S, Munoz N, Herrero R, Franceschi S, Peeling RW, Ashley R, Smith JS, Snijders PJ, et al. Worldwide human papillomavirus etiology of cervical adenocarcinoma and its cofactors: implications for screening and prevention. J Natl Cancer Inst 2006; 98:303-15; PMID:16507827; http://dx.doi.org/10.1093/jnci/djj067
- Munoz N, Bosch FX, de Sanjose S, Herrero R, Castellsague X, Shah KV, Snijders PJ, Meijer CJ. Epidemiologic classification of human papillomavirus types associated with cervical cancer. N Engl J Med 2003; 348:518-27; PMID:12571259; http://dx.doi.org/10.1056/NEJMoa021641
- Roden R, Wu TC. How will HPV vaccines affect cervical cancer? Nat Rev Cancer 2006; 6:753-63; PMID:16990853; http://dx.doi.org/10.1038/nrc1973
- Gardasil® ; [package insert]. United states food and drug administration. Initial approval 2006. Revised 03/2014. http://www.fda.gov/downloads/BiologicsBloodVaccines/Vaccines/ApprovedProducts/UCM111263.pdf. Accessed June 2014
- Cervarix® ; [package insert]. United states food and drug administration. Initial approval 2009. http://www.fda.gov/downloads/BiologicsBloodVaccines/Vaccines/ApprovedProducts/UCM186981.pdf. Accessed June 2014
- Schiller JT, Castellsague X, Garland SM. A review of clinical trials of human papillomavirus prophylactic vaccines. Vaccine 2012; 30 5:F123-38; PMID:23199956; http://dx.doi.org/10.1016/j.vaccine.2012.04.108
- Pastrana DV, Gambhira R, Buck CB, Pang YY, Thompson CD, Culp TD, Christensen ND, Lowy DR, Schiller JT, Roden RB. Cross-neutralization of cutaneous and mucosal Papillomavirus types with anti-sera to the amino terminus of L2. Virology 2005; 337:365-72; PMID:15885736; http://dx.doi.org/10.1016/j.virol.2005.04.011
- Gambhira R, Karanam B, Jagu S, Roberts JN, Buck CB, Bossis I, Alphs H, Culp T, Christensen ND, Roden RB. A protective and broadly cross-neutralizing epitope of human papillomavirus L2. J Virol 2007; 81:13927-31; PMID:17928339; http://dx.doi.org/10.1128/JVI.00936-07
- Jagu S, Karanam B, Gambhira R, Chivukula SV, Chaganti RJ, Lowy DR, Schiller JT, Roden RB. Concatenated multitype L2 fusion proteins as candidate prophylactic pan-human papillomavirus vaccines. J Natl Cancer Inst 2009; 101:782-92; PMID:19470949; http://dx.doi.org/10.1093/jnci/djp106
- Jagu S, Kwak K, Karanam B, Huh WK, Damotharan V, Chivukula SV, Roden RB. Optimization of multimeric human papillomavirus L2 vaccines. PLoS One 2013; 8:e55538; PMID:23383218; http://dx.doi.org/10.1371/journal.pone.0055538
- Bousarghin L, Combita-Rojas AL, Touze A, El Mehdaoui S, Sizaret PY, Bravo MM, Coursaget P. Detection of neutralizing antibodies against human papillomaviruses (HPV) by inhibition of gene transfer mediated by HPV pseudovirions. J Clin Microbiol 2002; 40:926-32; PMID:11880418; http://dx.doi.org/10.1128/JCM.40.3.926-932.2002
- Touze A, Coursaget P. In vitro gene transfer using human papillomavirus-like particles. Nucleic Acids Res 1998; 26:1317-23; PMID:9469843; http://dx.doi.org/10.1093/nar/26.5.1317
- Unckell F, Streeck RE, Sapp M. Generation and neutralization of pseudovirions of human papillomavirus type 33. J Virol 1997; 71:2934-9; PMID:9060652
- Stauffer Y, Raj K, Masternak K, Beard P. Infectious human papillomavirus type 18 pseudovirions. J Mol Biol 1998; 283:529-36; PMID:9784363; http://dx.doi.org/10.1006/jmbi.1998.2113
- Rossi JL, Gissmann L, Jansen K, Muller M. Assembly of human papillomavirus type 16 pseudovirions in saccharomyces cerevisiae. Hum Gene Ther 2000; 11:1165-76; PMID:10834618; http://dx.doi.org/10.1089/10430340050015211
- Kawana K, Yoshikawa H, Taketani Y, Yoshiike K, Kanda T. In vitro construction of pseudovirions of human papillomavirus type 16: incorporation of plasmid DNA into reassembled L1/L2 capsids. J Virol 1998; 72:10298-300; PMID:9811779
- Roden RB, Greenstone HL, Kirnbauer R, Booy FP, Jessie J, Lowy DR, Schiller JT. In vitro generation and type-specific neutralization of a human papillomavirus type 16 virion pseudotype. J Virol 1996; 70:5875-83; PMID:8709207
- White WI, Wilson SD, Bonnez W, Rose RC, Koenig S, Suzich JA. In vitro infection and type-restricted antibody-mediated neutralization of authentic human papillomavirus type 16. J Virol 1998; 72:959-64; PMID:9444988
- Smith LH, Foster C, Hitchcock ME, Leiserowitz GS, Hall K, Isseroff R, Christensen ND, Kreider JW. Titration of HPV-11 infectivity and antibody neutralization can be measured in vitro. J Invest Dermatol 1995; 105:438-44; PMID:7665926; http://dx.doi.org/10.1111/1523-1747.ep12321173
- Christensen ND, Kreider JW, Shah KV, Rando RF. Detection of human serum antibodies that neutralize infectious human papillomavirus type 11 virions. J Gen Virol 1992; 73 (Pt 5):1261-7; PMID:1316943; http://dx.doi.org/10.1099/0022-1317-73-5-1261
- Kemp TJ, Hildesheim A, Safaeian M, Dauner JG, Pan Y, Porras C, Schiller JT, Lowy DR, Herrero R, Pinto LA. HPV16/18 L1 VLP vaccine induces cross-neutralizing antibodies that may mediate cross-protection. Vaccine 2011; 29:2011-4; PMID:21241731; http://dx.doi.org/10.1016/j.vaccine.2011.01.001
- Kemp TJ, Safaeian M, Hildesheim A, Pan Y, Penrose KJ, Porras C, Schiller JT, Lowy DR, Herrero R, Pinto LA. Kinetic and HPV infection effects on cross-type neutralizing antibody and avidity responses induced by cervarix((R)). Vaccine 2012; 31:165-70; PMID:23123024; http://dx.doi.org/10.1016/j.vaccine.2012.10.067
- Safaeian M, Porras C, Pan Y, Kreimer A, Schiller JT, Gonzalez P, Lowy DR, Wacholder S, Schiffman M, Rodriguez AC, et al. Durable antibody responses following one dose of the bivalent human papillomavirus L1 virus-like particle vaccine in the costa rica vaccine trial. Cancer Prev Res (Phila) 2013; 6:1242-50; PMID:24189371; http://dx.doi.org/10.1158/1940-6207.CAPR-13-0203
- Kemp TJ, Garcia-Pineres A, Falk RT, Poncelet S, Dessy F, Giannini SL, Rodriguez AC, Porras C, Herrero R, Hildesheim A, et al. Evaluation of systemic and mucosal anti-HPV16 and anti-HPV18 antibody responses from vaccinated women. Vaccine 2008; 26:3608-16; PMID:18541349; http://dx.doi.org/10.1016/j.vaccine.2008.04.074
- Pastrana DV, Buck CB, Pang YY, Thompson CD, Castle PE, FitzGerald PC, Kruger Kjaer S, Lowy DR, Schiller JT. Reactivity of human sera in a sensitive, high-throughput pseudovirus-based papillomavirus neutralization assay for HPV16 and HPV18. Virology 2004; 321:205-16; PMID:15051381; http://dx.doi.org/10.1016/j.virol.2003.12.027
- Buck CB, Pastrana DV, Lowy DR, Schiller JT. Generation of HPV pseudovirions using transfection and their use in neutralization assays. Methods Mol Med 2005; 119:445-62; PMID:16350417
- Buck CB, Pastrana DV, Lowy DR, Schiller JT. Efficient intracellular assembly of papillomaviral vectors. J Virol 2004; 78:751-7; PMID:14694107; http://dx.doi.org/10.1128/JVI.78.2.751-757.2004
- Sehr P, Rubio I, Seitz H, Putzker K, Ribeiro-Muller L, Pawlita M, Muller M. High-throughput pseudovirion-based neutralization assay for analysis of natural and vaccine-induced antibodies against human papillomaviruses. PLoS One 2013; 8:e75677; PMID:24124504; http://dx.doi.org/10.1371/journal.pone.0075677
- LaMontagne DS, Barge S, Le NT, Mugisha E, Penny ME, Gandhi S, Janmohamed A, Kumakech E, Mosqueira NR, Nguyen NQ, et al. Human papillomavirus vaccine delivery strategies that achieved high coverage in low- and middle-income countries. Bull World Health Organ 2011; 89:821-30B; PMID:22084528; http://dx.doi.org/10.2471/BLT.11.08986
- Safaeian M, Kemp TJ, Pan DY, Porras C, Rodriguez AC, Schiffman M, Cortes B, Katki H, Wacholder S, Schiller JT, et al. Cross-protective vaccine efficacy of the bivalent HPV vaccine against HPV31 is associated with humoral immune responses: results from the costa rica vaccine trial. Hum Vaccin Immunother 2013; 9:1399-406; PMID:23571174; http://dx.doi.org/10.4161/hv.24340
- LaMontagne DS, Mugisha E, Pan Y, Kumakech E, Ssemaganda A, Kemp TJ, Cover J, Pinto LA, Safaeian M. Immunogenicity of bivalent HPV vaccine among partially vaccinated young adolescent girls in uganda. Vaccine 2014; PMID:25218297; doi: 10.1016/j.vaccine.2014.08.071
- Day PM, Pang YY, Kines RC, Thompson CD, Lowy DR, Schiller JT. A human papillomavirus (HPV) in vitro neutralization assay that recapitulates the in vitro process of infection provides a sensitive measure of HPV L2 infection-inhibiting antibodies. Clin Vaccine Immunol 2012; 19:1075-82; PMID:22593236; http://dx.doi.org/10.1128/CVI.00139-12
- Wu WH, Gersch E, Kwak K, Jagu S, Karanam B, Huh WK, Garcea RL, Roden RB. Capsomer vaccines protect mice from vaginal challenge with human papillomavirus. PLoS One 2011; 6:e27141; PMID:22069498; http://dx.doi.org/10.1371/journal.pone.0027141
- Buck CB, Thompson CD, Pang YY, Lowy DR, Schiller JT. Maturation of papillomavirus capsids. J Virol 2005; 79:2839-46; PMID:15709003; http://dx.doi.org/10.1128/JVI.79.5.2839-2846.2005
- Dessy FJ, Giannini SL, Bougelet CA, Kemp TJ, David MP, Poncelet SM, Pinto LA, Wettendorff MA. Correlation between direct ELISA, single epitope-based inhibition ELISA and pseudovirion-based neutralization assay for measuring anti-HPV-16 and anti-HPV-18 antibody response after vaccination with the AS04-adjuvanted HPV-16/18 cervical cancer vaccine. Hum Vaccin 2008; 4:425-34; PMID:18948732; http://dx.doi.org/10.4161/hv.4.6.6912
