Abstract
To better understand the role of radial glial (RG) cells in the evolution of the mammalian cerebral cortex, we investigated the role of RG cells in the dorsal cortex and dorsal ventricular ridge of the turtle, Trachemys scripta elegans. Unlike mammals, the glial architecture of adult reptile consists mainly of ependymoradial glia, which share features with mammalian RG cells, and which may contribute to neurogenesis that continues throughout the lifespan of the turtle. To evaluate the morphology and proliferative capacity of ependymoradial glia (here referred to as RG cells) in the dorsal cortex of embryonic and adult turtle, we adapted the cortical electroporation technique, commonly used in rodents, to the turtle telencephalon. Here, we demonstrate the morphological and functional characteristics of RG cells in the developing turtle dorsal cortex. We show that cell division occurs both at the ventricle and away from the ventricle, that RG cells undergo division at the ventricle during neurogenic stages of development, and that mitotic Tbr2+ precursor cells, a hallmark of the mammalian SVZ, are present in the turtle cortex. In the adult turtle, we show that RG cells encompass a morphologically heterogeneous population, particularly in the subpallium where proliferation is most prevalent. One RG subtype is similar to RG cells in the developing mammalian cortex, while 2 other RG subtypes appear to be distinct from those seen in mammal. We propose that the different subtypes of RG cells in the adult turtle perform distinct functions.
Abbreviations
| RG | = | radial glia |
| VZ | = | ventricular zone |
| SVC | = | subventricular zone |
| DVR | = | ventricular ridge |
| EGFP | = | enhanced green fluorescent protein |
Introduction
The turtle is considered the most closely related living animal to the ‘stem’ amniote that was the common ancestor of mammals and reptiles.Citation1 The adult cortex in turtle has a simple tri-laminar organization. The outer molecular layer contains thalamic and brainstem afferents and some inhibitory interneurons. The pyramidal cell layer consists mostly of excitatory pyramidal neurons. The subcellular layer, which is adjacent to the lateral ventricle, contains interneurons and some displaced pyramidal cells.Citation2,3 Glial cells are also present in the subcellular layer, and occupy the same approximate position as RG cells in the ventricular zone (VZ) of the developing mammalian neocortex. The turtle glial cells express GFAP, as do RG cells in prenatal primate,Citation4,61 and extend processes through the overlying layers, following a radial trajectory that is reminiscent of the organization of the embryonic mammalian cortex. However, free astrocytes are not present in the turtle cortex as in mammals.
The VZ of the developing mammalian telencephalon contains primary precursor cells that generate most of the cells that constitute the adult telencephalon. Descriptions of neuroepithelial cells, as well as the names used to describe these cells, have evolved over the last century (for review see ref.Citation5). Initially, 2 separate classes of cells were assumed to exist: neural precursor cells and neuroepithelial cells – now called RG cells. Neural precursor cells in the developing telencephalon were identified over one century ago by their round morphology as they divided at the ventricular surface.Citation6 In contrast, RG cells were defined by their characteristic bipolar morphology including a process contacting the ventricular surface, an oval nucleus in the VZ or subventricular zone (SVZ), and a long pial fiber extending to the pial limitans. RG cells were also identified by abundant glycogen granules in the pial endfeet.Citation7 Seminal work showed that RG cells guide neuronal migration,Citation8 undergo mitotis,Citation9 and that RG division coincides with sites of neurogenesis.Citation10 Conclusive evidence demonstrated that RG cells produce neurons.Citation5,Citation11-16 The evidence demonstrating that RG cells are neuronal precursor cells was made possible by new technology, specifically fluorescent protein-expressing retroviruses combined with time-lapse analysis in live slice cultures.Citation13 Previously established techniques including Golgi impregnation, thymidine analogs, electron microscopy, and even retroviruses expressing a non-fluorescent tag, did not pinpoint the neurogenic activity of mammalian RG cells, although it provided evidence that some clones contained both RG cells and neurons in the chick optic tectum.Citation17 Since RG cells were identified as neural precursor cells in rodents, the identity of neural precursor cells in other vertebrate species has followed, and inferences that RG cells are neurogenic in all vertebrates rests on conclusive evidence from rodent, ferret, non-human primate, and human.
There are reports of extensive postnatal and adult neurogenesis deriving from the ventricular wall of non-mammalian species both constitutively and in injured animals including fish,Citation18-20 birdsCitation10,21 and reptiles.Citation22,23 These studies have begun to integrate the discovery that mammalian embryonic RG cells generate neurons. Ependymal cells with long radial processes persist throughout the lifespan of some non-mammalian species including fish,Citation24,25,26 birds,Citation27 amphibia,Citation28 and reptiles.Citation25,Citation29-36 In adult birds, ‘RG cells’ coincide spatially with regions of high proliferative activity referred to as “hotspots," an observation that suggested these avian RG cells generate neurons.Citation10 In a few reptiles, including lizards and turtles, VZ cells with long radial processes throughout the telencephalon are not superseded by free astrocytes. These radially oriented glial cells have been referred as tanycytes,Citation37 RG cells,Citation38 surface contact glial cells,Citation39 and radial ependymoglial cells.Citation34,35,40,41 In birds and lizard, they have been subclassified by electron microscopy as types “B” (RG cells) and “E” (ependymal) based on the features of their cilia and mitotic abilities.Citation42
The presence of RG-like cells in the telencephalon of adult vertebrates that exhibit continual growth throughout life prompted us to ask whether RG cells produce daughter cells during development and adulthood also in select vertebrates. In turtle, as in other reptiles, postnatal neurogenesis that derives largely from cells located in the ventricular wall of the striatum and dorsal ventricular ridge (DVR) has been reported.Citation43 We hypothesized that RG cells are neuronal precursors in the embryonic and adult turtle. To date, the most detailed morphologic descriptions of adult turtle RG cells have been performed by Golgi impregnation and are primarily restricted to the dorsal cortex.Citation44–46 In the adult cortex of turtles, a layer of roughly cuboidal cell bodies, similar to ependymal cells, forms a limiting epithelium at the ventricular surface. RG cell processes are coated by innumerable lamellate excrescences to the extent that it is difficult to see the main fiber.Citation47 These cells have been referred to variously as ‘lamellate ependymal cells’, ‘ependymoglia cells’, and ‘RG cells’.Citation34,35,40 It has been speculated that RG cells are neuronal precursor cells in fish, lizard, and turtle.Citation48–50 However, it is not clear that adult cells referred to as RG cells are derived from, or share features with, embryonic RG cells. The dorsal cortex of the adult turtle shows low levels of constitutive proliferation, and glia with radial morphologies are found throughout the turtle central nervous system including regions that are not proliferative. RG cells in the adult turtle that have been described in previous studies resemble embryonic RG cells at a basic level, but appear to be much more differentiated. The radial organization of the adult turtle cortex may facilitate the migration of newly generated neurons to the pyramidal cell layer, but the identity of the precursor cells that produce neurons in the adult turtle remains in question.
To evaluate the morphology and proliferative activity of cells in the embryonic and adult VZ, we adapted in utero electroporation of plasmidsCitation51 for use on the turtle telencephalon. We show that VZ cells are proliferative in the embryonic turtle and express markers that label RG cells. We also find a second proliferative cell population consisting of Tbr2-expressing cells. In the adult turtle, we describe a morphologically heterogeneous population of cells in the VZ, particularly in the striatum and DVR where proliferation is most active. Our results suggest that while turtle embryonic RG cells resemble those in developing mammalian cortex, RG like cells in the adult turtle telencephalon encompass a heterogeneous population. We describe 3 subclasses of adult RG cells and speculate that undifferentiated RG cells with simple radial processes likely represent the population of proliferative cells, while the more differentiated populations serve functions that are performed by astrocytes in the adult mammalian cortex.
Materials and Methods
Animals
After obtaining a license from the Louisiana Department of Agriculture and Forestry, turtles and eggs were purchased from Harvey Kliebert's Reptile Farm in Louisiana. All experimental protocols were approved and in accord with IACUC regulations and guidelines. Upon arrival, turtle eggs were incubated at 30°C in Hova-Bator thermal incubators with circulating fans. For histological analysis, embryos were removed from the egg shell and rapidly decapitated. Brains were removed and fixed in 4% paraformaldehyde in 0.1 mM PBS, (4°C, pH 7.4) overnight, then stored in PBS until histologic processing. Embryos were staged according to Yntema.Citation52,53 The development of the telencephalic vesicle in stage 16–17 turtle embryos is comparable to that of mouse at E11.5, with postmitotic neurons first detected in a subpial position in the pallium. By stage 19, the DVR begins to protrude into the lateral ventricle, and by stage 23 the rapid expansion of the telencephalon slows.Citation54,55 We used turtle embryos from stages 15–23, which includes the majority of the corticogenic period. Turtle eggs were incubated for no more than one week before use. For electroporation procedures, juvenile/adult turtles were anaesthetized by chilling on ice for 15 minutes, before the brain was removed. For immunohistochemistry in fixed tissue, turtles were anaesthetized with ketamine (90 mg/kg) and xylazine (10 mg/kg, Henry Schein, Port Washington) before transcardial perfusion with phosphate buffered saline (PBS) and 4% paraformaldehyde. The majority of animals used had carapace length of 1–2 inches, which correlates roughly to 1 y of age. Three animals with 4–5 inch carapaces were also used.
Immunohistochemistry
For Nissl analysis, cryostat sections (20–30 microns) were prepared after cryoprotection in 20% sucrose in PBS and embedding in Tissue-Tek (Sakura, Torrance, CA) before freezing. For immunohistochemistry, fixed coronal slices were cut at 100–200 μm on a Vibratome (Ted Pella). For immunohistochemical analysis, sections were rinsed in PBS for 30 min, blocking for 1 hr (10% normal goat or horse serum as appropriate, 0.1% Triton X-100, and 0.2% gelatin in PBS), washed for 10 min in PBS, and incubated overnight at 4°C in primary antibody (dilutions as described below in 2% normal goat or horse serum as appropriate, 0.1% Triton X-100, and 0.04% gelatin in PBS). The next day sections were washed 3 times for 20 min in PBS, and incubated in secondary antibody for 1–2 hr (dilutions as described below in 2% normal goat or horse serum as appropriate, 0.1% Triton X-100, and 0.04% gelatin in PBS). Primary antibodies were obtained from the following sources and used at the indicated dilutions: fluorescein-conjugated rat monoclonal anti-BrdU (Accurate Chemical; 1:10), Cy2 or Cy3-conjugated monoclonal anti-BrdU (Molecular Probes; 1:10), mouse monoclonal 4A4 (Abcam; 1:1000), mouse anti-PCNA (Millipore, 1:200), rabbit anti-Tbr2 (Abcam, 1:500) and mouse monoclonal anti-vimentin (Sigma, 1:40). Secondary antibodies were obtained from the following source and used at the indicated dilution: Cy2, Cy3, or Cy5 conjugated goat anti-mouse (Jackson ImmunoResearch, West Grove, PA; 1:50). For BrdU immunohistochemistry, 100 μm sections were pretreated with 2 N HCl in PBS for 30 min at 37°C before blocking, and a fluorescently tagged primary antibody (see above). Fluorescent labeling of chromatin with Syto-11 (Molecular Probes, Eugene, OR) was performed by incubation of sections with 5 μM Syto-11 and 0.1% Triton X-100 in PBS for 30–45 min followed by washing 3 times for 10 min in PBS.
BrdU labeling
Two protocols of 5-bromo-2-deoxyuridine (BrdU) pulsing immunohistochemistry were used, in ovo and ex ovo. For in ovo BrdU application, an injection of BrdU (Sigma, St. Louis, MO) (25 mg/kg in 0.9% NaCl) was administered to incubated eggs 3 hrs before they were opened. After removing the embryo, the brain was dissected and incubated in 200 ml of oxygenated PBS for 1.5 hours to allow for washout. For ex ovo BrdU application in electroporated embryos, whole embryos in culture were incubated for 8 hours in medium containing 5 mg/ml BrdU. Medium without BrdU was then substituted for 5 hours for washout.
Electroporation
Electroporation of an EGFP-expressing plasmid was employed in both embryos and adults. The plasmid (pEGFP-N1, Clontech, Palo Alto, CA) contains the gene for EGFP under the control the CMV IE promoter/enhancer element. Plasmids were diluted to working concentrations of 2.0–5.0 μg/μL in PBS colored with India ink. Voltage was set at 50 V, duration, frequency, and total number of pulses was maintained at 5 pulses of 50 msec each with a 1-sec gap between pulses using a square-wave pulse generator (BTX ECM 830, San Diego). Ten mm diameter Tweezertrodes were used in both embryos and adults (BTX, San Diego).
Embryonic Electroporation (): For culturing embryonic brain slices, eggs were rinsed several times in EtOH, scrubbed with iodine prep pads, and bathed in UV light for ∼30 minutes. Embryos were then removed from the shell and rinsed momentarily in EtOH before transfer to PBS. Thorough cleaning was critical for successful culturing, as was a separate set of dissection tools used to handle sterilized embryos. Electroporations were performed ex ovo in the whole embryo, resting in PBS. DNA was injected into the lateral ventricle and/or adjoining ventricle of the large optic tecta. After several minutes, pulses were delivered with the anode/cathode oriented ventral/dorsal, respectively. This approach labeled cells in the dorsal cortex. After an additional incubation of 10 minutes, embryos were either cultured whole, or slice-cultured (see below). Expression of EGFP was visible 8 hours later.
Figure 1. Individual RG cells were labeled using electroporation. (A) In embryonic animals, ex ovo electroporation was used. Subsequently, brains were either cultured as coronally sectioned slices or cultured in the whole embryo. Electrodes were applied following a dorso-ventral orientation to label cells in the dorsal cortex. (B) In adult animals, the brain was removed, hemisected and embedded in agarose. Electrode positioning following a medio-lateral orientation to label cells in the DVR and striatum. After electroporation, brains were cut into slices and organotypic slice cultures prepared.
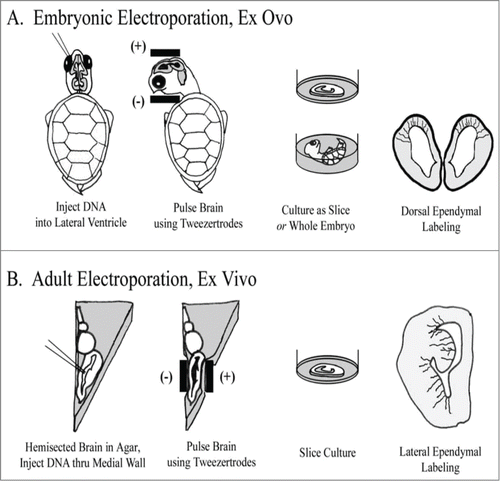
Adult Electroporation (): The adult forebrain was removed, hemisected, and embedded in low melt temperature agarose (Sigma, St. Louis, MO; 1.5% agarose in PBS, pH 7.4). Embedding in agarose served to support the brain and prevent leakage of the plasmid from the brain ex vivo. After chilling on ice, an agarose block containing the forebrain was cut out, roughly in the shape of a triangle, taking care to trim closely on the sides for electrode contact. Plasmid was injected into the lateral ventricle through the medial wall. The anode was positioned at the lateral wall, and the electrode at the medial wall, so that the vector was aimed primarily into the DVR (). A few minutes was allowed for the plasmid to spread. The embedded brain was then incubated in oxygenated ACSF for approximately 15 minutes before being sectioned coronally and transferred to slice culture. The first expression of EGFP was visible after 8 hours, with brightness increasing considerably thereafter.
Cell culture
Slice Culture (embryonic and adult): Brains were embedded in low-melt agarose (Fisher Biotech, Fair Lawn, NJ) and cut coronally at 300–400 μm using a Leica VT100S vibrating blade microtome (Nussloch, Germany). Slices were placed temporarily in PBS bubbled with 95% CO2 and 5% O2. Coronal slices were incubated at 30°C in a humidified 5% CO2 incubator, on Millicell-CM culture inserts (Millipore, Bedford) in 6-well culture trays (Corning, Inc., Corning) with medium containing 66% BME, 25% Hanks, 5% FBS, 1% Pen/Strep/Glutamine (each from Gibco) and 0.66% d-(+)-glucose (Sigma). Cultures were removed briefly for time-lapse imaging.
Whole Embryo Culture: Whole embryo cultures were established because slice cultures, which successfully expressed the EGFP reporter, showed minimal migration and mitoses. Briefly, whole embryos were placed in 6-well culture trays in pre-warmed medium. The fluid level was adjusted to nearly submerge the embryo. The wells were sealed with parafilm to allow for gas exchange while preserving moisture. Embryos were incubated at 30°C in a humidified 5% CO2 incubator, and left for up to 5 d. In general, cultures seemed healthy for up to 3 d before quality began to deteriorate.
Confocal imaging
All imaging was performed on an inverted Olympus Fluoview laser-scanning confocal microscope. Excitation/emission wavelengths used were 488/515 nm (EGFP, Cy2), 568/590 nm (Cy3), and 685/690 nm (Cy5). Z-series images were collected at 0.6-3 μm steps on a PC running Fluoview v3.3 software (Olympus). Images were contrast enhanced and assembled into montages.
Results
Radial glial cells are neurogenic in the embryonic turtle telencephalon
We labeled individual ependymal cells in the developing turtle telencephalon. We used ex ovo electroporation of the turtle brain with a plasmid containing the gene for EGFP under the control of the CMV promoter. Our protocol was similar to that used in utero to label cells in the rodent dorsal telencephalon.Citation51,56,57 In some cases we rotated the electrodes to target medio-ventral areas of the turtle telencephalon such as the DVR (). After electroporation, brains were either cultured as slices or as intact embryos. Slices were generally cultured for up to one week, but in several cases we incubated slice cultures for up to 4 weeks. We performed time-lapse in slice culture. We also used whole embryonic cultures. Electroporated areas in whole embryo cultures ranging from stage 17 to stage 23 could be maintained in culture for 3–5 d.
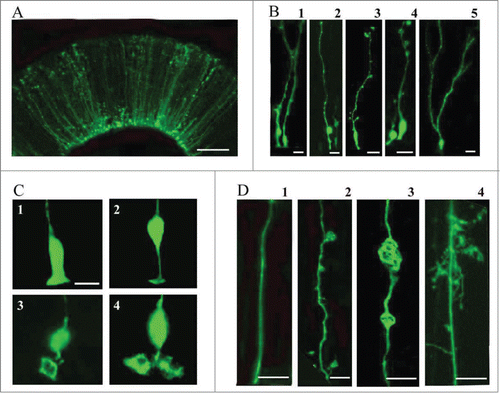
Electroporation of the embryonic turtle telencephalon with an EGFP-expressing plasmid labeled many cells with a mammalian RG-like morphology (, stage 20)Citation57. We refer to these cells as RG cells. Most of the EGFP+ cells were bipolar, maintaining one process contacting the ventricle and a pial process extending toward the pial surface (). RG cells had cell bodies both at and away from the ventricular surface (B1 and B2). There were often small expansions or processes that extended from the main shaft of the pial process (B3 and B4). In more developed embryos (stage 23), RG cells often exhibited pial processes with bifurcated and ‘hairy’ or lamellate fibers (B5). The ventricular contacting process of embryonic RG cells showed heterogeneity (). Some ventricular contacting processes were similar to classical descriptions of mammalian embryonic RG cells (C1 and C2), while others were large and complex (C3 and C4), enough so that they could be mistaken for additional cell bodies at lower magnification. Electroporation labeling of VZ cells demonstrated that pial fibers exhibited heterogeneous morphology (). Smooth processes, as commonly seen in embryonic rodents, were more prevalent in younger animals where most VZ cells exhibited a simple bipolar morphology (D1). In addition, we observed RG cells that showed expansions budding off of the pial process (D2), or within the process (D3). In some instances, the process expansions were the size of cell bodies and could be mistakenly interpreted as newborn neurons migrating along the RG fiber (D3). Finally, pial processes with complex ‘lamellate’ extensions became more common in more mature embryos (D4).
Figure 2. Electroporation of the embryonic turtle telencephalon with an EGFP-expressing plasmid labels many cells with RG morphology. (A) At 24 hours post-electroporation of a stage 20 embryo, an array of EGFP+ cells in the dorsal cortex show RG morphology. (B) At higher magnification a variety of morphologies are seen. RG cells in different phases of the cell cycle are apparent (B1, B2). Small expansions are often present along the length of the pial process (B3, B4). A smaller number of EGFP+ cells in the VZ do not appear to have a pial process. These may be newly generated neurons, intermediate progenitor cells, or RG cells. At later stages (stage 23), cells are found with bifurcated or branched pial processes, and ‘hairy’ or more lamellate fibers (B5). (C) A variety of ventricular contacting processes are seen. Some processes are typical for embryonic RG cells seen in rodents (C1, C2) while others are more complex (C3, C4). (D) RG cells exhibit different heterogeneous morphologies. D1, RG cells in younger animals often have a smooth pial process. In many cases expansions bud off of the process (D2) or in line with the process (D3). As development proceeds RG cell pial fibers exhibit more mature ‘lamellate’ fibers (D4). Scale bars: A, 50 μm; B, 10 μm; C,D, 10 μm.
We examined the activity of electroporated cells in slice cultures and whole embryo cultures through time-lapse observation and/or pulse labeling with BrdU. In slice culture, we imaged labeled cells every 2–3 hours for up to 5 d. The example in is a representative example of electroporation labeled cells after 12 hours and 36 hours in slice culture. We observed cell movement in slice culture. Electroporated cells often exhibited RG morphology with a process contacting the ventricle at early timepoints (A1). However, at later time points the RG cell bodies had moved away from the ventricular surface either through migration or perhaps translocation. Translocation of proliferative RG cells out of the VZ has been shown in time-lapse experiments in rat,Citation15,16 and more recently in human,Citation58 and mouse.Citation59 In some cases the turtle RG cells that moved away from the ventricle still maintained contact with the ventricle (A2). When we incubated slices for up to 4 weeks, EGFP+ cells with mature neuronal morphologies appeared in the pyramidal cell layer ( and ). shows a cell with neuronal morphology located superficial to the proliferating ependymal cells in the VZ. It is possible that over weeks, labeled cells underwent division in the slice culture but we did not capture the mitoses. The appearance of neurons in the pyramidal cell layer is consistent with this idea.
Figure 3. RG cells migrate, exhibit cellular movements, and proliferate in organotypic slice cultures and in whole embryo cultures. (A) RG cells in the embryonic turtle telencephalon labeled through electroporation. Shown are 2 time points, 12 hours and 36 hours. Electroporated cells were adjacent to the ventricle at early time points (A1), but at later time points, some RG cell bodies had translocated away from the ventricular surface (A2). (B and C) Slice cultures allowed to incubate for up to 4 weeks contained cells with mature neuronal morphologies. In C, the neuron (arrow) is located just superficial to the proliferating area of ependymal cells (arrowhead). (D and E) Representative examples of dorsal cortex that were incubated in whole embryos cultures for 12 hours (D) and 3 d (E). At 12 hours, RG cell bodies were close to the ventricle. At 72 hours, many electroporated cells had migrated or translocated into the cortex (arrows in E). (F) Whole embryos cultured with a pulse of BrdU. Left panel shows a single electroporated RG cell (green). The second panel is a higher power image of the same cell. BrdU staining revealed many mitotic cells (red), including GFP+ RG cells that were BrdU+ (green, arrowheads). The right panel is a negative control BrdU staining. DAPI stain (blue) labels all nuclei. Scale bars: A, B, C, 20 μm; D, E, 100 μm; F, 5 μm.
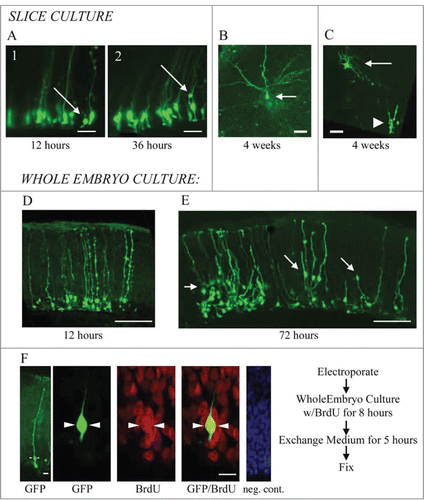
We next tested the activity of electroporated cells in whole embryo cultures. Electroporations were carried out as described above, but rather than prepare slice cultures, whole embryos were maintained in culture conditions for 3 d or more. The whole embryo cultures appeared to be well preserved for 3 d or longer. We found higher levels of cell migration among electroporated cells in the whole embryos, perhaps, in part, because endogenous growth factors and cell-cell signaling pathways are better preserved in the intact preparation. Twelve hours after electroporation, EGFP-labeled RG cells were close to the ventricular surface (), but after 72 hours, many labeled cells were located in the cortex (arrows in ), occasionally retaining contact with cells in the VZ (arrow at far right in ). These data suggests the production of neuronal cells that migrate from the VZ to the pyramidal cell layer. To determine if RG cells were indeed proliferative in the whole embryo cultures, we added the thymidine analog BrdU to culture media for 8 hours. We noted intense BrdU labeling in the culture, demonstrating high levels of proliferation in the whole embryo culture. Furthermore, we found examples of EGFP-labeled RG cells colabeled with BrdU (), demonstrating specifically that RG cells divided in the whole embryo culture. Less than 20% of the EGFP+ cells showed BrdU immunoreactivity after the BrdU pulse. However, this approach to gauging in vivo proliferation likely underestimates RG cell proliferation in the developing turtle telencephalon.
Mitotic cells express 4A4 and divide within the VZ and SVZ
To determine what proportion of the BrdU+ dividing cells we observed in the whole embryo cultures might be RG cells, we co-stained freshly fixed embryonic cortical tissue with syto-11 and anti-phosphorylated vimentin antibodies. Vimentin is phosphorylated by the cell cycle kinase cdc2 during cytokinesis.Citation60 Antibodies raised against the cdc2 phosphorylation site on vimentin, termed 4A4, label M-phase RG cells,Citation5 intermediate progenitor cells in the SVZ,Citation57 and translocating RG cells,Citation57 but not other dividing cell populations such as microglia.Citation61
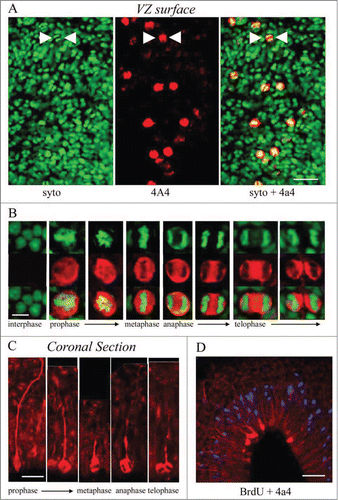
We labeled cortical slabs of the turtle telencephalon, in which the ventricular surface is exposed, with the chromatin dye syto-11 and 4A4. Syto-11 staining of the ventricular surface revealed M-phase cells (). In all cases, dividing cells strongly colabeled with 4A4 (). Similarly, M-phase cells from prophase through telophase could be identified dividing at the surface of the ventricle in coronal sections (). In coronal sections of the embryonic turtle cortex, the radial pial fibers of the 4A4+ cells were labeled brightly as we have shown in embryonic rat cortexCitation5. In addition, the 4A4 immunostaining showed that dividing RG cells in the turtle cortex maintained the pial process throughout division as reported in mammals.Citation5,13,15,16,57,61 In telophase, the pial fiber associated with one of the 2 daughter cells, indicating which daughter cell inherits the pial fiber, as shown in embryonic rat.Citation16 After a brief 3 hour in ovo BrdU pulse, all S-phase cells were located in an abventricular position, several cell bodies away from the ventricle (), while most 4A4 labeled cells were directly at the ventricular surface, consistent with the occurrence of interkinetic nuclear migration in the developing turtle VZ. Importantly, these data demonstrate that cell division occurs at a much higher rate in whole embryo culture of the turtle brain, than in coronal slice cultures.
Figure 4. M-phase cells in the embryonic turtle VZ express the RG lineage marker phosphorylated vimentin (4A4) and reveal RG cell morphology. (A) En face cortical slab stained with syto-11 (green) and 4A4 (red). All M-phase cells at the ventricular surface express 4A4. (B) 4A4 robustly labels M-phase cells throughout mitosis. (C) In coronal sections 4A4 labeled mitotic cells lining the ventricle that possessed pial fibers coursing out through the parenchyma. Pial fibers were most robust in prophase, but a thin process remained throughout division. (D) A brief BrdU (blue) pulse shows that S-phase cells are found in an abventricular position at the top of the VZ while 4A4+ cells mitotic cells were located at the ventricular surface, indicating interkinetic nuclear migration in the developing turtle telencephalon. Scale bars: A, 20 μm; B, 5 μm; C, 10 μm; D, 20 μm.
Tbr2 expressing intermediate progenitor cells are present in the embryonic turtle cortex
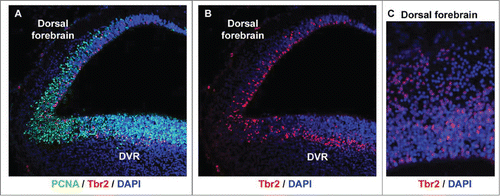
Since we observed mitotic cells both at the ventricle and away from the ventricle, we asked whether intermediate progenitor cells were present in the developing turtle telencephalon. Tbr2 expressing neural precursor cells in the SVZ were originally described by Hevner and colleagues in mouse.Citation62 The Tbr2+ precursor cells define the SVZ and have since been shown in many species including rat,Citation16 human,Citation58,63 ferret,Citation57,63,64 and monkey.Citation57 Time-lapse imaging studies of embryonic rodent slice cultures indicate that Tbr2+ progenitors, referred to as intermediate or basal progenitorsCitation15,65,66 arise from asymmetric divisions of RG cells.Citation15,16 We stained turtle tissue with anti-Tbr2 antibodies and found that many Tbr2+ cells were present in the proliferative zones of the dorsal cortex and the DVR during neurogenic periods. Counterstaining with the mitotic cell marker PCNA confirmed that the Tbr2+ cells were proliferative, as in the mammalian SVZ. Of special interest we noted that the Tbr2+ cells were scattered throughout the VZ in the dorsal cortex of the turtle (), similar to the distribution of Tbr2+ cells in embryonic rat cortex before the appearance of the SVZ.Citation16 In contrast, the distribution of Tbr2+ cells in the DVR resembled that of species that possess a true SVZ: Tbr2 cells were concentrated in a band superficial to the VZ (). These data demonstrate that turtles such as Trachemys scripta elegans possess components of the SVZ.
Figure 5. Tbr2-expressing intermediate progenitor cells are present in the developing turtle telencephalon. (A) A coronal section of a stage 20 turtle embryo immunostained with anti-PCNA (green) and anti-Tbr2 (red), and costained with DAPI (blue). (B and C) The distribution of Tbr2+ cells (red) differs in the dorsal cortex versus the dorsal DVR. Tbr2+ cells are distributed throughout the VZ in the dorsal cortex, but concentrate in a band superficial to the VZ in the DVR, as is seen in the developing mammalian neocortex.
Three subtypes of adult RG cells in the turtle VZ: Lamellate, protoplasmic, and undifferentiated
In the postnatal turtle VZ cells continue to proliferate, and most proliferation occurs in the ventricular wall underlying the DVR and striatum.Citation43 We therefore examined the morphology of individual RG cells in proliferative areas. We used electroporation to label individual RG cells in the adult telencephalon. We observed RG cells with heterogeneous morphologies that we grouped into 3 categories based on the classification scheme of Stensaas & Stensaas in turtle and bird.Citation67 The 3 categories, defined primarily by morphology of the pial fiber, are lamellate (L, Aa and Ab), protoplasmic (P, Ac–Ae), and undifferentiated (U, Af). Previous work has described lamellate cells among ependymal cell types in the turtle telencephalon.Citation67 However, the few Golgi studies performed in turtle have found a very dense labeling of cell bodies, compared to other vertebrates, which have been difficult to interpret. This may be due in part to the prevalence of the ‘hairy’ lamellate fibers that obscure nearby cells. Our labeling technique suggests that lamellar and protoplasmic RG cells constituted the majority of RG cell morphological types in the adult turtle, with a minority of cells, approximately 10%, exhibiting the undifferentiated phenotype that is more common in the embryonic turtle brain.
Figure 6. Electroporation of the adult turtle telencephalon reveals heterogeneous RG cells that we grouped into 3 categories distinguishable by their pial fiber morphology. Lamellate RG cells (L, Aa, Ab); Protoplasmic RG cells (P, Ac−Ae); and Undifferentiated RG cells (U, Af). (B) Further examples of the 3 cell types identified by letter under each image. (C) Lamellate RG cells have pial fibers possessing ‘hairy’ fine extensions, and a pial fiber that in some cases had multiple branches within the parenchyma. (D) Comparison of the pial fiber of the 3 cell types in higher magnification images. Protoplasmic fibers had many smooth expansions. Cell bodies were located at the ventricle and away from the ventricle. Protoplasmic RG cells had the most diverse cellular morphologies. Undifferentiated fiber types were smooth and traceable through the pyramidal cell layer and for several hundred micrometers into the parenchyma. They arose from smaller cell bodies most frequently found close to the ventricle (B and C). (E) Schematic showing the 3 classes of RG cells and their overlapping distribution (Ed). We hypothesize that undifferentiated RG cells retain the capacity for proliferation. Scale bars: A, B, C, 10 μm; D, 3 μm.
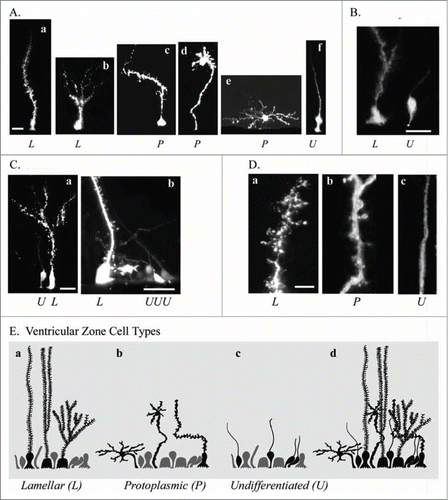
Lamellate RG cells (Da) were ‘hairy’ – their pial fibers possessed many fine lateral extensions. Lamellar cells either extended a single radial fiber to the pia, or had bifurcated or multiple branched processes within the parenchyma (Db) that terminated before reaching the pia (Ab and Ca). Protoplasmic RG cells had many smooth rounded expansions along the pial fiber. Protoplasmic RG cell bodies were located both at the ventricular surface (Ac) and away from the ventricle (Ad and Ae). Protoplasmic RG cells exhibited the most diverse cellular morphologies, with cellular processes appearing to follow fiber tracts or associate with synapses, as in some other species.Citation68 Undifferentiated RG cells in the turtle resembled interphase RG cells in the embryonic rodent (Dc). Undifferentiated RG cells had smooth pial fibers that could be traced through the pyramidal cell layer and for several hundred microns, but not all the way to the pia. Undifferentiated RG cells were bipolar, possessed both pial and ventricular contacting processes, had smaller cell bodies, and were frequently positioned at least one cell body away from the ventricular surface ( and Ca,b). The undifferentiated RG cells may be similar to cells with this morphology that have been functionally and physiologically characterized in the turtle spinal cord.Citation29,69 The schematic in shows the 3 classes of cells (Ea,b,c), as well as the overlapping distribution of these cell types in the adult VZ (Ed).
Discussion
We used BrdU and M-phase labeling to confirm that RG cells proliferate, and to show that RG cells constitute the major dividing cell class in the embryonic turtle brain. We show that precursor cells divide in abventricular positions in the embryonic turtle telencephalon, and that Tbr2+ cells are present in both the dorsal cortex and DVR of the developing turtle brain. Previous studies have suggested that the turtle lacks a true anatomically defined SVZ,Citation70 while our previous work indicated the presence of rudimentary elements of the SVZ in lateral portions of the turtle cortex.72 Our present results support our previous suggestions by showing that Tbr2+ cells are present in the turtle cortex. This shows that elements of the mammalian SVZ are present in the turtle cortex, and indicates that Tbr2+ precursor cells may have been present in the common ancestor of mammals and reptiles. Furthermore, the presence of mitotic Tbr2-expressing cells in the developing turtle brain suggests that the cellular mechanism by which cortical neurons are generated through intermediate progenitor cells evolved prior to its appearance in mammals. Importantly, since the turtle cortex is a simplified structure with a single layer of pyramidal neurons, our data provide further support for the idea that Tbr2 cells contribute neurons to this cortical layer. Indeed, previous work in mammals has suggested that Tbr2+ intermediate progenitor cells produce cortical neurons destined for each layer.Citation71 Our data are consistent with this view and suggest the possibility that Tbr2+ precursor cells produce the majority of excitatory cortical neurons. Finally, we show that in the adult turtle RG cells are heterogeneous, particularly in the subpallium where most proliferation occurs.Citation43
Embryonic RG cells
Our results demonstrate that embryonic RG cells in the turtle telencephalon share several key features with those in the embryonic mammalian telencephalon. Embryonic RG cells in the turtle exhibit the same bipolar morphology as do mammalian RG cells, and undergo division at the surface of the ventricle. Mitotic RG cells could also be labeled with antibodies directed against phosphorylated vimentin. We can also infer that embryonic turtle RG cells produce Tbr2+ intermediate progenitor cells and, indirectly, cortical neurons (). The demonstration that turtle RG cells share key characteristics with mammalian RG cells suggests that the basic paradigm for neuron production in the developing brain is widely present throughout vertebrates. Our data indicate that the 2-step pattern of neurogenesis first described in rodents,Citation72 RG cell > Intermediate Progenitor cell > Neuron, was either present in the common ancestor of reptiles and mammals, or evolved independently in each clade.
Adult RG cells
In mammals RG cells transform into astrocytes at the end of the neurogenic period,Citation15,Citation73-77 but a subset of radial glia-like GFAP-expressing astrocytes function as adult neural precursor cells in both the cortical SVZCitation78 and hippocampal dentate gyrus.Citation79 While mammalian RG cells exhibit robust neurogenic potential embryonically, the potential of adult astrocytes is reduced in cell type, cell number, and level of proliferation.Citation80 In rodents, neurogenic astrocytes in the SVZ contact the lateral ventricle,Citation81 while the neurogenic astrocytes in the subgranular zone of the dentate gyrus, which derives from the SVZ, no longer appear to maintain an association with the ventricle.Citation82,83
In several non-mammalian vertebrates, new neurons are generated in adulthood, and migrate to most or all regions of the telencephalon.Citation23,43,84 In turtles, new neurons are found in abundance in the olfactory bulbs and in lower numbers in the dorsal cortex.Citation43 Nevertheless, all major regions of the adult turtle telencephalon appear to incorporate new neurons. However, in adult mammals, only the olfactory bulb and hippocampus are widely acknowledged to be sites that normally permit incorporation of new neurons. It is unclear what makes the turtle cortex permissive to receiving new neurons, but following cortical injury, the amount of adult neurogenesis is increased leading to replacement of the lost neurons.Citation48,85
It will be interesting to examine the factors regulating the neurogenic potential of neural precursor cells, and the genetic or epigenetic factors that make a region receptive to new neurons. In the mammalian hippocampus, for instance, aspects of tissue architecture and glia-vascular organization have been proposed to serve as pro-neurogenic factors.Citation86
One classic feature of RG cells is their role as migrational guides for newborn neurons. There is a tight association between RG cell processes and migrating neurons.Citation87,88 In the adult turtle, the highly differentiated pial process of lamellar and protoplasmic RG cells may interfere with close associations between migrating neurons and radial glia fibers. In contrast, the smooth radial processes of undifferentiated RG cells may serve this function in the adult turtle, as in embryonic mammals.
Preceding the discovery that RG cells are neural precursor cells in mammals, evidence suggested that they also play this role in other species. It was demonstrated that mitotic ‘hotspots’ in the adult bird brain were neurogenic niches containing many RG cells.Citation89 There were also reports of adult neurogenesis in teleost, with proposals that RG cells serve as neural precursor cells and guide neuronal migration in the intact and damaged brain.Citation90 RG cells are thought to persist throughout adult life in the central nervous system of many, if not all, cartilaginous and bony fishes.Citation24,26,Citation91-98 The first and most convincing evidence of neurogenic RG cells in adult non-mammalian vertebrates came from studies in birds. In adult birds, vimentin-positive radial cells send long undivided fibers that reach most parts of the telencephalic gray matter, resembling RG cells in embryonic mammalian cortex.Citation84 These radial cells divide and produce young neurons,Citation99 indicating that embryonic traits are retained in adulthood, in some species.
We show that RG cells in the adult turtle comprise a heterogeneous population. Adult ependymal cells in the VZ of both avian and lizard have been examined closely by electron microscopy. In these species the apparent heterogeneity has led to descriptions of subclasses of ependymal cells based on key features of the cell body. Russo et al. have also described several classes of RG cells in turtle spinal cord.Citation29 One class of cells expresses the glial cell marker S100 and displays morphological and electrophysiological features of RG cells, while a second class of S100+ cells exhibit higher input resistance and outward rectification. In addition, some cells contacting the central canal of the spinal cord express HuC/D - a marker of immature neurons - and fire action potentials. The same group reported that the cells surrounding the central canal of the turtle spinal cord formed radial conglomerates in such a way that the RG lamellae envelop immature neurons. They also used electron microscopy to show the presence of gap junctions interconnecting clusters of RG cells. Furthermore, Russo, Trujillo-Cenóz and colleagues showed the presence of a single cilium associated with a conspicuous centrosomal complex in the cell apical zone.Citation69 They reasoned that the coexistence of cells with functional properties of precursor-like cells and immature neurons suggests that the region surrounding the central canal in adult turtle is a site of active neurogenesis.Citation29,100 Lazzari and Franceschini have also described a heterogeneous pattern of astroglial cells in the spinal cord of the soft-shell turtle.Citation101 We identified 3 categories of RG cells based primarily on pial fiber morphology. These distinct RG cell types suggest diversity of RG cell function in the adult brain. Based on our current findings we hypothesize that: undifferentiated RG cells retain the capacity for neurogenic proliferation and migration guidance; that lamellate cells are similar to fibrous astrocytes; and that protoplasmic ependymoglia are similar to protoplasmic astrocytes in mammals and birds. Furthermore, we propose that lamellate and protoplasmic RG cells perform functions characteristic of non-neurogenic astrocytes in the adult mammalian brain, such as regulating synapse function. Supporting this idea, previous work has reported that lamellate Bergmann glia in the developing cerebellum regulate synapse formation.Citation102 Previous work has highlighted the finding that the turtle telencephalon is devoid of astrocytes.Citation35,40 The diverse RG cell types of the adult brain therefore likely represent the functional equivalent of astroglial cells in the mammalian brain.
Conclusion
We have provided evidence consistent with the concept that RG cells are neuronal precursor cells in the developing turtle telencephalon. We have also shown that elements of the SVZ, specifically mitotic Tbr2+ cells, are present in the developing turtle telencephalon. Furthermore, we have presented evidence consistent with the hypothesis that RG cells remain proliferative in the adult turtle. We also demonstrated 3 basic subclasses of RG cells in the adult turtle. The turtle may prove to be a useful model for studying adult neurogenesis based on several features. First, reptiles have extended growing periods, with brain size expanding over decades. Secondly, freshwater species are resistant to anoxia, facilitating in vitro experimentation.Citation104,105 Thirdly, turtles are generally thought to stem from the common ancestor of reptiles and mammals.Citation106,107 Certainly, the neuroanatomy of the turtle suggests that it has maintained an early representation of the laminated cortex, unique even among reptiles. Free astrocytes, which are widespread in other reptiles such as the crocodile, are limited in lizards and snakes, but not present in the turtle.Citation108 The turtle's adult RG cell architecture and lack of free astrocytes may be a shared feature with the stem reptile, and one that is recapitulated at early stages in the developing mammalian brain. The turtle has much to contribute to our understanding of telencephalic evolution, adult neurogenesis, and the potential for reclaiming this ability in the future.
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Funding
This work was supported by the Shriners Hospitals, the NIH (MH 094681 to VMC; MH 101188 to SCN; NS 21223 to ARK), and the UC Davis MIND Institute.
References
- Cruce WL, Nieuwenhuys R. The cell masses in the brain stem of the turtle Testudo hermanni; a topographical and topological analysis. J Comp Neurol 1974; 156:277-306; PMID:4418301; http://dx.doi.org/10.1002/cne.901560303
- Connors BW, Kriegstein AR. Cellular physiology of the turtle visual cortex: distinctive properties of pyramidal and stellate neurons. J Neurosci 1986; 6:164-177; PMID:3944618
- Blanton MG, Shen JM, Kriegstein AR. Evidence for the inhibitory neurotransmitter gamma-aminobutyric acid in aspiny and sparsely spiny nonpyramidal neurons of the turtle dorsal cortex. J Comp Neurol 1987; 259:277-297; PMID:2438317; http://dx.doi.org/10.1002/cne.902590208
- Levitt P, Rakic P. Immunoperoxidase localization of glial fibrillary acidic protein in radial glial cells and astrocytes of the developing rhesus monkey brain. J Comp Neurol 1980; 193:815-840; PMID:7002963; http://dx.doi.org/10.1002/cne.901930316
- Noctor SC, Flint AC, Weissman TA, Wong WS, Clinton BK, Kriegstein AR. Dividing precursor cells of the embryonic cortical ventricular zone have morphological and molecular characteristics of radial glia. J Neurosci 2002; 22:3161-3173; PMID:11943818
- His W. Die Neuroblasten und deren Entstehung im embryonalen Mark. Abh Kgl sachs Ges Wissensch math phys 1889; 15:311-372.
- Rakic P. In: Kettenmann H & Ransom BR, eds. Radial gial cells: Scaffolding for brain construction. Neuroglia. Oxford, UK: Oxford University Press, 1995, 746-762.
- Rakic P. Mode of cell migration to the superficial layers of fetal monkey neocortex. J Comp Neurol 1972; 145:61-83; PMID:4624784; http://dx.doi.org/10.1002/cne.901450105
- Misson JP, Edwards MA, Yamamoto M, Caviness VS, Jr. Mitotic cycling of radial glial cells of the fetal murine cerebral wall: a combined autoradiographic and immunohistochemical study. Brain Res 1988; 466:183-190; PMID:3359310; http://dx.doi.org/10.1016/0165-3806(88)90043-0
- Alvarez-Buylla A, Theelen M, Nottebohm F. Proliferation “hot spots” in adult avian ventricular zone reveal radial cell division. Neuron 1990; 5:101-109; PMID:2369518; http://dx.doi.org/10.1016/0896-6273(90)90038-H
- Malatesta P, Hartfuss E, Gotz M. Isolation of radial glial cells by fluorescent-activated cell sorting reveals a neuronal lineage. Development 2000; 127:5253-5263; PMID:11076748
- Miyata T, Kawaguchi A, Okano H, Ogawa M. Asymmetric inheritance of radial glial fibers by cortical neurons. Neuron 2001; 31:727-741; PMID:11567613; http://dx.doi.org/10.1016/S0896-6273(01)00420-2
- Noctor SC, Flint AC, Weissman TA, Dammerman RS, Kriegstein AR. Neurons derived from radial glial cells establish radial units in neocortex. Nature 2001; 409:714-720; PMID:11217860; http://dx.doi.org/10.1038/35055553
- Tamamaki N, Nakamura K, Okamoto K, Kaneko T. Radial glia is a progenitor of neocortical neurons in the developing cerebral cortex. Neurosci Res 2001; 41:51-60; PMID:11535293; http://dx.doi.org/10.1016/S0168-0102(01)00259-0
- Noctor SC, Martinez-Cerdeno V, Ivic L, Kriegstein AR. Cortical neurons arise in symmetric and asymmetric division zones and migrate through specific phases. Nat Neurosci 2004; 7, 136-144pii ; PMID:14703572; http://dx.doi.org/10.1038/nn1172
- Noctor SC, Martinez-Cerdeno V, Kriegstein AR. Distinct behaviors of neural stem and progenitor cells underlie cortical neurogenesis. J Comp Neurol 2008; 508:28-44; PMID:18288691; http://dx.doi.org/10.1002/cne.21669
- Gray GE, Glover JC, Majors J, Sanes JR. Radial arrangement of clonally related cells in the chicken optic tectum: lineage analysis with a recombinant retrovirus. Proc Natl Acad Sci U S A 1988; 85:7356-7360; PMID:3174639; http://dx.doi.org/10.1073/pnas.85.19.7356
- Zikopoulos B, Kentouri M, Dermon CR. Proliferation zones in the adult brain of a sequential hermaphrodite teleost species (Sparus aurata). Brain Behav Evol 2000; 56:310-322; http://dx.doi.org/10.1159/000047215
- Zupanc GK, Horschke I. Proliferation zones in the brain of adult gymnotiform fish: a quantitative mapping study. J Comp Neurol 1995; 353:213-233; PMID:7745132; http://dx.doi.org/10.1002/cne.903530205
- Zupanc GK, Zupanc MM. Birth and migration of neurons in the central posterior/prepacemaker nucleus during adulthood in weakly electric knifefish (Eigenmannia sp.). Proc Natl Acad Sci U S A 1992; 89:9539-9543; PMID:1409663; http://dx.doi.org/10.1073/pnas.89.20.9539
- Ling C, Zuo M, Alvarez-Buylla A, Cheng MF. Neurogenesis in juvenile and adult ring doves. J Comp Neurol 1997; 379:300-312; PMID:9050792; http://dx.doi.org/10.1002/(SICI)1096-9861(19970310)379:2%3c300::AID-CNE10%3e3.0.CO;2-T
- Ramirez-Castillejo C, Nacher, J, Molowny A, Ponsoda X, Lopez-Garcia C. PSA-NCAM immunocytochemistry in the cerebral cortex and other telencephalic areas of the lizard Podarcis hispanica: differential expression during medial cortex neuronal regeneration. J Comp Neurol 2002; 453:145-156; PMID:12373780; http://dx.doi.org/10.1002/cne.10390
- Perez-Canellas MM, Garcia-Verdugo JM. Adult neurogenesis in the telencephalon of a lizard: a ; 3Hthymidine autoradiographic and bromodeoxyuridine immunocytochemical study. Brain Res Dev Brain Res 1996; 93:49-61; PMID:8804691; http://dx.doi.org/10.1016/0165-3806(96)00014-4
- Kalman M. Astroglial architecture of the carp (Cyprinus carpio) brain as revealed by immunohistochemical staining against glial fibrillary acidic protein (GFAP). Anat Embryol (Berl) 1998; 198:409-433; PMID:9801060; http://dx.doi.org/10.1007/s004290050193
- Yanes C, Monzon-Mayor M, Ghandour MS, de Barry J, Gombos G. Radial glia and astrocytes in developing and adult telencephalon of the lizard Gallotia galloti as revealed by immunohistochemistry with anti-GFAP and anti-vimentin antibodies. J Comp Neurol 1990; 295:559-568; PMID:2358521; http://dx.doi.org/10.1002/cne.902950405
- Manso MJ, Becerra M, Becerra M, Anadon R. Expression of a low-molecular-weight (10 kDa) calcium binding protein in glial cells of the brain of the trout (Teleostei). Anat Embryol (Berl) 1997; 196:403-416; PMID:9406842; http://dx.doi.org/10.1007/s004290050108
- Kalman M, Szekely AD, Csillag A. Distribution of glial fibrillary acidic protein and vimentin-immunopositive elements in the developing chicken brain from hatch to adulthood. Anat Embryol (Berl) 1998; 198:213-235; PMID:9764976
- Mathieu M, Bruzzone F, Chartrel N, Serra GP, Spiga S, Vallarino M, Vaudry H. Somatostatin in the brain of the cave salamander, Hydromantes genei (Amphibia, Plethodontidae): immunohistochemical localization and biochemical characterization. J Comp Neurol 2004; 475:163-176; PMID:15211458; http://dx.doi.org/10.1002/cne.20175
- Russo RE, Fernandez A, Reali C, Radmilovich M, Trujillo-Cenoz O. Functional and molecular clues reveal precursor-like cells and immature neurones in the turtle spinal cord. J Physiol 2004; 560:831-838; PMID:15331672; http://dx.doi.org/10.1113/jphysiol.2004.072405
- Ahboucha S, Laalaoui A, Didier-Bazes M, Montange M, Cooper HM, Gamrani H. Differential patterns of glial fibrillary acidic protein-immunolabeling in the brain of adult lizards. J Comp Neurol 2003; 464:159-171; PMID:12898609; http://dx.doi.org/10.1002/cne.10781
- Monzon-Mayor M, Yanes C, Renau-Piqueras J. Distribution of neurofilaments in the telencephalon and mesencephalon of the adult and developing Gallotia galloti lizard. Eur J Histochem : EJH 1998; 42:213-226 ; PMID:9857247
- Bodega G, Suarez I, Rubio M, Fernandez B. Distribution and characteristics of the different astroglial cell types in the adult lizard (Lacerta lepida) spinal cord. Anat Embryol (Berl) 1990; 181:567-575; PMID:2396756; http://dx.doi.org/10.1007/BF00174628
- Onteniente B, Kimura H, Maeda T. Comparative study of the glial fibrillary acidic protein in vertebrates by PAP immunohistochemistry. J Comp Neurol 1983; 215:427-436; PMID:6408144; http://dx.doi.org/10.1002/cne.902150407
- Kalman M, Kiss A, Majorossy K. Distribution of glial fibrillary acidic protein-immunopositive structures in the brain of the red-eared freshwater turtle (Pseudemys scripta elegans). Anat Embryol (Berl) 1994; 189:421-434; PMID:7522421; http://dx.doi.org/10.1007/BF00185437
- Kalman M, Martin-Partido G, Hidalgo-Sanchez M, Majorossy K. Distribution of glial fibrillary acidic protein-immunopositive structures in the developing brain of the turtle Mauremys leprosa. Anat Embryol (Berl) 1997; 196:47-65; PMID:9242888; http://dx.doi.org/10.1007/s004290050079
- Kriegstein AR, Shen JM, Eshhar N. Monoclonal antibodies to the turtle cortex reveal neuronal subsets, antigenic cross-reactivity with the mammalian neocortex, and forebrain structures sharing a pallial derivation. J Comp Neurol 1986; 254:330-340; PMID:2432104; http://dx.doi.org/10.1002/cne.902540306
- Peters A, Palay S, Webster H. The fine strucutre of the nervous system. Oxford, UK: Oxford University Press, 1991.
- Varon SS, Somjen GG. Neuron-glia interactions. Neurosci Res Prog Bull 1979; 17:1-239; PMID:39273
- Hajos F, Basco E. The surface-contact glia. Adv Anat Embryol Cell Biol 1984; 84:1-79; PMID:6377846; http://dx.doi.org/10.1007/978-3-642-69623-7_1
- Kriegstein AR, Shen JM, Eshhar N. Monoclonal antibodies to the turtle cortex reveal neuronal subsets, antigenic cross-reactivity with the mammalian neocortex, and forebrain structures sharing a pallial derivation. J Comp Neurol 1986; 254:330-340; PMID:2432104; http://dx.doi.org/10.1002/cne.902540306
- Lazzari M, Franceschini V. Glial fibrillary acidic protein and vimentin immunoreactivity of astroglial cells in the central nervous system of adult Podarcis sicula (Squamata, Lacertidae). J Anat 2001; 198:67-75; PMID:11215769; http://dx.doi.org/10.1046/j.1469-7580.2001.19810067.x
- Garcia-Verdugo JM, Ferrón S, Flames N, Collado L, Desfilis E, Font E. The proliferative ventricular zone in adult vertebrates: a comparative study using reptiles, birds, and mammals. Brain Res Bull 2002; 57:765-775; PMID:12031273; http://dx.doi.org/10.1016/S0361-9230(01)00769-9
- Perez-Canellas MM, Font E, Garcia-Verdugo JM. Postnatal neurogenesis in the telencephalon of turtles: evidence for nonradial migration of new neurons from distant proliferative ventricular zones to the olfactory bulbs. Brain Res Dev Brain Res 1997; 101:125-137; PMID:9263587; http://dx.doi.org/10.1016/S0165-3806(97)00058-8
- Stensaas LJ, Stensaas SS. Astrocytic neuroglial cells, oligodendrocytes and microgliacytes in the spinal cord of the toad. I. Light microscopy. Zeitschrift fur Zellforschung und mikroskopische Anatomie 1968; 84:473-489; PMID:4178203; http://dx.doi.org/10.1007/BF00320863
- Ebner FF, Colonnier M. Synaptic patterns in the visual cortex of turtle: an electron microscopic study. J Comp Neurol 1975; 160:51-79; PMID:1112922; http://dx.doi.org/10.1002/cne.901600105
- Connors BW, Ransom BR. Electrophysiological properties of ependymal cells (radial glia) in dorsal cortex of the turtle, Pseudemys scripta. J Physiol 1987; 385:287-306; PMID:3116210
- Stensaas LJ, Stensaas SS. Astrocytic neuroglial cells, oligodendrocytes and microgliacytes in the spinal cord of the toad. II. Electron microscopy. Zeitschrift fur Zellforschung und mikroskopische Anatomie 1968; 86:184-213; PMID:5707643; http://dx.doi.org/10.1007/BF00348524
- Font E, Desfilis E, Perez-Canellas MM, Garcia-Verdugo JM. Neurogenesis and neuronal regeneration in the adult reptilian brain. Brain Behav Evol 2001; 58:276-295; PMID:11978946; http://dx.doi.org/10.1159/000057570
- Weissman T, Noctor SC, Clinton BK, Honig LS, Kriegstein AR. Neurogenic radial glial cells in reptile, rodent and human: from mitosis to migration. Cereb Cortex 2003; 13:550-559; PMID:12764028; http://dx.doi.org/10.1093/cercor/13.6.550
- Zupanc GK. Neurogenesis, cell death and regeneration in the adult gymnotiform brain. J Exp Biol 1999; 202 (Pt 10), 1435-1446; PMID:10210684
- Saito T, Nakatsuji N. Efficient gene transfer into the embryonic mouse brain using in vivo electroporation. Dev Biol 2001; 240:237-246; PMID:11784059; http://dx.doi.org/10.1006/dbio.2001.0439
- Yntema CL. A series of stages in the embryonic development of Chelydra serpentina. J Morphol 1968; 125:219-251; PMID:5681661; http://dx.doi.org/10.1002/jmor.1051250207
- Cordery P, Molnar Z. Embryonic development of connections in turtle pallium. J Comp Neurol 1999; 413:26-54; PMID:10464368; http://dx.doi.org/10.1002/(SICI)1096-9861(19991011)413:1%3c26::AID-CNE2%3e3.0.CO;2-N
- Blanton MG, Kriegstein AR. Appearance of putative amino acid neurotransmitters during differentiation of neurons in embryonic turtle cerebral cortex. J Comp Neurol 1991; 310:571-592; PMID:1682348; http://dx.doi.org/10.1002/cne.903100406
- Fernandez AS, Pieau C, Reperant J, Boncinelli E, Wassef M. Expression of the Emx-1 and Dlx-1 homeobox genes define three molecularly distinct domains in the telencephalon of mouse, chick, turtle and frog embryos: implications for the evolution of telencephalic subdivisions in amniotes. Development 1998; 125:2099-2111; PMID:9570774
- Saito T, Fujimoto T, Maegawa S, Inoue K, Tanaka M, Arai K, Yamaha E. Visualization of primordial germ cells in vivo using GFP-nos1 3’UTR mRNA. Int J Dev Biol 2006; 50:691-699; PMID:17051479; http://dx.doi.org/10.1387/ijdb.062143ts
- Martinez-Cerdeno V, Cunningham CL, Camacho J, Antczak JL, Prakash AN, Cziep ME, Walker AI, Noctor SC. Comparative analysis of the subventricular zone in rat, ferret and macaque: evidence for an outer subventricular zone in rodents. PLoS One 2012; 7:e30178; PMID:22272298; http://dx.doi.org/10.1371/journal.pone.0030178
- Hansen DV, Lui JH, Parker PR, Kriegstein AR. Neurogenic radial glia in the outer subventricular zone of human neocortex. Nature 2010; 464:554-561; PMID:20154730; http://dx.doi.org/10.1038/nature08845
- Wang X, Tsai JW, LaMonica B, Kriegstein AR. A new subtype of progenitor cell in the mouse embryonic neocortex. Nat Neurosci 2011; 14:555-561; PMID:21478886; http://dx.doi.org/10.1038/nn.2807
- Kamei Y, Inagaki N, Nishizawa M, Tsutsumi O, Taketani Y, Inagaki M. Visualization of mitotic radial glial lineage cells in the developing rat brain by Cdc2 kinase-phosphorylated vimentin. Glia 1998; 23:191-199; PMID:9633804; http://dx.doi.org/10.1002/(SICI)1098-1136(199807)23:3%3c191::AID-GLIA2%3e3.0.CO;2-8
- Cunningham CL, Martinez-Cerdeno V, Noctor SC. Diversity of neural precursor cell types in the prenatal macaque cerebral cortex exists largely within the astroglial cell lineage. PLoS One 2013; 8:e63848; PMID:23724007; http://dx.doi.org/10.1371/journal.pone.0063848
- Englund C, Fink A, Lau C, Pham D, Daza RA, Bulfone A, Kowalczyk T, Hevner RF. Pax6, Tbr2, and Tbr1 are expressed sequentially by radial glia, intermediate progenitor cells, and postmitotic neurons in developing neocortex. J Neurosci 2005; 25:247-251; PMID:15634788; http://dx.doi.org/10.1523/JNEUROSCI.2899-04.2005
- Fietz SA, Kelava I, Vogt J, Wilsch-Bräuninger M, Stenzel D, Fish JL, Corbeil D, Riehn A, Distler W, Nitsch R, et al. OSVZ progenitors of human and ferret neocortex are epithelial-like and expand by integrin signaling. Nat Neurosci 2010; 13:690-699; PMID:20436478; http://dx.doi.org/10.1038/nn.2553
- Reillo I, Borrell V. Germinal zones in the developing cerebral cortex of ferret: ontogeny, cell cycle kinetics, and diversity of progenitors. Cereb Cortex 2012; 22:2039-2054; PMID:21988826; http://dx.doi.org/10.1093/cercor/bhr284
- Haubensak W, Attardo A, Denk W, Huttner WB. Neurons arise in the basal neuroepithelium of the early mammalian telencephalon: a major site of neurogenesis. Proc Natl Acad Sci U S A 2004; 101:3196-3201; PMID:14963232; http://dx.doi.org/10.1073/pnas.0308600100
- Miyata T, Kawaguchi A, Saito K, Kawano M, Muto T, Ogawa M. Asymmetric production of surface-dividing and non-surface-dividing cortical progenitor cells. Development 2004; 131:3133-3145; PMID:15175243; http://dx.doi.org/10.1242/dev.01173
- Stensaas LJ, Stensaas SS. Light microscopy of glial cells in turtles and birds. Zeitschrift fur Zellforschung und mikroskopische Anatomie 1968; 91:315-340; PMID:4894065; http://dx.doi.org/10.1007/BF00440762
- Bushong EA, Martone ME, Jones YZ, Ellisman MH. Protoplasmic astrocytes in CA1 stratum radiatum occupy separate anatomical domains. J Neurosci 2002; 22:183-192; PMID:11756501
- Trujillo-Cenoz O, Fernandez A, Radmilovich M, Reali C, Russo RE. Cytological organization of the central gelatinosa in the turtle spinal cord. J Comp Neurol 2007; 502:291-308; PMID:17348014; http://dx.doi.org/10.1002/cne.21306
- Cheung AF, Pollen AA, Tavare A, DeProto J, Molnar Z. Comparative aspects of cortical neurogenesis in vertebrates. J Anat 2007; 211:164-176; PMID:17634059; http://dx.doi.org/10.1111/j.1469-7580.2007.00769.x
- Sessa A, Mao CA, Hadjantonakis AK, Klein WH, Broccoli V. Tbr2 directs conversion of radial glia into basal precursors and guides neuronal amplification by indirect neurogenesis in the developing neocortex. Neuron 2008; 60:56-69; pii 10. 1016/j.neuron.2008.09.028 (); PMID:18940588; http://dx.doi.org/10.1016/j.neuron.2008.09.028
- Martinez-Cerdeno V, Noctor SC, Kriegstein AR. The role of intermediate progenitor cells in the evolutionary expansion of the cerebral cortex. Cereb Cortex 2006; 16:i152-i161; PMID:16766701; http://dx.doi.org/10.1093/cercor/bhk017
- Alves JA, Barone P, Engelender S, Froes MM, Menezes JR. Initial stages of radial glia astrocytic transformation in the early postnatal anterior subventricular zone. J Neurobiol 2002; 52:251-265; PMID:12210108; http://dx.doi.org/10.1002/neu.10087
- Choi BH, Lapham LW. Radial glia in the human fetal cerebrum: a combined Golgi, immunofluorescent and electron microscopic study. Brain Res 1978; 148:295-311; PMID:77708; http://dx.doi.org/10.1016/0006-8993(78)90721-7
- Schmechel DE, Rakic P. A Golgi study of radial glial cells in developing monkey telencephalon: morphogenesis and transformation into astrocytes. Anat Embryol (Berl) 1979; 156:115-152; PMID:111580
- Takahashi T, Misson JP, Caviness VS., Jr. Glial process elongation and branching in the developing murine neocortex: a qualitative and quantitative immunohistochemical analysis. J Comp Neurol 1990; 302:15-28; PMID:2086612; http://dx.doi.org/10.1002/cne.903020103
- Voigt T. Development of glial cells in the cerebral wall of ferrets: direct tracing of their transformation from radial glia into astrocytes. J Comp Neurol 1989; 289:74-88; PMID:2808761; http://dx.doi.org/10.1002/cne.902890106
- Doetsch F, Caille I, Lim DA, Garcia-Verdugo JM, Alvarez-Buylla A. Subventricular zone astrocytes are neural stem cells in the adult mammalian brain. Cell 1999; 97:703-716; PMID:10380923; http://dx.doi.org/10.1016/S0092-8674(00)80783-7
- Seri B, Garcia-Verdugo JM, McEwen BS, Alvarez-Buylla A. Astrocytes give rise to new neurons in the adult mammalian hippocampus. J Neurosci 2001; 21:7153-7160; PMID:11549726
- Doetsch F Scharff C. Challenges for brain repair: insights from adult neurogenesis in birds and mammals. Brain Behav Evol 2001; 58:306-322; http://dx.doi.org/10.1159/000057572
- Tramontin AD, Garcia-Verdugo JM, Lim DA, Alvarez-Buylla A. Postnatal development of radial glia and the ventricular zone (VZ): a continuum of the neural stem cell compartment. Cereb Cortex 2003; 13:580-587; PMID:12764031; http://dx.doi.org/10.1093/cercor/13.6.580
- Altman J Das GD. Autoradiographic and histological evidence of postnatal hippocampal neurogenesis in rats. J Comp Neurol 1965; 124:319-335; PMID:5861717; http://dx.doi.org/10.1002/cne.901240303
- Bayer SA, Altman J. Neocortical Development. New York: Raven Press, 1991.
- Nottebohm F. Neuronal replacement in adult brain. Brain Res Bull 2002; 57:737-749; PMID:12031270; http://dx.doi.org/10.1016/S0361-9230(02)00750-5
- Font E, Desfilis E, Perez-Canellas M, Alcantara S, Garcia-Verdugo JM. Three-Acetylpyridine-induced degeneration and regeneration in the adult lizard brain: a qualitative and quantitative analysis. Brain Res 1997; 754:245-259; PMID:9134982; http://dx.doi.org/10.1016/S0006-8993(97)00085-1
- Seki T. Microenvironmental elements supporting adult hippocampal neurogenesis. Anatomical science international 2003; 78:69-78; PMID:12828419; http://dx.doi.org/10.1046/j.0022-7722.2003.00043.x
- Elias LA, Wang DD, Kriegstein AR. Gap junction adhesion is necessary for radial migration in the neocortex. Nature 2007; 448:901-907; PMID:17713529; http://dx.doi.org/10.1038/nature06063
- Rakic S, Zecevic N. Emerging complexity of layer I in human cerebral cortex. Cereb Cortex 2003; 13:1072-1083; PMID:12967924; http://dx.doi.org/10.1093/cercor/13.10.1072
- Alvarez-Buylla A. Mechanism of neurogenesis in adult avian brain. Experientia 1990; 46:948-955; PMID:2209804; http://dx.doi.org/10.1007/BF01939388
- Zupanc GK. Adult neurogenesis and neuronal regeneration in the central nervous system of teleost fish. Brain Behav Evol 2001; 58:250-275; http://dx.doi.org/10.1159/000057569
- Stevenson JA, Yoon MG. Morphology of radial glia, ependymal cells, and periventricular neurons in the optic tectum of goldfish (Carassius auratus). J Comp Neurol 1982; 205:128-138; PMID:7076888; http://dx.doi.org/10.1002/cne.902050204
- Stevenson JA, Yoon MG. Mitosis of radial glial cells in the optic tectum of adult goldfish. J Neurosci 1981; 1:862-875; PMID:7346591
- Rubio M, Suarez I, Bodega G, Fernandez B. Glial fibrillary acidic protein and vimentin immunohistochemistry in the posterior rhombencephalon of the Iberian barb (Barbus comiza). Neurosci Lett 1992; 134:203-206; PMID:1589147; http://dx.doi.org/10.1016/0304-3940(92)90517-B
- Bodega G, Suarez I, Rubio M, Villalba RM, Fernandez B. Astroglial pattern in the spinal cord of the adult barbel (Barbus comiza). Anat Embryol (Berl) 1993; 187:385-398; PMID:8512091; http://dx.doi.org/10.1007/BF00185897
- Wasowicz M, Ward R, Reperant J. An investigation of astroglial morphology in torpedo and scyliorhinus. J Neurocytol 1999; 28:639-653; PMID:10851343; http://dx.doi.org/10.1023/A:1007004714712
- Chiba A. S-100 protein-immunoreactive structures in the brains of the elasmobranchs Scyliorhinus torazame and Mustelus manazo. Neurosci Lett 2000; 279:65-68; PMID:10670789; http://dx.doi.org/10.1016/S0304-3940(99)00949-0
- Tomizawa K, Inoue Y, Nakayasu H. A monoclonal antibody stains radial glia in the adult zebrafish (Danio rerio) CNS. J Neurocytol 2000; 29:119-128; PMID:11068340; http://dx.doi.org/10.1023/A:1007156529390
- Kalman M, Gould RM. GFAP-immunopositive structures in spiny dogfish, Squalus acanthias, and little skate, Raia erinacea, brains: differences have evolutionary implications. Anat Embryol (Berl) 2001; 204:59-80; PMID:11506433
- Alvarez-Buylla A, Theelen M, Nottebohm F. Birth of projection neurons in the higher vocal center of the canary forebrain before, during, and after song learning. Proc Natl Acad Sci U S A 1988; 85:8722-8726; PMID:3186755; http://dx.doi.org/10.1073/pnas.85.22.8722
- Russo RE, Reali C, Radmilovich M, Fernandez A, Trujillo-Cenoz O. Connexin 43 delimits functional domains of neurogenic precursors in the spinal cord. J Neurosci 2008; 28:3298-3309; PMID:18367597; http://dx.doi.org/10.1523/JNEUROSCI.5736-07.2008
- Lazzari M, Franceschini V. Glial cytoarchitecture in the central nervous system of the soft-shell turtle, Trionyx sinensis, revealed by intermediate filament immunohistochemistry. Anat Embryol (Berl) 2006; 211:497-506; PMID:16763812; http://dx.doi.org/10.1007/s00429-006-0101-5
- Yamada K, Fukaya M, Shibata T, Kurihara H, Tanaka K, Inoue Y, Watanabe M. Dynamic transformation of Bergmann glial fibers proceeds in correlation with dendritic outgrowth and synapse formation of cerebellar Purkinje cells. J Comp Neurol 2000; 418:106-120; PMID:10701759; http://dx.doi.org/10.1002/(SICI)1096-9861(20000228)418:1%3c106::AID-CNE8%3e3.0.CO;2-N
- Perez-Pinzon MA, Lutz PL, Sick TJ, Rosenthal M. Metabolic mechanisms of anoxia tolerance in the turtle brain. Advances in experimental medicine and biology 1997; 411:75-81; PMID:9269413; http://dx.doi.org/10.1007/978-1-4615-5865-1_9
- Blanton MG, Kriegstein AR. Morphological differentiation of distinct neuronal classes in embryonic turtle cerebral cortex. J Comp Neurol 1991; 310:558-570; PMID:1719040
- Hylland P, Nilsson GE, Lutz PL. Time course of anoxia-induced increase in cerebral blood flow rate in turtles: evidence for a role of adenosine. Journal of cerebral blood flow and metabolism : official journal of the International Society of Cerebral Blood Flow and Metabolism 1994; 14:877-881; PMID:8063883; http://dx.doi.org/10.1038/jcbfm.1994.110
- Bar I, Lambert de Rouvroit C, Goffinet AM. The evolution of cortical development. An hypothesis based on the role of the Reelin signaling pathway. Trends Neurosci 2000; 23:633-638; PMID:11137154; http://dx.doi.org/10.1016/S0166-2236(00)01675-1
- Gans C, Parsons T. Biology of the Reptilia: Morphology F. New York: Academic Press; 1981.
- Kalman M, Pritz M. B. Glial fibrillary acidic protein-immunopositive structures in the brain of a Crocodilian, Caiman crocodilus, and its bearing on the evolution of astroglia. J Comp Neurol 2001; 431:460-480; PMID:11223815; http://dx.doi.org/10.1002/1096-9861(20010319)431:4%3c460::AID-CNE1083%3e3.0.CO;2-H
