Abstract
We have recently explored the in vivo functional and oncologic relevance of Lon protease (LONP1), an enzyme involved in mitochondrial quality control. We found that LONP1 is an essential protein for life and that it also performs a critical function in tumorigenesis by regulating the bioenergetics of cancer cells.
Abbreviations
| ACO2 | = | aconitase 2 |
| LONP1 | = | Lon protease |
| OXPHOS | = | oxidative phosphorylation system |
| TFAM | = | mitochondrial transcription factor A |
Lon protease, or LONP1, is one of the major mitochondrial quality control proteases involved in maintaining mitochondrial function and proteostasis. This serine protease is highly conserved through evolution from bacteria to eukaryotic cells, and its function has been widely studied for many years.Citation1 Previous studies have suggested that LONP1 might play a key role in the regulation of mitochondrial function because of its ability to perform proteolytic processing of essential proteins such as ACO2 and TFAM.Citation2 Furthermore, changes in the expression of LONP1 have been described during aging and in different pathological conditions, suggesting that the function of this proteolytic enzyme may be important for cellular and organismal fitness.Citation1 However, despite all this previous research, the functional relevance of LONP1 in mammals remains largely unknown.
To unveil the physiological and pathological relevance of this enzyme, we recently generated mice deficient in Lon protease.Citation3 Using these mutant mice, we have demonstrated that LONP1 is essential for viability as animals deficient in this protease show embryonic lethality during the gastrulation period. In addition, LONP1-deficient embryos exhibit a marked loss of mitochondrial DNA and are unable to develop in vitro. These results clearly demonstrate the essential role of this proteolytic enzyme for cell viability.
The importance of Lon protease has also been shown in other organisms and cell lines, in which absence or silencing of this enzyme generates multiple cellular defects and even accelerated aging.Citation4 We have further shown that downregulation of Lon protease in tumor cell lines decreases their growth rates and tumorigenic potential, indicating that LONP1 is also required for the viability of cancer cells.Citation3 In addition, a deeper analysis using melanoma cells has allowed us to conclude that the absence of Lon protease induces a mitochondrial catastrophe, which is characterized by a loss of mitochondrial structure and respiration complexes, and an increase in fragmentation and reactive oxygen species levels. These defects result in a decrease in mitochondrial respiration and function, and in a shift to a glycolytic metabolism to try to counteract the mitochondrial defects. However, despite these cellular attempts to overcome the shortfall in energy, the severe mitochondrial alterations induce a DNA-damage response that ultimately triggers the activation of a senescence phenotype, one of the hallmarks of aging and a barrier to cancer developmentCitation5 ().
Figure 1. Effects of changing levels of Lon protease in malignant cells. Schematic model summarizing the principal effects of knockdown and overexpression of LONP1 in tumor cells.
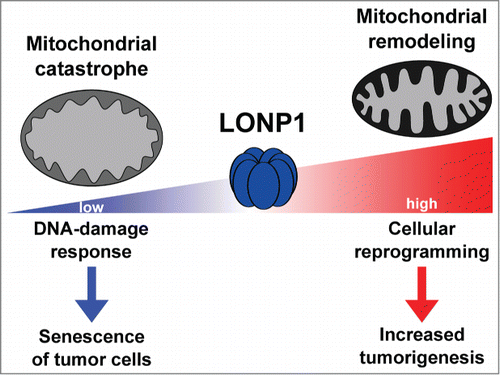
Previous data have shown that the expression of LONP1 is increased in some tumor cell lines, and that inactivation of this proteolytic enzyme could serve as a treatment for some human malignant tumors including lymphomas.Citation6 Now, using haploinsufficient mice and cellular models of gain-of-function and loss-of-function, we have shown that LONP1 activity is critical for cancer development.Citation3 Thus, a decrease in levels of the Lon protease in mice protected them against colorectal and skin tumors. Interestingly, expression data in human tumors have shown a similar correlation, with high levels of LONP1 being related to a worse prognosis in both tumor types. Furthermore, we have demonstrated that downregulation of Lon protease decreases the tumorigenic properties of cancer cells, whereas its overexpression enhances tumorigenesis. Therefore, these results allowed us to establish a link between expression and activity of Lon protease and oncogenic properties of cancer cells.Citation3 However, it was not easy to explain how changes in LONP1 expression might affect the properties of cancer cells.
In this regard, previous studies have described many substrates for LONP1, and its function has been associated with the turnover of oxidized and misfolded proteins.Citation7 However, our hypothesis addressing the functional relevance of LONP1 in cancer was related to the putative occurrence of bioenergetic alterations and mitochondrial reprogramming caused by dysregulation of this protease in cancer cells. Over the last few years, changes in mitochondrial function have attracted great interest in cancer research because of the relationship between this organelle and the metabolic reprogramming of tumor cells, one of the new hallmarks of cancer.Citation8 Tumor cells, like normal cells, adapt their metabolism depending on the energy requirements for each particular circumstance. This adaptation includes changes in mitochondrial function and respiration, which require remodeling of mitochondrial complexes and supercomplexes.Citation9 The most general metabolic reprogramming observed in tumor cells is the glycolytic switch, which induces cells to change from oxidative respiration to a glycolytic metabolism. Under these glycolytic conditions, mitochondria do not need the oxidative phosphorylation system (OXPHOS) because the cells obtain energy through glycolysis. Accordingly, the mitochondria remain as an anabolic machine generating precursors of nucleic acids and proteins.
Our experimental work in this context has revealed that LONP1 upregulation in tumor cells induces profound changes in mitochondrial complexes and supercomplexes, leading to inactivation of mitochondrial respiration and favoring the glycolytic switch.Citation3 However, the decrease in the OXPHOS complexes is not general, as LONP1 upregulation also induces upregulation of some structural subunits of these complexes. These changes suggest that the observed metabolic remodeling would allow re-formation of the OXPHOS complexes and supercomplexes, depending on the nutritional circumstances and requirements. In addition to these changes, we have also detected the occurrence of a series of cellular reprogramming events that are mainly characterized by a remarkable increase in the levels of proteins related to gene expression, translation, and protein metabolism.Citation3 Furthermore, and contrary to the situation found in LONP1-deficient cells, we have observed protection against senescence which, together with the metabolic reprogramming, would contribute to explaining the increase in tumorigenesis observed in cells overexpressing Lon protease ().Citation3 These changes suggest a coordinated response from the mitochondria to nucleus, similar to the previously described mitochondrial retrograde signal,Citation10 based on the reprogramming of mitochondrial function.
In conclusion, we have demonstrated in vivo that Lon protease is essential for cell viability and proliferation, and that it plays a critical function in tumor cells by controlling bioenergetics. These findings, together with similar data derived from analysis of human tumors, suggest that inhibition or inactivation of LONP1 could serve as a potential treatment for melanoma and colorectal cancer.
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
References
- Venkatesh S, Lee J, Singh K, Lee I, Suzuki CK. Multitasking in the mitochondrion by the ATP-dependent Lon protease. Biochim Biophys Acta 2012; 1823:56-66; PMID:22119779; http://dx.doi.org/10.1016/j.bbamcr.2011.11.003
- Lu B, Lee J, Nie X, Li M, Morozov YI, Venkatesh S, Bogenhagen DF, Temiakov D, Suzuki CK. Phosphorylation of human TFAM in mitochondria impairs DNA binding and promotes degradation by the AAA+ Lon protease. Mol Cell 2013; 49:121-32; PMID:23201127
- Quiros PM, Espanol Y, Acin-Perez R, Rodriguez F, Barcena C, Watanabe K, Calvo E, Loureiro M, Fernandez-Garcia MS, Fueyo A, et al. ATP-dependent Lon protease controls tumor bioenergetics by reprogramming mitochondrial activity. Cell Rep 2014; 8:542-56; PMID:25017063; http://dx.doi.org/10.1016/j.celrep.2014.06.018
- Erjavec N, Bayot A, Gareil M, Camougrand N, Nystrom T, Friguet B, Bulteau AL. Deletion of the mitochondrial Pim1/Lon protease in yeast results in accelerated aging and impairment of the proteasome. Free Radic Biol Med 2013; 56:9-16; PMID:23220263; http://dx.doi.org/10.1016/j.freeradbiomed.2012.11.019
- Lopez-Otin C, Blasco MA, Partridge L, Serrano M, Kroemer G. The hallmarks of aging. Cell 2013; 153:1194-217; PMID:23746838; http://dx.doi.org/10.1016/j.cell.2013.05.039
- Bernstein SH, Venkatesh S, Li M, Lee J, Lu B, Hilchey SP, Morse KM, Metcalfe HM, Skalska J, Andreeff M, et al. The mitochondrial ATP-dependent Lon protease: a novel target in lymphoma death mediated by the synthetic triterpenoid CDDO and its derivatives. Blood 2012; 119:3321-9; PMID:22323447; http://dx.doi.org/10.1182/blood-2011-02-340075
- Bota DA, Davies KJ. Lon protease preferentially degrades oxidized mitochondrial aconitase by an ATP-stimulated mechanism. Nat Cell Biol 2002; 4:674-80; PMID:12198491; http://dx.doi.org/10.1038/ncb836
- Hanahan D, Weinberg RA. Hallmarks of cancer: the next generation. Cell 2011; 144:646-74; PMID:21376230; http://dx.doi.org/10.1016/j.cell.2011.02.013
- Lapuente-Brun E, Moreno-Loshuertos R, Acin-Perez R, Latorre-Pellicer A, Colas C, Balsa E, Perales-Clemente E, Quiros PM, Calvo E, Rodriguez-Hernandez MA, et al. Supercomplex assembly determines electron flux in the mitochondrial electron transport chain. Science 2013; 340:1567-70; PMID:23812712; http://dx.doi.org/10.1126/science.1230381
- Guha M, Avadhani NG. Mitochondrial retrograde signaling at the crossroads of tumor bioenergetics, genetics and epigenetics. Mitochondrion 2013; 13:577-91; PMID:24004957; http://dx.doi.org/10.1016/j.mito.2013.08.007
