Abstract
Selective autophagic degradation of cellular components underlies many of the important physiological and pathological implications that autophagy has for mammalian cells. Cytoplasmic vesicles, just like other intracellular items, can be subjected to conventional autophagic events where double-membrane autophagosomes specifically isolate and deliver them for lysosomal destruction. However, intracellular membranes appear to constitute common platforms for unconventional versions of the autophagic pathway, a notion that has become apparent during the past few years. For instance, in many cases of autophagy directed against bacterial phagosomes, subversion of the process results in multimembrane vacuoles that promote bacterial replication instead of the usual degradative outcome. In a different atypical modality, single-membrane vesicles can be labeled with LC3 to direct their contents for lysosomal degradation. In fact, single-membrane compartments of various kinds often provide an assembly site for the autophagic machinery to perform unanticipated nondegradative activities that range from localized secretion of lysosomal contents to melanosome function. Interestingly, many of these unconventional processes seem to be initiated through engagement of relevant nodes of the autophagic signaling network that, once activated, promote LC3 decoration of the targeted membrane, and some cases of inducer/receptor proteins that specifically engage those important signaling hubs have recently been described. Here we review the available examples of all autophagic variants involving membranous compartments, with a main focus on the more recently discovered unconventional phenomena where the usual degradation purpose of autophagy or its canonical mechanistic features are not completely conserved.
Autophagy
Macroautophagy (generally referred to as autophagy) is a complex cellular route that promotes the regulated degradation of cytoplasmic components and is highly conserved in all eukaryotic organisms.Citation1,Citation2 The molecular pathways that control autophagy have been well studied in yeast,Citation3 and the relevance that this phenomenon has in a wide variety of human pathophysiological processes has also fueled its detailed analysis in mammalian systems.Citation4-Citation6 This route involves cargo isolation into canonical double-membrane vesicles called autophagosomes, which in essence constitute sacs of disposable elements that eventually fuse with lysosomes to degrade their contents.Citation7 Therefore, autophagy at the minimum must involve the action of molecular machinery that promotes isolation of the item targeted for destruction and also mechanisms to label the final “garbage bag” for fusion with the lysosomal compartment.
Autophagy was initially characterized as an adaptive response to starvation.Citation4 In this biological context, it mainly acts to obtain basic constituents from random cytoplasmic components, thus redirecting nutrients to feed essential metabolic pathways.Citation8,Citation9 However, the autophagic process is rapidly upregulated as a common adaptation response to a variety of stressful situations, not only starvation.Citation10 In a number of these conditions, like organelle malfunction, genotoxic stress, or the presence of foreign invaders, autophagy functions as a degradation mechanism for specific, obsolete, or potentially harmful components, irrespective of the recycling consequences of the process.Citation11,Citation12 Autophagic destruction also plays critical housekeeping roles by proceeding at a low, constitutive level under basal conditions, a situation where it can also act with some degree of specificity by removing, for example, superfluous or damaged organelles.Citation11 Therefore, although traditionally thought to be a fairly nonspecific process that responds to the lack of nutrients, it is now known that autophagy can target specific cellular items for timely degradation.
Autophagic Machinery
Early work in yeast has facilitated discovery of the core molecular machinery involved in the autophagic response to starvation, a collection of molecules known as autophagy-related (Atg) proteins.Citation2,Citation13,Citation14 Subsequent work in higher eukaryotes has led to identification of many Atg orthologs, thus producing a relatively clear picture of how autophagy is regulated in mammalian cells.Citation4 Briefly, a protein complex containing ULK1/ATG1-MTOR-ATG13-RB1CC1/FIP200 senses starvation or stress signals and derepresses the autophagic process that is downregulated by constitutively active MTOR. In a process not fully understood, this complex promotes the translocation of a second molecular complex including PIK3C3/VPS34-BECN1/Beclin 1-ATG14 to the endoplasmic reticulum (ER) where it generates phosphatidylinositol 3-phosphate (PtdIns3P), an uncommon lipid in this compartment.Citation14 The local presence of PtdIns3P initiates autophagosome nucleation through formation of prototypical ER-associated structures called cradles or omegasomes,Citation15,Citation16 a phenomenon that may involve PtdIns3P-binding proteins like members of the WIPI family or ZFYVE1/DFCP1.Citation17,Citation18 The ER itself likely acts as a membrane source in this case, but other organelles such as mitochondria,Citation19 mitochondrial associated membranes,Citation20 or the plasma membraneCitation21 have also been suggested as membrane donors for autophagosome formation.Citation7,Citation22
Following these initiating events, membrane elongation and phagophore closure are driven by 2 ubiquitin-like modification systems that promote the covalent addition of phosphatidylethanolamine (PE) to LC3, thus producing a membrane-bound form named LC3-II.Citation2,Citation23 In these conjugation systems ATG12 and LC3 behave as ubiquitin-like modifiers, and ATG7 separately acts as an E1 enzyme for both molecules. In the next step, ATG10 functions as the E2 enzyme for the subsequent formation of an ATG12–ATG5 covalent complex in the absence of a proper E3 conjugation system, whereas ATG3 behaves as an E2 module for LC3-I. ATG12–ATG5 then binds ATG16L1 (the mammalian ortholog of yeast Atg16) to assemble a final E3 system for the conjugation of PE to the pool of LC3 brought to the vicinity through interaction between ATG3-LC3-I and ATG12.Citation24 Interestingly, whereas the ATG12–ATG5 complex suffices for this E3-ligase activity,Citation25 ATG16L1 defines the site for LC3-II generation.Citation24
LC3 associates with the autophagosomal compartment at all stages of the process, and therefore it is used as the major autophagic reporter system.Citation26 However, its precise molecular function is still unclear. Although most of the available mechanistic information has been obtained by studying LC3B, a total of 8 LC3 or LC3-like homologs are predicted to exist: LC3A (which includes 2 splicing isoforms), LC3B, LC3C, GABARAP, GABARAPL1, GABARAPL2, and GABARAPL3. It has been clearly established that LC3 lipidation is essential for elongation of the phagophore membrane and its final closure.Citation27,Citation28 Notably, the GABARAP subgroup seems to function mainly in autophagosome maturation.Citation29 However, since LC3 was originally discovered as a microtubule-associated protein,Citation30 a role for this family in directing the mature autophagosomes to fusion with the lysosomes through the cytoskeletal tracks has been invoked.Citation23 In addition, LC3 labeling of both canonical autophagosomes and single-membrane vesicles has been proposed to directly facilitate fusion with the lysosome,Citation31,Citation32 a function that might have been overlooked due to the critical upstream activity that LC3 has in autophagosome formation and maturation. This possible involvement in lysosomal fusion could explain why certain nonautophagosomal compartments (phagosomes or endosomes, for example) are targeted to the lysosome more efficiently when becoming coated with LC3.Citation33,Citation34 However, definitive proof for any of these alternative activities is still missing. Whatever the case, LC3 labeling seems to be the end-point event that commits a given vesicle to fusion with the lysosomal compartment. This notion may be more clearly illustrated by unconventional autophagic processes where LC3 mediates nondegradative activities that still involve lysosomal targeting, like the extracellular secretion of lysosomal contents required for bone resorption (see below).
Although the fundamental molecular mechanisms involved in autophagosome formation and maturation in mammalian cells have been reasonably well described, important issues remain to be clarified. For example, a full consensus is yet to be reached regarding the main membrane source for autophagosome generation and whether or not different compartments act as membrane donors depending on the initial autophagic stimulus. Mechanistically, how exactly the preinitiation complexes containing ULK1 and BECN1 interact to initiate autophagosome formation, the role of the PtdIns3P-binding proteins WIPI and ZFYVE1 in this process, how the 2 ubiquitin-like protein modification cascades that culminate with LC3-II synthesis promote membrane elongation and final phagophore closure, and whether or not there are autophagy-specific mechanisms that promote fusion of the mature autophagosome with the lysosome (and the role of LC3 in this process), are key issues still waiting to be fully resolved.
Functions of Autophagy and Relevance of Selective Autophagy
Judging from the phenotypes of Atg-deficient mice, the autophagic process has wide implications in a number of physiological and pathological processes, including tumor suppression, neurodegeneration, inflammation, and native and adaptive immunity.Citation11,Citation35,Citation36 For example, the absence of BECN1 or ATG4 increases susceptibility to tumorigenesis,Citation37-Citation39 likely because these animals are unable to fight the deleterious effects of cellular stress.Citation40 Brain-specific deletion of Atg5 or Atg7 provokes early neurodegeneration caused by accumulation of insoluble protein aggregates.Citation41,Citation42 Absence of ATG16L1 causes an intestinal inflammatory phenotype that resembles Crohn disease,Citation43-Citation45 an observation that correlates well with the existence of a human ATG16L1 allele that increases susceptibility to this pathology.Citation46-Citation48 Other roles include participation in developmental processes,Citation49 or in adaptive immunity by promoting efficient antigen presentation.Citation50 Autophagy also constitutes an innate mechanism that helps fight intracellular pathogens, such as viruses and bacteria.Citation51
Interestingly, selective autophagy has a relevant role in many of these phenotypes, and therefore is now a matter of active investigation. For example, the cancer susceptibility phenotype might be due to poor elimination of damaged organelles and the consequent overproduction of pro-inflammatory and DNA-damaging reactive oxygen species.Citation52-Citation54 Neurodegeneration in Atg-deficient mice is mediated by the toxic effects of protein aggregates that are selectively targeted to the autophagic pathway in normal conditions.Citation55,Citation56 Elimination of foreign invaders also involves their specific recognition by the autophagic machinery, whether they are loose in the cytoplasmCitation57,Citation58 or enclosed in conventional phagosomes.Citation33,Citation59
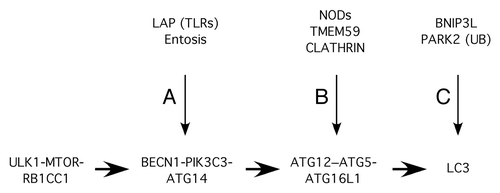
Mechanistically, these different modalities of selective autophagy seem to be executed through the action of inducer or receptor proteins that link the item targeted for destruction with certain nodes of the core autophagic machinery with the apparent final purpose of promoting LC3 labeling of the selected cargo. One of these nodes seems to be LC3 itself (). Thus, both insoluble protein aggregates and cytosolic bacteria become heavily ubiquitinated, and receptor proteins able to simultaneously bind ubiquitin and LC3 target them for autophagic degradation.Citation60 NBR1,Citation61 SQSTM1/p62,Citation62 CALCOCO2/NDP52,Citation63 and OPTN/OPTINEURINCitation64 are examples of these receptors. In addition, damaged or obsolete mitochondria become ubiquitinated by the E3-ligase PARK2/PARKIN that is recruited to the target organelle by PINK1,Citation65-Citation67 a kinase that becomes stabilized on damaged mitochondria and thus promotes PARK2-mediated ubiquitination of a wide array of mitochondrial proteins.Citation68 The labeled mitochondria are then recognized by SQSTM1/p62 for autophagic delivery.Citation69 Mitochondria can also be directed to autophagic destruction by recruiting BNIP3L/NIX, a BCL2-family protein able to bind LC3 thus acting as a receptor module.Citation70 In all these cases, interaction between the receptors and LC3 is mediated by a common LC3-interacting motif (LIR), whether it is the canonical signature (WXXL)Citation71 or an atypical motif recognized by LC3C.Citation72 Other examples of autophagic signaling nodes that are engaged (either directly or indirectly) by specific inducers are the BECN1-PIK3C3 and ATG12–ATG5-ATG16L1 complexes (). Evidence supporting such roles is provided throughout this review.
Figure 1. Relevant nodes of the canonical autophagic pathway that are specifically engaged in different modalities of unconventional autophagy against membranous compartments. The canonical autophagic pathway flows from the ULK1-MTOR-RB1CC1 complex to LC3-II synthesis (horizontal arrows). The BECN1-PIK3C3-ATG14 complex is directly or indirectly activated during LAP and entosis (A). NODs and TMEM59 (and perhaps CLATHRIN) induce the ATG12–ATG5-ATG16L1 complex by interacting with ATG16L1 (B). BNIP3L/NIX and PARK2 (through ubiquitination, UB) engage the pathway at the level of LC3 (C). The end result in all cases is LC3 labeling of the targeted membranous structure.
Selective Autophagy Against Membrane Compartments with Canonical Degradation Purposes
Membrane-bound intracellular organelles can be subjected to conventional autophagic processes where regular autophagosomes form around them (or fuse with them) thus producing multimembrane vesicles destined for lysosomal destruction (). For instance, phagosomes whose natural maturation pathway is stalled by Mycobacteria are targeted by regular autophagosomal compartments that fuse with the bacteria-containing sac, an autophagic response that suppresses survival of the invader.Citation59 A similar phenomenon involving multiple-membrane vesicular structures targets the Salmonella-containing vacuole in epithelial cells, thus inhibiting bacterial replication. An atypical route is involved here, since this process shows independency of the classical mediators PIK3C3, RB1CC1, and ATG9,Citation73 and entails recognition of damaged phagosomes by carbohydrate-binding proteins.Citation74 As mentioned above, damaged mitochondria can also be eliminated by canonical autophagy mediated by ubiquitination. Stressed portions of the endoplasmic reticulum are degraded in regular autophagosomes containing lamellar structures that stain positive for reticular markers.Citation75,Citation76 Similarly, excess peroxisomesCitation77 or damaged lysosomesCitation78 in mammalian cells are cleared through an autophagic process that encloses these organelles within double-membrane autophagosomes.
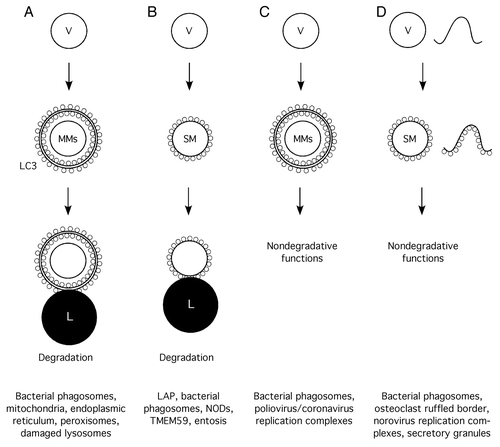
Figure 2. Scheme of the different modalities of autophagy involving membrane compartments and their functional consequences (bottom). (A) Regular, single-membrane vesicles are targeted by conventional autophagy to produce multimembrane vacuoles that fuse with lysosomes for degradation of their contents. (B) Regular, single-membrane vesicles become directly labeled with LC3-II and eventually fuse with lysosomes for degradation. (C) Regular, single-membrane vesicles are targeted by conventional autophagy producing multimembrane vacuoles with nondegradative functions. (D) Regular, single-membrane vesicles or other membranous structures become directly labeled with LC3-II for a variety of nondegradative functions. (V, vesicle; MMs, multiple membranes; SM, single membrane; L, lysosome).
However, while autophagic targeting of conventional cytoplasmic substrates needs formation of a membranous compartment that isolates the doomed item, autophagy against the contents of a vesicular structure could conceivably spare cargo isolation and rely only on LC3 labeling of an already existing vesicle. In fact, some recently described examples support this novel idea (). In the following we review the new developments on this topic.
Phagosomes containing stimulated TLR2 (toll-like receptor 2) become rapidly decorated with BECN1 to promote LC3 labeling of this nonautophagic single-membrane compartment, a process that promotes more efficient acquisition of lysosomal features.Citation33 This phenomenon has been called LC3-associated phagocytosis (LAP) and it has since been observed in different settings, including engulfment of dead cells,Citation79 phagocytosis of Burkholderia pseudomallei,Citation80 stimulation of phagosomal Fcγ-receptors,Citation81 efficient formation of phagosomal IRF7 (interferon regulatory factor 7)-signaling compartments in response to TLR9 stimulation by DNA immune complexes,Citation82 circadian degradation of phagocytosed photoreceptors by retinal pigment epithelial cells,Citation83 or various phagocytic phenomena in dendritic cells that favor presentation of major histocompatibility complex class II-restricted antigens.Citation50 However, the notion that certain TLRs activate autophagy to fight intracellular invaders has also been linked to the induction of general autophagy.Citation59,Citation84,Citation85 Therefore, some controversy has arisen as to whether LAP could in fact involve distant generation of autophagosomes through the regular route followed by a quick fusion event with the phagosome. This possibility could explain why induction of general autophagy might have some of the beneficial effects of selective autophagy in this context, as has been previously noted.Citation10 However, translocation of BECN1 to the phagosome and labeling with LC3 are extremely rapid events in LAP, and no evidence of multiple membranes is detected.Citation33 These observations argue against the fusion model and favor the involvement of an in situ LC3-II synthesis event. Still, unequivocally ruling out that activated TLR2 first induces formation of canonical autophagosomes that then rapidly fuse with the targeted phagosome may not be easy. Perhaps the most convincing evidence is that LAP proceeds independently of some of the initiating autophagic machinery (ULK1)Citation79 and so it seems mechanistically different from other examples where stimulation of general autophagy (which does involve ULK1) has antibacterial effects.Citation59 It is entirely possible, however, that both autophagic mechanisms contribute to the same goal in this physiological context.
In any case, the notion that LC3-II can be synthesized in situ to label single-membrane structures has been raised again in other experimental systems where different inducer molecules act by directly engaging ATG16L1. As mentioned above, ATG16L1 defines the site of LC3 lipidation by bringing both LC3 and the LC3-II-synthesis complex (ATG12–ATG5 and ATG3-LC3) close to a membrane source,Citation24 so it is conceivable that ATG16L1 may have a relevant role in autophagy directed to membranous compartments. Thus, ATG16L1 might constitute an additional node for selective autophagy with a particularly important function in autophagy directed against intracellular membranes. In fact, the existence of specific factors that recruit ATG16L1 to the proper sites for LC3-II synthesis has been suggested before.Citation14,Citation24,Citation73 Examples of such factors have recently become available.
For instance, the native immune receptors NOD1 and NOD2 recognize bacteria (Shigella) at the entry site and engage ATG16L1 to promote LC3 labeling of the nascent phagosome.Citation86 Although it is unclear if acquisition of LC3 in this context results from fusion with preexisting autophagosomes or from local LC3-II synthesis, NODs probably constitute the first description of an ATG16L1 receptor molecule (). In an additional example, it has recently been described that the transmembrane molecule TMEM59 holds a minimal 19 amino acid stretch in its intracellular domain that promotes LC3 labeling of the same single-membrane endosomes where the peptide becomes activated by aggregation.Citation34 This minimal peptide can be reduced to a 4-amino acid motif whose integrity is necessary for the activity. The LC3-labeled endosomes undergo more efficient lysosomal degradation of their contents, thus recapitulating some of the main functional consequences of LAP. Mechanistically, the active peptide interacts with ATG16L1 through the minimal motif, suggesting that TMEM59 may be another example of an ATG16L1 receptor involved in selective autophagy against single-membrane vesicular compartments (). Again, distinguishing between mechanisms involving fusion with canonical autophagic vesicles vs. local LC3 lipidation is a relevant issue. The LC3-labeled vesicles do not show evidence of multiple membranes and appear devoid of cellular material in electron microscopy (EM) studies, suggesting that they do not undergo fusion events with preexisting autophagosomes to become LC3-positive. Consistent with this, there is a very tight colocalization between GFP-LC3 and endocytosed TMEM59, and distal formation of autophagosomes in response to TMEM59 activation is not detected. The process relies on the canonical autophagic effectors ATG5 and ATG7,Citation34 but appears to proceed independently of BECN1 (unpublished observations), arguing that TMEM59 bypasses the upstream autophagic machinery, just like LAP does at a different level. From a mechanistic point of view, this independence of upstream activators is the logical consequence of directly engaging ATG16L1 and, with it, the LC3-II synthesis machinery (ATG12–ATG5 and ATG3-LC3) that is normally assembled with ATG16L1 in a macromolecular complex.Citation87 Proximity between this machinery and a membrane source would favor local (in situ) lipidation of LC3 and, therefore, LC3 labeling of the same vesicle where activation of TMEM59 takes place.
In an effort to evaluate a possible interaction of TMEM59 with the canonical autophagic pathway, the authors tested if TMEM59 overexpression (or forced aggregation of chimeric transmembrane molecules) was able to influence the clearance rate of established autophagic substrates like SQSTM1/p62, NBR1 or insoluble protein aggregates formed by a pathological form (Q74) of HTT/huntingtin. The rationale here is that any possible intersection with the conventional route should result in altered levels of molecules that are normally cleared through the same pathway. While TMEM59 activation provokes substantial LC3-II accumulation, it does not alter at all the levels of these substrates, indicating that TMEM59 neither blocks nor accelerates the canonical autophagic flux. This is actually somewhat surprising since, even assuming that TMEM59 does not participate at all in the regular route, one would expect that its overexpression could sequester at least a fraction of the autophagic machinery (the ATG16L1 complex, in this case) to cause some depression of the canonical autophagic flux. Why such an effect (even a marginal one) does not occur is unclear, but the notion that separate pools of critical effectors (ATG16L1) are involved in different autophagic-like processes might be worth considering in future studies. In any event, such tight segregation between the 2 autophagic events may provide a methodological advantage to clearly show dissociation of an atypical autophagic phenomenon from the canonical pathway. Thus, apart from establishing whether or not it involves single membranes (EM) or different autophagic machinery (independence of upstream effectors), a routine assay could involve testing a possible impact on recognized substrates of canonical autophagy.
ATG16L1 is substantially larger than the yeast ortholog,Citation87 raising the notion that the molecule has evolved structurally to acquire new functions. In particular, a C-terminal domain containing 7 WD40-type repetitions has been added during evolution, the function of which has been unclear. This domain is dispensable for conventional autophagy, because a version of ATG16L1 lacking this portion fully sustains the pathway in mammalian cells.Citation88,Citation89 Notably, the ATG16L1-binding motif found in TMEM59 recognizes the WD40 domain of ATG16L1, thus ascribing for the first time a molecular function to this region.Citation34 Since different versions of the motif are conserved with the same ATG16L1-binding functionality in different unrelated proteins,Citation34 the existence of a whole family of ATG16L1 receptors that share this structural feature and engage ATG16L1 in response to different stimuli appears as a likely possibility. Accessing the whole collection of such receptors would help expand the field of selective autophagy against membranous compartments, and might shed light into the role of ATG16L1 in some of the nonautophagic biological situations where it has been involved (see below).
Endogenous TMEM59 mediates the autophagic response triggered by Staphylococcus aureus at early infection times when the bacteria are enclosed in single-membrane phagosomes.Citation34 These data are consistent with a relevant role of ATG16L1 in the autophagic burst elicited by bacterial invasion, a notion further underscored by the established function of ATG16L1 in the antibacterial activity of NOD1 and NOD2 (see above). In fact, ATG16L1 seems to be important in intestinal homeostasis, a biological environment that needs proper management of the bacterial flora naturally inhabiting the digestive tract. This idea was initially suggested by the fact that a polymorphic form of ATG16L1 (T300A) increases the risk of suffering Crohn disease,Citation46-Citation48 and is further supported by the Crohn disease-like defects exhibited by Atg16l1-deficient mice.Citation43-Citation45 Thus, these results point to a relevant role of ATG16L1 in antibacterial immunity, and raise the notion that its WD40 domain may have arisen during evolution to fulfill more sophisticated autophagic functions that are exclusive to multicellular organisms, like native immunity. More generally, a similar principle may have driven the increased complexity of the autophagic machinery that is observed in higher eukaryotes compared with yeast cells, with additional ATGs, more ATG isoforms and structural changes in certain ATGs (like addition of the WD40 domain in ATG16L1), all of which may have facilitated adaptation of the autophagic process to new functions. Although TMEM59 mediates autophagy induced by Staphylococcus aureus, it does not seem to have a general role in autophagy directed to bacterial phagosomes, because only an effect during infection with this bacteria, but not other strains, was found (unpublished observations). Interestingly, TMEM59 was identified in a proteomics study of phagosomes containing inert latex beads,Citation90 so the described function in Staphylococcus phagosomes might reflect a more general role in vesicle trafficking.
Entosis constitutes an additional example of single-membrane vesicles that become labeled with LC3. This is a specialized form of phagocytosis where whole live cells are engulfed by neighboring cells in a striking process that eventually causes nonapoptotic death of the internalized cell.Citation91 This phenomenon involves formation of single-membrane phagosomes that quickly become LC3-positive independently of conventional autophagosomes.Citation31 Thus, no evidence of fusion with autophagic compartments or multiple membranes was detected, and labeling with GFP-LC3 is achieved at the expense of the diffuse pool of the marker but not preexisting GFP-LC3-positive autophagosomes. Again, this is surprising, since one would expect that engagement of the autophagic machinery by the phagosome would remove important signaling effectors from the main autophagic route. LC3 recruitment to the phagosome is dependent on PIK3C3, ATG5 and ATG7, but independent of RB1CC1, suggesting mechanistic differences with canonical autophagy and similarities with LAP. The fact that both LAP and entosis engage the autophagic route at the level of BECN1-PIK3C3 indicates that this complex may constitute an additional relevant node for selective autophagy of membranous compartments, aside from ATG16L1 (). Engulfment of apoptotic bodies and macropinosomes trigger a similar process.Citation31 Because all these entotic-like, LC3-associated phagocytic events involve internalization of self material, it is unlikely that they are triggered by TLR activation, suggesting that TLR-independent mechanisms contribute to LC3 activation in entosis. It would be very interesting to know the mechanisms involved in recruitment of the autophagic machinery to the entotic vesicle. Of note, ATG16L1 interacts with CLATHRIN,Citation21 so a direct interaction between the endocytic and autophagic machineries could explain how entotic-like vesicles become LC3-positive. CLATHRIN might, therefore, constitute an additional example of ATG16L1 receptor ().
Selective Autophagic Labeling of Membrane Compartments with Unconventional, Nondegradative Purposes
From a purely etymological point of view, the processes described in the previous section fit into the general category of “autophagy,” since, for the most part, they result in degradation (phagy) of intracellular components (auto) ( and ). However, in this section we now include an expanding collection of phenomena where intracellular membranes become decorated with LC3 or other autophagic effectors without resulting in digestion of the labeled compartment ( and ). Whether or not these constitute autophagic processes is debatable, and their recent discovery may invoke the need for a more strict definition of what the term autophagy really defines. As discussed below, some of these phenomena may be particularly informative regarding the functional roles of some autophagic effectors, LC3 in particular.
Bacterial phagosomes are in some cases targeted by regular autophagy to produce multimembrane LC3-positive compartments that, instead of promoting degradation of the invader, appear to provide a niche for bacterial replication ().Citation58 For example, Yersinia resides in autophagosomal, multimembrane, LC3-positive, nonacidic vesicles that sustain bacterial proliferation.Citation92,Citation93 In addition, phagosomes containing Staphylococcus aureus are surrounded by multiple membranous structures that become LC3-positive around 3 h post infection, an autophagic phenomenon that favors replication of the pathogen.Citation94 Similarly, Brucella abortus traffics to multimembrane vesicles whose formation depends on the upstream autophagic machinery (BECN1, ULK1), but not downstream ATGs. These structures favor cell-to-cell spreading of the bacteria instead of its killing.Citation95 Poliovirus utilizes LC3-positive, double-membrane vesicles as replication sites, and inhibition of the autophagic pathway decreases viral yield.Citation96 Coronaviruses assemble their replicative complexes on the surface of double-membrane vesicles that become decorated with nonlipidated LC3 without the involvement of canonical autophagic mediators, and this unconventional LC3 labeling serves viral replication.Citation97
Single-membrane phagosomes containing a number of bacteria also acquire autophagic features that promote survival of the enclosed bacterium, rather than its elimination (). For example, Coxiella burnetii is internalized in a spacious phagosome whose maturation is delayed by the bacteria. This structure ends up acquiring autophagic traits that are reminiscent of LAP (decoration with BECN1 and LC3) and help increase replication of the pathogen.Citation98,Citation99 Similarly, a fraction of intracellular Listeria reside in LC3-positive single-membrane compartments that facilitate its proliferation, thus causing persistent infection.Citation100 The mechanisms explaining how these single-membrane phagosomes become LC3-positive, and why this process results in bacterial survival, remain to be investigated in detail.
Examples involving assembly of the autophagic machinery on single-membrane structures for additional nondegradative outcomes have recently been described (), and the diversity of processes where this phenomenon has been observed was actually unexpected. For instance, bone resorption entails the localized secretion of lysosomal contents through a specialized “ruffled border” of polarized osteoclasts that is apposed to the bone surface, and recent evidence indicates that some autophagic mediators play a critical role in this phenomenon.Citation101 Thus, the ruffled border becomes labeled with LC3 in a process mediated by conventional autophagic effectors (ATG5, ATG7 or ATG4). No evidence of double membranes was found in this specialized membrane domain, whose full maturation actually depends on the autophagic machinery. The nature of the initiating signal that promotes specific LC3 labeling of this region is unknown, but the involvement of specific inducers or receptors that locally engage nodal autophagic mediators appears as a plausible mechanism.
What the exact role of LC3 might be at this location is currently unclear. However, it is tempting to speculate that LC3 labels the ruffled border to promote the directed secretion of lysosomal contents. This possibility would point to a direct role of LC3 in promoting fusion with the lysosome that might have remained unidentified in the canonical route owing to the critical activities that LC3 family members play in previous steps of phagophore and autophagosome biogenesis (membrane elongation and closure, and autophagosome maturation). A phenomenon that is implicitly involved in the autophagic pathway is the final fusion of the mature autophagosome with the lysosomal compartment, so it is conceivable that the same machinery mediating this final step works in other cellular processes where lysosome fusion with membranous structures is also required.Citation31,Citation32,Citation102 This possible function of LC3 is particularly well supported by examples where LC3 labeling has unconventional, nonautophagic outcomes that still require lysosomal delivery, as happens in osteoclasts.
The antiviral activity provided by IFNG during norovirus infection involves a nondegradative role of the ATG12–ATG5-ATG16L1 complex that was recently identified.Citation103 This unconventional function proceeds with ATG16L1 localization in membranous structures that lack an obvious resemblance to autophagosomes and provide a niche for assembly of the norovirus replication complex.Citation103,Citation104 Although the precise role of ATG16L1 at this location remains to be determined, the notion that a specific factor recruits this mediator for a nondegradative function is again conceivable.
A role of some autophagic effectors in secretion processes can be inferred from additional recent data. Single-membrane secretory granules of the neuroendocrine PC12 cell line stain positive for ATG16L1, and depletion of this effector results in defective hormone secretion.Citation105 Since LC3 does not associate with the same granules, the function of ATG16L1 is probably unrelated to the orthodox autophagic pathway in this context. This report is in line with a previous one showing that both ATG16L1 hypomorphic mice and Crohn disease patients harboring the risk allele ATG16L1-T300A exhibit secretory defects in Paneth cells, a specialized intestinal cell that discharges protective peptides to the intestinal lumen.Citation43 The defective cells emit less lysozyme and show a hypervacuolated cytoplasm owing to a disorganized secretory granule compartment. Whether or not ATG16L1 itself and/or LC3 are present in the secretory vesicles of these cells has not been carefully examined, but the phenotype is similar to the one caused by ATG16L1 depletion in PC12 cells, so both experimental systems might share the same mechanism to target ATG16L1 to the relevant vesicles. A role of autophagy in the unconventional secretion of IL1B has also recently been proposed.Citation106 Although whether or not this process is linked to single-membrane vesicles has not been explored, it does involve colocalization between the IL1B-containing structures and LC3. The involvement of autophagy in unconventional secretion of otherwise cytoplasmic molecules was first described in yeast cells,Citation107,Citation108 suggesting that this is an evolutionarily conserved process.
In a different example, melanosomes are single-membrane vesicles that contain the pigment melanin and colocalize with ATG5 and LC3. In fact, proper function of these vacuoles seems to rely on autophagic effectors such as WIPI1, BECN1 or ATG5, as revealed by identification of these proteins in a siRNA-based screen for melanosome activity.Citation109 The mechanisms that promote assembly of the autophagic machinery on these membranes are unknown, but the existence of specific inducer molecules involved in this job appears as a likely possibility once again.
Taken together, the examples mentioned in this section illustrate that intracellular membranes appear to constitute common assembly sites for the autophagic machinery to carry out biological functions not directly linked to the canonical degradation route. In addition, many of these processes appear to share a common link to vesicle trafficking, a finding that may reflect a more general role of the autophagic machinery in this basic cellular process.Citation110,Citation111 Since autophagy is, in essence, a vesicular trafficking phenomenon, the alternative use of some autophagic effectors in other nonautophagic trafficking events is perhaps not surprising.
Summary and Perspectives
Here we have reviewed a collection of examples showing that intracellular membranes constitute a particularly flexible stage for the action of the autophagic machinery with both degradative and nondegradative purposes. In the most conventional version of the process, intracellular vesicles can be subject to canonical autophagic phenomena where the relevant vacuole is treated just like any other autophagic substrate, and the end product is a multimembrane compartment that eventually undergoes fusion with the lysosome (). However, an expanding range of examples show, first, that noncanonical single-membrane vesicles can be dominantly committed to lysosomal fusion through the action of autophagic machinery (), second, that canonical multimembrane autophagosomes may serve nondegradative purposes () and, third, that unconventional membranous structures often constitute assembly sites for functional autophagic modules that promote a wide variety of nondegradative activities (). Why intracellular membranes constitute such a versatile platform is unclear, but it is conceivable that, in a way, the autophagic machinery just represents a subgroup of the membrane-trafficking machinery, and that all this complexity arises from the intersection between both basic pathways. A few key issues in the field deserve particular attention. One is the identification of the mechanisms that turn a theoretically degradative compartment into a cozy niche for microbial replication/survival. Reaching a full consensus on whether LC3 labeling of single-membrane compartments involves fusion with preexisting LC3-positive structures or is mainly due to local LC3-II synthesis, is also an important aspect. We anticipate that most of the unconventional processes included here are likely to be regulated by the action of molecular receptors that promote assembly of specific nodal cassettes of the autophagic machinery on the relevant membrane sites. Identification of these receptors constitutes an important and interesting matter. From the available literature, significant autophagic nodes in this context appear to be the BECN1-PIK3C3 and ATG12–ATG5-ATG16L1 complexes, and perhaps LC3 in some cases such as damaged mitochondria (). We predict that a wide variety of receptor/inducer molecules able to engage these or other yet to be discovered nodal effectors will be identified in the near future.
| Abbreviations: | ||
| ATG | = | autophagy-related |
| ATG16L1 | = | autophagy-related 16-like 1 |
| BECN1 | = | Beclin 1, autophagy-related |
| GABARAP | = | GABA(A) receptor-associated protein |
| GABARAPL | = | GABA(A) receptor-associated protein like |
| GFP | = | green fluorescent protein |
| LAP | = | LC3-associated phagocytosis |
| LC3 | = | microtubule-associated protein 1 light chain 3 |
| MTOR | = | mechanistic target of rapamycin |
| NOD | = | nucleotide-binding oligomerization domain containing |
| PARK2 | = | parkin RBR E3 ubiquitin protein ligase |
| PIK3C3 phosphatidylinositol 3-kinase | = | catalytic subunit type 3 |
| PtdIns3P | = | phosphatidylinositol 3-phosphate |
| RB1CC1 | = | RB1-inducible coiled-coil 1 |
| siRNA | = | small interfering RNA |
| SQSTM1 | = | sequestosome 1 |
| TLR | = | toll-like receptor |
| TMEM59 | = | transmembrane protein 59 |
| ULK1 | = | unc-51 like autophagy activating kinase 1 |
| WIPI | = | WD repeat domain, phosphoinositide interacting |
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Acknowledgments
Funding was provided by grants from the Ministerio de Ciencia e Innovación of the Spanish Government (Refs. SAF2008-00350 and SAF2011-23714), the Fundación Solórzano, the Junta de Castilla y León local government (Consejería de Educación, Ref. CSI001A10-2, and Consejería de Sanidad) and the Consejo Superior de Investigaciones Científicas (CSIC; Ref. 200720I026). Additional funding comes from the FEDER program of the European Union. EB is a graduate student funded by a predoctoral fellowship from the FPU program (Ministerio de Educación, MEC, Spanish Government). FXP holds a tenured position at the CSIC.
References
- Mizushima N. Autophagy: process and function. Genes Dev 2007; 21:2861 - 73; http://dx.doi.org/10.1101/gad.1599207; PMID: 18006683
- Yang Z, Klionsky DJ. An overview of the molecular mechanism of autophagy. Curr Top Microbiol Immunol 2009; 335:1 - 32; http://dx.doi.org/10.1007/978-3-642-00302-8_1; PMID: 19802558
- Inoue Y, Klionsky DJ. Regulation of macroautophagy in Saccharomyces cerevisiae. Semin Cell Dev Biol 2010; 21:664 - 70; http://dx.doi.org/10.1016/j.semcdb.2010.03.009; PMID: 20359542
- Yang Z, Klionsky DJ. Mammalian autophagy: core molecular machinery and signaling regulation. Curr Opin Cell Biol 2010; 22:124 - 31; http://dx.doi.org/10.1016/j.ceb.2009.11.014; PMID: 20034776
- Choi AM, Ryter SW, Levine B. Autophagy in human health and disease. N Engl J Med 2013; 368:651 - 62; http://dx.doi.org/10.1056/NEJMra1205406; PMID: 23406030
- Rubinsztein DC, Codogno P, Levine B. Autophagy modulation as a potential therapeutic target for diverse diseases. Nat Rev Drug Discov 2012; 11:709 - 30; http://dx.doi.org/10.1038/nrd3802; PMID: 22935804
- Rubinsztein DC, Shpilka T, Elazar Z. Mechanisms of autophagosome biogenesis. Curr Biol 2012; 22:R29 - 34; http://dx.doi.org/10.1016/j.cub.2011.11.034; PMID: 22240478
- Singh R, Cuervo AM. Autophagy in the cellular energetic balance. Cell Metab 2011; 13:495 - 504; http://dx.doi.org/10.1016/j.cmet.2011.04.004; PMID: 21531332
- Rabinowitz JD, White E. Autophagy and metabolism. Science 2010; 330:1344 - 8; http://dx.doi.org/10.1126/science.1193497; PMID: 21127245
- Kroemer G, Mariño G, Levine B. Autophagy and the integrated stress response. Mol Cell 2010; 40:280 - 93; http://dx.doi.org/10.1016/j.molcel.2010.09.023; PMID: 20965422
- Mizushima N, Komatsu M. Autophagy: renovation of cells and tissues. Cell 2011; 147:728 - 41; http://dx.doi.org/10.1016/j.cell.2011.10.026; PMID: 22078875
- Komatsu M, Ichimura Y. Selective autophagy regulates various cellular functions. Genes Cells 2010; 15:923 - 33; http://dx.doi.org/10.1111/j.1365-2443.2010.01433.x; PMID: 20670274
- Nakatogawa H, Suzuki K, Kamada Y, Ohsumi Y. Dynamics and diversity in autophagy mechanisms: lessons from yeast. Nat Rev Mol Cell Biol 2009; 10:458 - 67; http://dx.doi.org/10.1038/nrm2708; PMID: 19491929
- Mizushima N, Yoshimori T, Ohsumi Y. The role of Atg proteins in autophagosome formation. Annu Rev Cell Dev Biol 2011; 27:107 - 32; http://dx.doi.org/10.1146/annurev-cellbio-092910-154005; PMID: 21801009
- Axe EL, Walker SA, Manifava M, Chandra P, Roderick HL, Habermann A, Griffiths G, Ktistakis NT. Autophagosome formation from membrane compartments enriched in phosphatidylinositol 3-phosphate and dynamically connected to the endoplasmic reticulum. J Cell Biol 2008; 182:685 - 701; http://dx.doi.org/10.1083/jcb.200803137; PMID: 18725538
- Hayashi-Nishino M, Fujita N, Noda T, Yamaguchi A, Yoshimori T, Yamamoto A. A subdomain of the endoplasmic reticulum forms a cradle for autophagosome formation. Nat Cell Biol 2009; 11:1433 - 7; http://dx.doi.org/10.1038/ncb1991; PMID: 19898463
- Itakura E, Mizushima N. Characterization of autophagosome formation site by a hierarchical analysis of mammalian Atg proteins. Autophagy 2010; 6:764 - 76; http://dx.doi.org/10.4161/auto.6.6.12709; PMID: 20639694
- Polson HE, de Lartigue J, Rigden DJ, Reedijk M, Urbé S, Clague MJ, Tooze SA. Mammalian Atg18 (WIPI2) localizes to omegasome-anchored phagophores and positively regulates LC3 lipidation. Autophagy 2010; 6:506 - 22; http://dx.doi.org/10.4161/auto.6.4.11863; PMID: 20505359
- Hailey DW, Rambold AS, Satpute-Krishnan P, Mitra K, Sougrat R, Kim PK, Lippincott-Schwartz J. Mitochondria supply membranes for autophagosome biogenesis during starvation. Cell 2010; 141:656 - 67; http://dx.doi.org/10.1016/j.cell.2010.04.009; PMID: 20478256
- Hamasaki M, Furuta N, Matsuda A, Nezu A, Yamamoto A, Fujita N, Oomori H, Noda T, Haraguchi T, Hiraoka Y, et al. Autophagosomes form at ER-mitochondria contact sites. Nature 2013; 495:389 - 93; http://dx.doi.org/10.1038/nature11910; PMID: 23455425
- Ravikumar B, Moreau K, Jahreiss L, Puri C, Rubinsztein DC. Plasma membrane contributes to the formation of pre-autophagosomal structures. Nat Cell Biol 2010; 12:747 - 57; http://dx.doi.org/10.1038/ncb2078; PMID: 20639872
- Hamasaki M, Shibutani ST, Yoshimori T. Up-to-date membrane biogenesis in the autophagosome formation. Curr Opin Cell Biol 2013; 25:455 - 60; http://dx.doi.org/10.1016/j.ceb.2013.03.004; PMID: 23578367
- Noda T, Fujita N, Yoshimori T. The late stages of autophagy: how does the end begin?. Cell Death Differ 2009; 16:984 - 90; http://dx.doi.org/10.1038/cdd.2009.54; PMID: 19424283
- Fujita N, Itoh T, Omori H, Fukuda M, Noda T, Yoshimori T. The Atg16L complex specifies the site of LC3 lipidation for membrane biogenesis in autophagy. Mol Biol Cell 2008; 19:2092 - 100; http://dx.doi.org/10.1091/mbc.E07-12-1257; PMID: 18321988
- Hanada T, Noda NN, Satomi Y, Ichimura Y, Fujioka Y, Takao T, Inagaki F, Ohsumi Y. The Atg12-Atg5 conjugate has a novel E3-like activity for protein lipidation in autophagy. J Biol Chem 2007; 282:37298 - 302; http://dx.doi.org/10.1074/jbc.C700195200; PMID: 17986448
- Mizushima N, Yoshimori T, Levine B. Methods in mammalian autophagy research. Cell 2010; 140:313 - 26; http://dx.doi.org/10.1016/j.cell.2010.01.028; PMID: 20144757
- Fujita N, Hayashi-Nishino M, Fukumoto H, Omori H, Yamamoto A, Noda T, Yoshimori T. An Atg4B mutant hampers the lipidation of LC3 paralogues and causes defects in autophagosome closure. Mol Biol Cell 2008; 19:4651 - 9; http://dx.doi.org/10.1091/mbc.E08-03-0312; PMID: 18768752
- Sou YS, Waguri S, Iwata J, Ueno T, Fujimura T, Hara T, Sawada N, Yamada A, Mizushima N, Uchiyama Y, et al. The Atg8 conjugation system is indispensable for proper development of autophagic isolation membranes in mice. Mol Biol Cell 2008; 19:4762 - 75; http://dx.doi.org/10.1091/mbc.E08-03-0309; PMID: 18768753
- Weidberg H, Shvets E, Shpilka T, Shimron F, Shinder V, Elazar Z. LC3 and GATE-16/GABARAP subfamilies are both essential yet act differently in autophagosome biogenesis. EMBO J 2010; 29:1792 - 802; http://dx.doi.org/10.1038/emboj.2010.74; PMID: 20418806
- Kuznetsov SA, Gelfand VI. 18 kDa microtubule-associated protein: identification as a new light chain (LC-3) of microtubule-associated protein 1 (MAP-1). FEBS Lett 1987; 212:145 - 8; http://dx.doi.org/10.1016/0014-5793(87)81574-0; PMID: 3803603
- Florey O, Kim SE, Sandoval CP, Haynes CM, Overholtzer M. Autophagy machinery mediates macroendocytic processing and entotic cell death by targeting single membranes. Nat Cell Biol 2011; 13:1335 - 43; http://dx.doi.org/10.1038/ncb2363; PMID: 22002674
- Florey O, Overholtzer M. Autophagy proteins in macroendocytic engulfment. Trends Cell Biol 2012; 22:374 - 80; http://dx.doi.org/10.1016/j.tcb.2012.04.005; PMID: 22608991
- Sanjuan MA, Dillon CP, Tait SW, Moshiach S, Dorsey F, Connell S, Komatsu M, Tanaka K, Cleveland JL, Withoff S, et al. Toll-like receptor signalling in macrophages links the autophagy pathway to phagocytosis. Nature 2007; 450:1253 - 7; http://dx.doi.org/10.1038/nature06421; PMID: 18097414
- Boada-Romero E, Letek M, Fleischer A, Pallauf K, Ramón-Barros C, Pimentel-Muiños FX. TMEM59 defines a novel ATG16L1-binding motif that promotes local activation of LC3. EMBO J 2013; 32:566 - 82; http://dx.doi.org/10.1038/emboj.2013.8; PMID: 23376921
- Cecconi F, Levine B. The role of autophagy in mammalian development: cell makeover rather than cell death. Dev Cell 2008; 15:344 - 57; http://dx.doi.org/10.1016/j.devcel.2008.08.012; PMID: 18804433
- Levine B, Kroemer G. Autophagy in the pathogenesis of disease. Cell 2008; 132:27 - 42; http://dx.doi.org/10.1016/j.cell.2007.12.018; PMID: 18191218
- Mariño G, Salvador-Montoliu N, Fueyo A, Knecht E, Mizushima N, López-Otín C. Tissue-specific autophagy alterations and increased tumorigenesis in mice deficient in Atg4C/autophagin-3. J Biol Chem 2007; 282:18573 - 83; http://dx.doi.org/10.1074/jbc.M701194200; PMID: 17442669
- Liang XH, Jackson S, Seaman M, Brown K, Kempkes B, Hibshoosh H, Levine B. Induction of autophagy and inhibition of tumorigenesis by beclin 1. Nature 1999; 402:672 - 6; http://dx.doi.org/10.1038/45257; PMID: 10604474
- Qu X, Yu J, Bhagat G, Furuya N, Hibshoosh H, Troxel A, Rosen J, Eskelinen EL, Mizushima N, Ohsumi Y, et al. Promotion of tumorigenesis by heterozygous disruption of the beclin 1 autophagy gene. J Clin Invest 2003; 112:1809 - 20; PMID: 14638851
- White E, Karp C, Strohecker AM, Guo Y, Mathew R. Role of autophagy in suppression of inflammation and cancer. Curr Opin Cell Biol 2010; 22:212 - 7; http://dx.doi.org/10.1016/j.ceb.2009.12.008; PMID: 20056400
- Komatsu M, Waguri S, Chiba T, Murata S, Iwata J, Tanida I, Ueno T, Koike M, Uchiyama Y, Kominami E, et al. Loss of autophagy in the central nervous system causes neurodegeneration in mice. Nature 2006; 441:880 - 4; http://dx.doi.org/10.1038/nature04723; PMID: 16625205
- Hara T, Nakamura K, Matsui M, Yamamoto A, Nakahara Y, Suzuki-Migishima R, Yokoyama M, Mishima K, Saito I, Okano H, et al. Suppression of basal autophagy in neural cells causes neurodegenerative disease in mice. Nature 2006; 441:885 - 9; http://dx.doi.org/10.1038/nature04724; PMID: 16625204
- Cadwell K, Liu JY, Brown SL, Miyoshi H, Loh J, Lennerz JK, Kishi C, Kc W, Carrero JA, Hunt S, et al. A key role for autophagy and the autophagy gene Atg16l1 in mouse and human intestinal Paneth cells. Nature 2008; 456:259 - 63; http://dx.doi.org/10.1038/nature07416; PMID: 18849966
- Cadwell K, Patel KK, Maloney NS, Liu TC, Ng AC, Storer CE, Head RD, Xavier R, Stappenbeck TS, Virgin HW. Virus-plus-susceptibility gene interaction determines Crohn’s disease gene Atg16L1 phenotypes in intestine. Cell 2010; 141:1135 - 45; http://dx.doi.org/10.1016/j.cell.2010.05.009; PMID: 20602997
- Saitoh T, Fujita N, Jang MH, Uematsu S, Yang BG, Satoh T, Omori H, Noda T, Yamamoto N, Komatsu M, et al. Loss of the autophagy protein Atg16L1 enhances endotoxin-induced IL-1beta production. Nature 2008; 456:264 - 8; http://dx.doi.org/10.1038/nature07383; PMID: 18849965
- Prescott NJ, Fisher SA, Franke A, Hampe J, Onnie CM, Soars D, Bagnall R, Mirza MM, Sanderson J, Forbes A, et al. A nonsynonymous SNP in ATG16L1 predisposes to ileal Crohn’s disease and is independent of CARD15 and IBD5. Gastroenterology 2007; 132:1665 - 71; http://dx.doi.org/10.1053/j.gastro.2007.03.034; PMID: 17484864
- Rioux JD, Xavier RJ, Taylor KD, Silverberg MS, Goyette P, Huett A, Green T, Kuballa P, Barmada MM, Datta LW, et al. Genome-wide association study identifies new susceptibility loci for Crohn disease and implicates autophagy in disease pathogenesis. Nat Genet 2007; 39:596 - 604; http://dx.doi.org/10.1038/ng2032; PMID: 17435756
- Hampe J, Franke A, Rosenstiel P, Till A, Teuber M, Huse K, Albrecht M, Mayr G, De La Vega FM, Briggs J, et al. A genome-wide association scan of nonsynonymous SNPs identifies a susceptibility variant for Crohn disease in ATG16L1. Nat Genet 2007; 39:207 - 11; http://dx.doi.org/10.1038/ng1954; PMID: 17200669
- Mizushima N, Levine B. Autophagy in mammalian development and differentiation. Nat Cell Biol 2010; 12:823 - 30; http://dx.doi.org/10.1038/ncb0910-823; PMID: 20811354
- Lee HK, Mattei LM, Steinberg BE, Alberts P, Lee YH, Chervonsky A, Mizushima N, Grinstein S, Iwasaki A. In vivo requirement for Atg5 in antigen presentation by dendritic cells. Immunity 2010; 32:227 - 39; http://dx.doi.org/10.1016/j.immuni.2009.12.006; PMID: 20171125
- Deretic V, Levine B. Autophagy, immunity, and microbial adaptations. Cell Host Microbe 2009; 5:527 - 49; http://dx.doi.org/10.1016/j.chom.2009.05.016; PMID: 19527881
- Karantza-Wadsworth V, Patel S, Kravchuk O, Chen G, Mathew R, Jin S, White E. Autophagy mitigates metabolic stress and genome damage in mammary tumorigenesis. Genes Dev 2007; 21:1621 - 35; http://dx.doi.org/10.1101/gad.1565707; PMID: 17606641
- Mathew R, Karp CM, Beaudoin B, Vuong N, Chen G, Chen HY, Bray K, Reddy A, Bhanot G, Gelinas C, et al. Autophagy suppresses tumorigenesis through elimination of p62. Cell 2009; 137:1062 - 75; http://dx.doi.org/10.1016/j.cell.2009.03.048; PMID: 19524509
- Mathew R, Kongara S, Beaudoin B, Karp CM, Bray K, Degenhardt K, Chen G, Jin S, White E. Autophagy suppresses tumor progression by limiting chromosomal instability. Genes Dev 2007; 21:1367 - 81; http://dx.doi.org/10.1101/gad.1545107; PMID: 17510285
- Komatsu M, Ueno T, Waguri S, Uchiyama Y, Kominami E, Tanaka K. Constitutive autophagy: vital role in clearance of unfavorable proteins in neurons. Cell Death Differ 2007; 14:887 - 94; PMID: 17332773
- Harris H, Rubinsztein DC. Control of autophagy as a therapy for neurodegenerative disease. Nat Rev Neurol 2012; 8:108 - 17; http://dx.doi.org/10.1038/nrneurol.2011.200; PMID: 22187000
- Deretic V. Autophagy in immunity and cell-autonomous defense against intracellular microbes. Immunol Rev 2011; 240:92 - 104; http://dx.doi.org/10.1111/j.1600-065X.2010.00995.x; PMID: 21349088
- Shahnazari S, Brumell JH. Mechanisms and consequences of bacterial targeting by the autophagy pathway. Curr Opin Microbiol 2011; 14:68 - 75; http://dx.doi.org/10.1016/j.mib.2010.11.001; PMID: 21112809
- Gutierrez MG, Master SS, Singh SB, Taylor GA, Colombo MI, Deretic V. Autophagy is a defense mechanism inhibiting BCG and Mycobacterium tuberculosis survival in infected macrophages. Cell 2004; 119:753 - 66; http://dx.doi.org/10.1016/j.cell.2004.11.038; PMID: 15607973
- Kirkin V, McEwan DG, Novak I, Dikic I. A role for ubiquitin in selective autophagy. Mol Cell 2009; 34:259 - 69; http://dx.doi.org/10.1016/j.molcel.2009.04.026; PMID: 19450525
- Kirkin V, Lamark T, Sou YS, Bjørkøy G, Nunn JL, Bruun JA, Shvets E, McEwan DG, Clausen TH, Wild P, et al. A role for NBR1 in autophagosomal degradation of ubiquitinated substrates. Mol Cell 2009; 33:505 - 16; http://dx.doi.org/10.1016/j.molcel.2009.01.020; PMID: 19250911
- Pankiv S, Clausen TH, Lamark T, Brech A, Bruun JA, Outzen H, Øvervatn A, Bjørkøy G, Johansen T. p62/SQSTM1 binds directly to Atg8/LC3 to facilitate degradation of ubiquitinated protein aggregates by autophagy. J Biol Chem 2007; 282:24131 - 45; http://dx.doi.org/10.1074/jbc.M702824200; PMID: 17580304
- Thurston TL, Ryzhakov G, Bloor S, von Muhlinen N, Randow F. The TBK1 adaptor and autophagy receptor NDP52 restricts the proliferation of ubiquitin-coated bacteria. Nat Immunol 2009; 10:1215 - 21; http://dx.doi.org/10.1038/ni.1800; PMID: 19820708
- Wild P, Farhan H, McEwan DG, Wagner S, Rogov VV, Brady NR, Richter B, Korac J, Waidmann O, Choudhary C, et al. Phosphorylation of the autophagy receptor optineurin restricts Salmonella growth. Science 2011; 333:228 - 33; http://dx.doi.org/10.1126/science.1205405; PMID: 21617041
- Matsuda N, Sato S, Shiba K, Okatsu K, Saisho K, Gautier CA, Sou YS, Saiki S, Kawajiri S, Sato F, et al. PINK1 stabilized by mitochondrial depolarization recruits Parkin to damaged mitochondria and activates latent Parkin for mitophagy. J Cell Biol 2010; 189:211 - 21; http://dx.doi.org/10.1083/jcb.200910140; PMID: 20404107
- Narendra DP, Jin SM, Tanaka A, Suen DF, Gautier CA, Shen J, Cookson MR, Youle RJ. PINK1 is selectively stabilized on impaired mitochondria to activate Parkin. PLoS Biol 2010; 8:e1000298; http://dx.doi.org/10.1371/journal.pbio.1000298; PMID: 20126261
- Vives-Bauza C, Zhou C, Huang Y, Cui M, de Vries RL, Kim J, May J, Tocilescu MA, Liu W, Ko HS, et al. PINK1-dependent recruitment of Parkin to mitochondria in mitophagy. Proc Natl Acad Sci U S A 2010; 107:378 - 83; http://dx.doi.org/10.1073/pnas.0911187107; PMID: 19966284
- Sarraf SA, Raman M, Guarani-Pereira V, Sowa ME, Huttlin EL, Gygi SP, Harper JW. Landscape of the PARKIN-dependent ubiquitylome in response to mitochondrial depolarization. Nature 2013; 496:372 - 6; http://dx.doi.org/10.1038/nature12043; PMID: 23503661
- Geisler S, Holmström KM, Skujat D, Fiesel FC, Rothfuss OC, Kahle PJ, Springer W. PINK1/Parkin-mediated mitophagy is dependent on VDAC1 and p62/SQSTM1. Nat Cell Biol 2010; 12:119 - 31; http://dx.doi.org/10.1038/ncb2012; PMID: 20098416
- Novak I, Kirkin V, McEwan DG, Zhang J, Wild P, Rozenknop A, Rogov V, Löhr F, Popovic D, Occhipinti A, et al. Nix is a selective autophagy receptor for mitochondrial clearance. EMBO Rep 2010; 11:45 - 51; http://dx.doi.org/10.1038/embor.2009.256; PMID: 20010802
- Noda NN, Ohsumi Y, Inagaki F. Atg8-family interacting motif crucial for selective autophagy. FEBS Lett 2010; 584:1379 - 85; http://dx.doi.org/10.1016/j.febslet.2010.01.018; PMID: 20083108
- von Muhlinen N, Akutsu M, Ravenhill BJ, Foeglein Á, Bloor S, Rutherford TJ, Freund SM, Komander D, Randow F. LC3C, bound selectively by a noncanonical LIR motif in NDP52, is required for antibacterial autophagy. Mol Cell 2012; 48:329 - 42; http://dx.doi.org/10.1016/j.molcel.2012.08.024; PMID: 23022382
- Kageyama S, Omori H, Saitoh T, Sone T, Guan JL, Akira S, Imamoto F, Noda T, Yoshimori T. The LC3 recruitment mechanism is separate from Atg9L1-dependent membrane formation in the autophagic response against Salmonella. Mol Biol Cell 2011; 22:2290 - 300; http://dx.doi.org/10.1091/mbc.E10-11-0893; PMID: 21525242
- Thurston TL, Wandel MP, von Muhlinen N, Foeglein A, Randow F. Galectin 8 targets damaged vesicles for autophagy to defend cells against bacterial invasion. Nature 2012; 482:414 - 8; http://dx.doi.org/10.1038/nature10744; PMID: 22246324
- Kamimoto T, Shoji S, Hidvegi T, Mizushima N, Umebayashi K, Perlmutter DH, Yoshimori T. Intracellular inclusions containing mutant alpha1-antitrypsin Z are propagated in the absence of autophagic activity. J Biol Chem 2006; 281:4467 - 76; http://dx.doi.org/10.1074/jbc.M509409200; PMID: 16365039
- Ogata M, Hino S, Saito A, Morikawa K, Kondo S, Kanemoto S, Murakami T, Taniguchi M, Tanii I, Yoshinaga K, et al. Autophagy is activated for cell survival after endoplasmic reticulum stress. Mol Cell Biol 2006; 26:9220 - 31; http://dx.doi.org/10.1128/MCB.01453-06; PMID: 17030611
- Iwata J, Ezaki J, Komatsu M, Yokota S, Ueno T, Tanida I, Chiba T, Tanaka K, Kominami E. Excess peroxisomes are degraded by autophagic machinery in mammals. J Biol Chem 2006; 281:4035 - 41; http://dx.doi.org/10.1074/jbc.M512283200; PMID: 16332691
- Maejima I, Takahashi A, Omori H, Kimura T, Takabatake Y, Saitoh T, Yamamoto A, Hamasaki M, Noda T, Isaka Y, et al. Autophagy sequesters damaged lysosomes to control lysosomal biogenesis and kidney injury. EMBO J 2013; 32:2336 - 47; http://dx.doi.org/10.1038/emboj.2013.171; PMID: 23921551
- Martinez J, Almendinger J, Oberst A, Ness R, Dillon CP, Fitzgerald P, Hengartner MO, Green DR. Microtubule-associated protein 1 light chain 3 alpha (LC3)-associated phagocytosis is required for the efficient clearance of dead cells. Proc Natl Acad Sci U S A 2011; 108:17396 - 401; http://dx.doi.org/10.1073/pnas.1113421108; PMID: 21969579
- Gong L, Cullinane M, Treerat P, Ramm G, Prescott M, Adler B, Boyce JD, Devenish RJ. The Burkholderia pseudomallei type III secretion system and BopA are required for evasion of LC3-associated phagocytosis. PLoS One 2011; 6:e17852; http://dx.doi.org/10.1371/journal.pone.0017852; PMID: 21412437
- Huang J, Canadien V, Lam GY, Steinberg BE, Dinauer MC, Magalhaes MA, Glogauer M, Grinstein S, Brumell JH. Activation of antibacterial autophagy by NADPH oxidases. Proc Natl Acad Sci U S A 2009; 106:6226 - 31; http://dx.doi.org/10.1073/pnas.0811045106; PMID: 19339495
- Henault J, Martinez J, Riggs JM, Tian J, Mehta P, Clarke L, Sasai M, Latz E, Brinkmann MM, Iwasaki A, et al. Noncanonical autophagy is required for type I interferon secretion in response to DNA-immune complexes. Immunity 2012; 37:986 - 97; http://dx.doi.org/10.1016/j.immuni.2012.09.014; PMID: 23219390
- Kim JY, Zhao H, Martinez J, Doggett TA, Kolesnikov AV, Tang PH, Ablonczy Z, Chan CC, Zhou Z, Green DR, et al. Noncanonical autophagy promotes the visual cycle. Cell 2013; 154:365 - 76; http://dx.doi.org/10.1016/j.cell.2013.06.012; PMID: 23870125
- Xu Y, Jagannath C, Liu XD, Sharafkhaneh A, Kolodziejska KE, Eissa NT. Toll-like receptor 4 is a sensor for autophagy associated with innate immunity. Immunity 2007; 27:135 - 44; http://dx.doi.org/10.1016/j.immuni.2007.05.022; PMID: 17658277
- Delgado MA, Elmaoued RA, Davis AS, Kyei G, Deretic V. Toll-like receptors control autophagy. EMBO J 2008; 27:1110 - 21; http://dx.doi.org/10.1038/emboj.2008.31; PMID: 18337753
- Travassos LH, Carneiro LA, Ramjeet M, Hussey S, Kim YG, Magalhães JG, Yuan L, Soares F, Chea E, Le Bourhis L, et al. Nod1 and Nod2 direct autophagy by recruiting ATG16L1 to the plasma membrane at the site of bacterial entry. Nat Immunol 2010; 11:55 - 62; http://dx.doi.org/10.1038/ni.1823; PMID: 19898471
- Mizushima N, Kuma A, Kobayashi Y, Yamamoto A, Matsubae M, Takao T, Natsume T, Ohsumi Y, Yoshimori T. Mouse Apg16L, a novel WD-repeat protein, targets to the autophagic isolation membrane with the Apg12-Apg5 conjugate. J Cell Sci 2003; 116:1679 - 88; http://dx.doi.org/10.1242/jcs.00381; PMID: 12665549
- Kuballa P, Huett A, Rioux JD, Daly MJ, Xavier RJ. Impaired autophagy of an intracellular pathogen induced by a Crohn’s disease associated ATG16L1 variant. PLoS One 2008; 3:e3391; http://dx.doi.org/10.1371/journal.pone.0003391; PMID: 18852889
- Fujita N, Saitoh T, Kageyama S, Akira S, Noda T, Yoshimori T. Differential involvement of Atg16L1 in Crohn disease and canonical autophagy: analysis of the organization of the Atg16L1 complex in fibroblasts. J Biol Chem 2009; 284:32602 - 9; http://dx.doi.org/10.1074/jbc.M109.037671; PMID: 19783656
- Shui W, Sheu L, Liu J, Smart B, Petzold CJ, Hsieh TY, Pitcher A, Keasling JD, Bertozzi CR. Membrane proteomics of phagosomes suggests a connection to autophagy. Proc Natl Acad Sci U S A 2008; 105:16952 - 7; http://dx.doi.org/10.1073/pnas.0809218105; PMID: 18971338
- Overholtzer M, Mailleux AA, Mouneimne G, Normand G, Schnitt SJ, King RW, Cibas ES, Brugge JS. A nonapoptotic cell death process, entosis, that occurs by cell-in-cell invasion. Cell 2007; 131:966 - 79; http://dx.doi.org/10.1016/j.cell.2007.10.040; PMID: 18045538
- Pujol C, Klein KA, Romanov GA, Palmer LE, Cirota C, Zhao Z, Bliska JB. Yersinia pestis can reside in autophagosomes and avoid xenophagy in murine macrophages by preventing vacuole acidification. Infect Immun 2009; 77:2251 - 61; http://dx.doi.org/10.1128/IAI.00068-09; PMID: 19289509
- Moreau K, Lacas-Gervais S, Fujita N, Sebbane F, Yoshimori T, Simonet M, Lafont F. Autophagosomes can support Yersinia pseudotuberculosis replication in macrophages. Cell Microbiol 2010; 12:1108 - 23; http://dx.doi.org/10.1111/j.1462-5822.2010.01456.x; PMID: 20180800
- Schnaith A, Kashkar H, Leggio SA, Addicks K, Krönke M, Krut O. Staphylococcus aureus subvert autophagy for induction of caspase-independent host cell death. J Biol Chem 2007; 282:2695 - 706; http://dx.doi.org/10.1074/jbc.M609784200; PMID: 17135247
- Starr T, Child R, Wehrly TD, Hansen B, Hwang S, López-Otin C, Virgin HW, Celli J. Selective subversion of autophagy complexes facilitates completion of the Brucella intracellular cycle. Cell Host Microbe 2012; 11:33 - 45; http://dx.doi.org/10.1016/j.chom.2011.12.002; PMID: 22264511
- Jackson WT, Giddings TH Jr., Taylor MP, Mulinyawe S, Rabinovitch M, Kopito RR, Kirkegaard K. Subversion of cellular autophagosomal machinery by RNA viruses. PLoS Biol 2005; 3:e156; http://dx.doi.org/10.1371/journal.pbio.0030156; PMID: 15884975
- Reggiori F, Monastyrska I, Verheije MH, Calì T, Ulasli M, Bianchi S, Bernasconi R, de Haan CA, Molinari M. Coronaviruses Hijack the LC3-I-positive EDEMosomes, ER-derived vesicles exporting short-lived ERAD regulators, for replication. Cell Host Microbe 2010; 7:500 - 8; http://dx.doi.org/10.1016/j.chom.2010.05.013; PMID: 20542253
- Vázquez CL, Colombo MI. Coxiella burnetii modulates Beclin 1 and Bcl-2, preventing host cell apoptosis to generate a persistent bacterial infection. Cell Death Differ 2010; 17:421 - 38; http://dx.doi.org/10.1038/cdd.2009.129; PMID: 19798108
- Gutierrez MG, Vázquez CL, Munafó DB, Zoppino FC, Berón W, Rabinovitch M, Colombo MI. Autophagy induction favours the generation and maturation of the Coxiella-replicative vacuoles. Cell Microbiol 2005; 7:981 - 93; http://dx.doi.org/10.1111/j.1462-5822.2005.00527.x; PMID: 15953030
- Birmingham CL, Canadien V, Kaniuk NA, Steinberg BE, Higgins DE, Brumell JH. Listeriolysin O allows Listeria monocytogenes replication in macrophage vacuoles. Nature 2008; 451:350 - 4; http://dx.doi.org/10.1038/nature06479; PMID: 18202661
- DeSelm CJ, Miller BC, Zou W, Beatty WL, van Meel E, Takahata Y, Klumperman J, Tooze SA, Teitelbaum SL, Virgin HW. Autophagy proteins regulate the secretory component of osteoclastic bone resorption. Dev Cell 2011; 21:966 - 74; http://dx.doi.org/10.1016/j.devcel.2011.08.016; PMID: 22055344
- Sanjuan MA, Milasta S, Green DR. Toll-like receptor signaling in the lysosomal pathways. Immunol Rev 2009; 227:203 - 20; http://dx.doi.org/10.1111/j.1600-065X.2008.00732.x; PMID: 19120486
- Hwang S, Maloney NS, Bruinsma MW, Goel G, Duan E, Zhang L, Shrestha B, Diamond MS, Dani A, Sosnovtsev SV, et al. Nondegradative role of Atg5-Atg12/ Atg16L1 autophagy protein complex in antiviral activity of interferon gamma. Cell Host Microbe 2012; 11:397 - 409; http://dx.doi.org/10.1016/j.chom.2012.03.002; PMID: 22520467
- Wobus CE, Karst SM, Thackray LB, Chang KO, Sosnovtsev SV, Belliot G, Krug A, Mackenzie JM, Green KY, Virgin HW. Replication of Norovirus in cell culture reveals a tropism for dendritic cells and macrophages. PLoS Biol 2004; 2:e432; http://dx.doi.org/10.1371/journal.pbio.0020432; PMID: 15562321
- Ishibashi K, Uemura T, Waguri S, Fukuda M. Atg16L1, an essential factor for canonical autophagy, participates in hormone secretion from PC12 cells independently of autophagic activity. Mol Biol Cell 2012; 23:3193 - 202; http://dx.doi.org/10.1091/mbc.E12-01-0010; PMID: 22740627
- Dupont N, Jiang S, Pilli M, Ornatowski W, Bhattacharya D, Deretic V. Autophagy-based unconventional secretory pathway for extracellular delivery of IL-1β. EMBO J 2011; 30:4701 - 11; http://dx.doi.org/10.1038/emboj.2011.398; PMID: 22068051
- Duran JM, Anjard C, Stefan C, Loomis WF, Malhotra V. Unconventional secretion of Acb1 is mediated by autophagosomes. J Cell Biol 2010; 188:527 - 36; http://dx.doi.org/10.1083/jcb.200911154; PMID: 20156967
- Manjithaya R, Anjard C, Loomis WF, Subramani S. Unconventional secretion of Pichia pastoris Acb1 is dependent on GRASP protein, peroxisomal functions, and autophagosome formation. J Cell Biol 2010; 188:537 - 46; http://dx.doi.org/10.1083/jcb.200911149; PMID: 20156962
- Ganesan AK, Ho H, Bodemann B, Petersen S, Aruri J, Koshy S, Richardson Z, Le LQ, Krasieva T, Roth MG, et al. Genome-wide siRNA-based functional genomics of pigmentation identifies novel genes and pathways that impact melanogenesis in human cells. PLoS Genet 2008; 4:e1000298; http://dx.doi.org/10.1371/journal.pgen.1000298; PMID: 19057677
- Subramani S, Malhotra V. Non-autophagic roles of autophagy-related proteins. EMBO Rep 2013; 14:143 - 51; http://dx.doi.org/10.1038/embor.2012.220; PMID: 23337627
- Deretic V, Jiang S, Dupont N. Autophagy intersections with conventional and unconventional secretion in tissue development, remodeling and inflammation. Trends Cell Biol 2012; 22:397 - 406; http://dx.doi.org/10.1016/j.tcb.2012.04.008; PMID: 22677446
