Abstract
SQSTM1/p62 (sequestosome 1) is a multifunctional signaling molecule, involved in a variety of cellular pathways. SQSTM1 is one of the best-known autophagic substrates, and is therefore widely used as an indicator of autophagic degradation. Here we report that the expression level of SQSTM1 can be restored during prolonged starvation. Upon starvation, SQSTM1 is initially degraded by autophagy. However, SQSTM1 is restored back to basal levels during prolonged starvation in mouse embryonic fibroblasts and HepG2 cells, but not in HeLa and HEK293 cells. Restoration of SQSTM1 depends on its transcriptional upregulation, which is triggered by amino acid starvation. Furthermore, amino acids derived from the autophagy–lysosome pathway are used for de novo synthesis of SQSTM1 under starvation conditions. The restoration of SQSTM1 is independent of reactivation of MTORC1 (mechanistic target of rapamycin complex 1). These results suggest that the expression level of SQSTM1 in starved cells is determined by at least 3 factors: autophagic degradation, transcriptional upregulation, and availability of lysosomal-derived amino acids. The results of this study also indicate that the expression level of SQSTM1 does not always inversely correlate with autophagic activity.
Keywords: :
Introduction
In response to environmental perturbations, cells undergo metabolic change. For instance, nutrient starvation triggers several adaptive changes, one of which is macroautophagy (hereafter referred to as autophagy), an intracellular protein degradation system.Citation1-Citation4 Under starvation conditions, especially depletion of amino acids or insulin/growth factors, MTORC1 is suppressed, and subsequently autophagy is activated. The phagophore encloses a portion of the cytoplasm, which generates a double-membraned structure, the autophagosome. The lysosome fuses with the outer autophagosomal membrane, leading to degradation of the enclosed materials by hydrolytic enzymes in the lysosome.
Autophagy has been considered to be essentially a nonselective process, but in fact it is partially selective.Citation5-Citation7 Selective substrates of autophagy include damaged mitochondria, intracellular pathogens, and even a subset of cytosolic proteins. One of the best known selective substrates is SQSTM1.Citation8,Citation9 SQSTM1 is a ubiquitously expressed cellular protein that is conserved in metazoa but not in plants and fungi. SQSTM1 has a Phox and Bem1 (PB1) domain for self-oligomerzation and a ubiquitin-associated (UBA) domain for binding to ubiqitinated proteins. SQSTM1 serves as a scaffolding hub in various cellular signaling pathways such as in NFKB/NFκB (nuclear factor of kappa light polypeptide gene enhancer in B-cells) activation, nerve growth factor signaling, and caspase activation. Thus, SQSTM1 has multiple physiological functions in bone metabolism, obesity, inclusion body formation, and tumorigenesis.Citation10-Citation13
Besides these signaling functions, SQSTM1 also interacts with MAP1LC3/LC3 (microtubule-associated protein 1 light chain 3) through its LC3-interacting region (LIR).Citation5-Citation7 The presence of the UBA domain enables SQSTM1 to serve as an adaptor for selective autophagy of ubiquitinated substrates. Additionally, SQSTM1 itself is able to translocate not only to the phagophore membrane but also to the autophagosome formation site even independently of LC3 binding.Citation14 Therefore, SQSTM1 is selectively incorporated into the autophagosome and then degraded.Citation8,Citation9 Accordingly, SQSTM1 is widely used as an indicator of autophagic flux.Citation15,Citation16
The expression level of SQSTM1 is strictly regulated by continuous degradation by basal autophagy. Impairment of autophagy causes massive accumulation of SQSTM1, followed by formation of aggregates containing SQSTM1 and ubiquitinated proteins.Citation17 Accumulation of SQSTM1 is toxic, at least in the liver, as excess SQSTM1 sequesters KEAP1, a component of the ubiquitin ligase complex for NFE2L2/Nrf2 (nuclear factor, erythroid 2-like 2), resulting in inappropriate activation of transcription of NFE2L2 target genes.Citation18,Citation19 Conversely, when autophagy is upregulated by starvation, SQSTM1 is rapidly degraded and its total expression level is reduced.Citation8,Citation20,Citation21 However, how SQSTM1 levels are regulated during prolonged starvation (i.e., a condition of sustained autophagy) remains largely unclear.
In this study we examined the determinants of SQSTM1 gene expression during long-term starvation. We showed that SQSTM1 levels are reduced during the first few hours, but restored to basal levels during prolonged starvation. This restoration is independent of MTORC1 reactivation, but requires transcriptional upregulation of SQSTM1 and de novo protein synthesis. We also provided evidence that amino acids derived from the autophagy–lysosome pathway are used for SQSTM1 synthesis during prolonged starvation.
Results
SQSTM1 expression level is restored during prolonged starvation
We first assessed the SQSTM1 expression levels in MEFs under starvation conditions (lacking both amino acids and serum). During the first 2 h of starvation, the SQSTM1 level was reduced in wild-type cells as previously observed ().Citation8,Citation20,Citation21 LC3 conversion from LC3-I to LC3-II and inhibition of MTORC1 (dephosphorylation of RPS6KB/p70 S6 kinase and EIF4EBP1/4E-BP1) were simultaneously observed. The starvation-induced decrease in SQSTM1 level was not observed in autophagy-deficient Rb1cc1/Fip200 KO and Atg5 KO MEFs, confirming that the initial decrease was due to autophagic degradation of SQSTM1.
Figure 1. SQSTM1 is restored during prolonged starvation. (A) Wild-type, Rb1cc1 KO and Atg5 KO MEFs were cultured in starvation medium lacking amino acids and serum for 1, 2, 4, 6, and 8 h. Cell lysates were analyzed by immunoblotting using the indicated antibodies. Densitometric quantification of SQSTM1 protein levels using ImageJ software is shown on the graph. Data represent the mean ± SEM of 4 independent experiments (including the data in , , and ) for wild-type cells. A representative result is shown for Rb1cc1 KO and Atg5 KO cells. (B) HepG2, HeLa and HEK293 cells were analyzed as in (A). (C) Wild-type immortalized and primary MEFs were analyzed as in (A). (D) Wild-type MEFs were cultured in starvation medium lacking amino acids and serum for 2, 4, and 8 h. Cells were fixed and stained with anti-SQSTM1/p62 antibody. Scale bar: 5 µm.
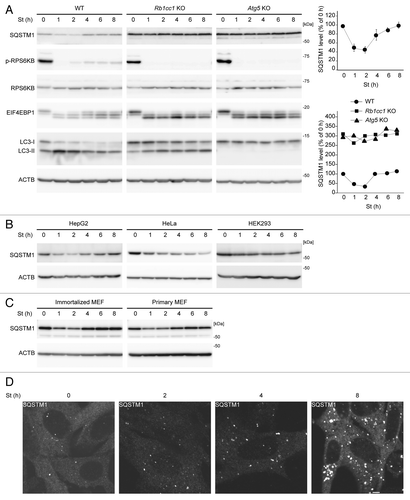
However, during prolonged starvation, we found that the SQSTM1 level was increased and restored to almost basal levels following starvation for 4 h (). Such restoration of SQSTM1 was also observed in the human hepatocellular carcinoma cell line HepG2 and primary MEFs (). By contrast, we did not observe any apparent restoration of the SQSTM1 level in HeLa cells or HEK293 cells during 8 h of starvation. These data suggest that, although SQSTM1 is rapidly degraded by autophagy, its expression level is restored during starvation for 4 to 8 h at least in MEFs and HepG2 cells.
Immunofluorescence microscopy of endogenous SQSTM1 in MEFs showed that the number of punctate structures, which represent LC3-positive autophagosome-associating SQSTM1, increased during the first 4 h of starvation, whereas cytosolic SQSTM1 signals decreased (; Fig. S1A). However, at 8 h of starvation, not only the number of the puncta but also the cytosolic signal of SQSTM1 was increased. Some of the puncta should represent autophagosomes because they were positive for the autophagosomal SNARE STX17 (syntaxin 17) (Fig. S1A),Citation22 but others might be ubiquitin-positive SQSTM1 aggregates, which may be generated by induction of SQSTM1 (Fig. S1B). These data confirm that the SQSTM1 level is restored during prolonged starvation.
SQSTM1 restoration requires de novo protein synthesis but is independent of MTORC1
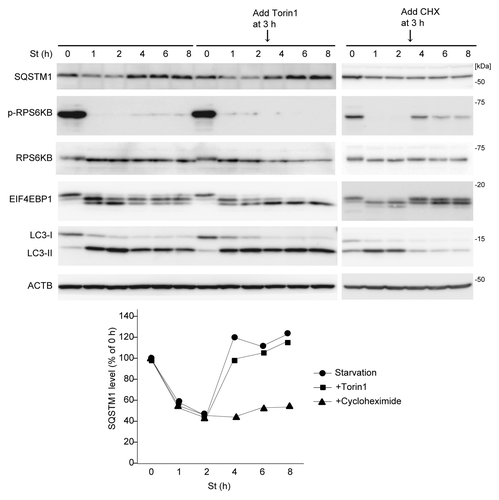
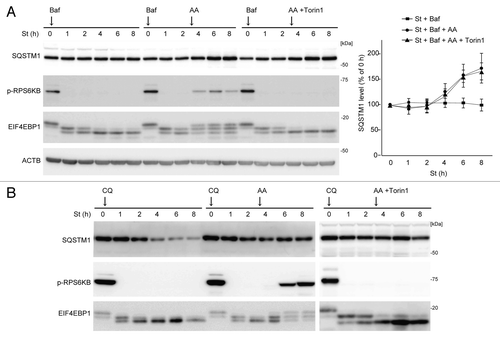
We investigated the mechanism underlying the restoration of SQSTM1 during prolonged starvation. We first determined the involvement of MTORC1, which can be reactivated by autophagy-derived amino acids during prolonged starvation.Citation23 Indeed, RPS6KB and EIF4EBP1, which were dephosphorylated in the early phase of starvation, were rephosphorylated to a small extent after 4 h in wild-type MEFs, but not in Rb1cc1 KO and Atg5 KO MEFs (; ). When we treated wild-type MEFs with the potent MTOR inhibitor Torin1 after 3 h of starvation,Citation24 Torin1 effectively suppressed rephosphorylation of RPS6KB and EIF4EBP1 during prolonged starvation (). However, restoration of SQSTM1 still occurred after 4 h of starvation even in the presence of Torin1, suggesting that it is independent of MTORC1 reactivation (). Even though MTOR does not have a role, the restoration of SQSTM1 requires new protein synthesis; treatment of wild-type MEFs with cycloheximide (CHX) after 3 h of starvation abolished SQSTM1 restoration (). As it is well known that CHX secondarily activates MTORC1, the inhibitory effect of CHX may be through MTORC1-mediated autophagy suppression. To rule out this possibility, we treated MEFs with CHX and the MTOR inhibitor Torin1, and confirmed that SQSTM1 restoration was still abolished independently of MTORC1 activity (Fig. S2). Together, these data suggest that SQSTM1 restoration is due to de novo synthesis of SQSTM1 protein in an MTOR-independent manner.
Figure 2. SQSTM1 restoration requires de novo protein synthesis but is independent of MTORC1. Wild-type MEFs were cultured in starvation medium lacking amino acids and serum for 1, 2, 4, 6, and 8 h. At 3 h after starvation, Torin1 (250 nM) or cycloheximide (CHX, 50 μg/ml) was added. Cell lysates were analyzed by immunoblotting using the indicated antibodies. Densitometric quantification of SQSTM1 protein levels using ImageJ software is shown on the graph.
Amino acid starvation-induced upregulation of Sqstm1 transcription is required for SQSTM1 restoration during prolonged starvation
Next we determined whether restoration of SQSTM1 required transcriptional upregulation. We measured the relative amount of Sqstm1 mRNA by quantitative PCR during starvation. We found that, in MEFs, Sqstm1 mRNA was greatly increased during amino acid and serum starvation (by approximately 20-fold after 8 h of starvation) using Actb (), and glyceraldehyde-3-phosphate dehydrogenase (Gapdh) (data not shown), as housekeeping internal controls. It was also significantly increased in HepG2 cells, but not in HeLa or HEK293 cells (), which could explain why SQSTM1 restoration was observed in MEFs and HepG2 cells but not in HeLa or HEK293 cells.
Figure 3. Amino acid starvation-induced upregulation of Sqstm1 transcription is required for SQSTM1 restoration during prolonged starvation. (A) Wild-type MEFs were cultured in DMEM lacking both amino acids and serum, or either amino acids or serum alone, or regular DMEM containing 250 nM Torin1 for 2, 4, and 8 h. Relative Sqstm1 mRNA is estimated by quantitative PCR using Actb as an internal control. Data represent the mean ± SEM of 3 independent experiments. (B) HepG2, HeLa and HEK293 cells were cultured in DMEM lacking both amino acids and serum and analyzed as in (A). (C) Wild-type MEFs were cultured as in (A). The SQSTM1 protein level was analyzed by immunoblotting. ACTB was used as a loading control. Densitometric quantification of SQSTM1 protein levels using ImageJ software is shown on the graph. Data represent the mean ± SEM of 3 independent experiments. (D) Wild-type MEFs stably expressing GFP-SQSTM1 were cultured in DMEM without amino acids and serum. Expression levels of endogenous SQSTM1 and GFP-SQSTM1 were analyzed as in (C). (E) Wild-type MEFs were pretreated with or without 20 µg/ml α-amanitin for 4 h, and then cultured in DMEM without serum and amino acids for 4 or 8 h in the presence or absence of 20 µg/ml α-amanitin. Sqstm1 mRNA was quantified as in (A). (F) Wild-type MEFs were treated as in (E) and the SQSTM1 protein level was analyzed by immunoblotting. ACTB was used as a loading control. Densitometric quantification of SQSTM1 protein levels using ImageJ software is shown on the graph.
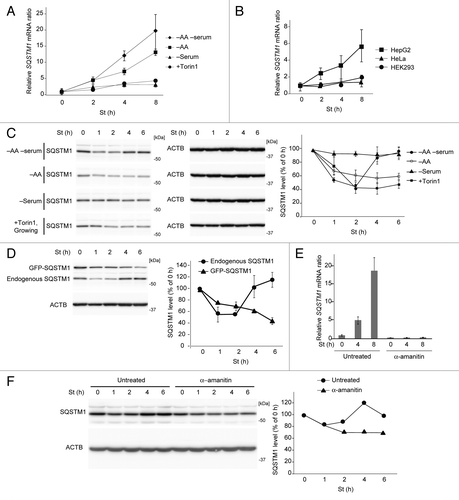
Amino acid starvation alone also increased the Sqstm1 mRNA level, but its effect was weaker than that of amino acid and serum starvation (). In contrast, serum starvation alone or Torin1 treatment (in growing medium) increased Sqstm1 mRNA only slightly (~3-fold). Accordingly, restoration of SQSTM1 protein levels was not significant under conditions of either amino acid starvation, serum starvation, or Torin1 treatment, although amino acid starvation and Torin1 treatment both induced autophagic degradation of SQSTM1 (). These data suggest that amino acid starvation effectively induces SQSTM1 transcription through an MTOR-independent pathway, and there is an additive effect of amino acid and serum starvation on SQSTM1 restoration.
To confirm that the endogenous SQSTM1 promoter is regulated by prolonged starvation, we monitored endogenous and exogenous SQSTM1 expression levels simultaneously in MEFs stably expressing green fluorescent protein (GFP)-tagged SQSTM1. These MEFs express GFP-SQSTM1 through an exogenous promoter. Although both endogenous and exogenous GFP-SQSTM1 were degraded during initial starvation, only endogenous SQSTM1, and not cytomegalovirus (CMV) promoter-driven exogenous GFP-SQSTM1, was restored after 4 h (). The mRNA level of GFP-Sqstm1 was estimated to be between the basal and induced levels of endogenous Sqstm1, and did not increase in response to starvation like that of endogenous Sqstm1 (Fig. S3), supporting the notion that the own promoter of SQSTM1 is important (). Furthermore, α-amanitin, a potent inhibitor of transcription, completely inhibited starvation-induced Sqstm1 mRNA upregulation (), and abolished restoration of the SQSTM1 protein level (). Based on these data, we conclude that transcriptional upregulation is required for restoration of SQSTM1 during prolonged starvation.
Amino acids generated by autophagy are used for SQSTM1 synthesis
Our data showed that synthesis of SQSTM1 protein is upregulated during prolonged starvation even though there are no available amino acids outside the cell. Next we evaluated how cells obtain amino acids for SQSTM1 synthesis. Given that starvation-induced autophagy supplies amino acids within cells, autophagy itself could contribute to the restoration of SQSTM1. In fact, the expression level of SQSTM1 was very stable during starvation in Rb1cc1 KO and Atg5 KO MEFs, with neither initial reduction nor later restoration of expression (). However, the accumulation of SQSTM1 in autophagy-deficient MEFs may mask a slight increase in SQSTM1 during prolonged starvation.
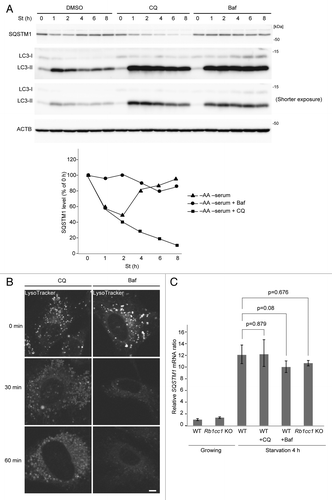
To better understand the contribution of the autophagy–lysosomal pathway, we inhibited lysosomal function using chloroquine (CQ) or bafilomycin A1 (Baf), a specific inhibitor of vacuolar-type H+ ATPase. Both treatments caused accumulation of LC3-II until at least 8 h after the beginning of starvation in wild-type MEFs, suggesting that the autophagy–lysosomal pathway is inhibited (). In Baf-treated cells, degradation of SQSTM1 during initial starvation (up to 2 h) was efficiently inhibited. Furthermore, the SQSTM1 level was virtually unchanged during prolonged starvation (), suggesting that autophagic degradation is critical for the increase in the SQSTM1 protein level. On the other hand, CQ did not inhibit SQSTM1 degradation (). This could be because the lysosome-inhibitory effect of CQ is weaker than that of Baf.Citation25 In fact, LysoTracker Red staining suggested that inhibitory effect of CQ on lysosomal acidification was weaker than that of Baf (). SQSTM1 may be more sensitive to lysosomal degradation compared with LC3-II, and the remaining lysosomal activity could be sufficient to degrade SQSTM1. Nonetheless, SQSTM1 restoration was also inhibited in CQ-treated cells (), suggesting that even though SQSTM1 degradation is not affected, overall lysosomal degradation is impaired, which results in amino acid shortage. The lack of prolonged starvation-induced SQSTM1 production in CQ- or Baf-treated cells () and autophagy-deficient cells () was not due to transcriptional failure; Sqstm1 mRNA was upregulated in these cells as in normal cells (). Together these data suggest that autophagy is critical for SQSTM1 restoration, likely through production of amino acids.
Figure 4. Inhibition of lysosomal function abolishes SQSTM1 restoration. (A) Wild-type MEFs were cultured in DMEM lacking both amino acids and serum in the presence of 20 µM chloroquine (CQ) or 0.1 µM bafilomycinA1 (Baf) for 1, 2, 4, 6, and 8 h. Densitometric quantification of SQSTM1 protein levels is shown on the graph. (B) Wild-type MEFs were cultured in regular DMEM medium. These MEFs were pretreated with 1 µM LysoTracker for 30 min and washed with PBS twice. Pretreated MEFs were treated with 20 µM CQ or 0.1 µM Baf for indicated times. Scale bar: 5 µm. (C) Wild-type and Rb1cc1 KO MEFs were cultured in regular DMEM medium or starvation medium (DMEM without amino acids and serum) for 4 h. Wild-type MEFs were also treated with 20 µM CQ or 0.1 µM Baf during the 4 h of starvation. Relative Sqstm1 mRNA was estimated by quantitative PCR. Data represent mean ± SEM of 3 independent experiments.
If amino acid limitation is indeed a cause of the defect in starvation induced-SQSTM1 synthesis in Baf-treated cells, exogenous amino acid supplementation should increase the SQSTM1 level. We added amino acids at 3 h after starvation to Baf- (or CQ-) treated cells and found that the SQSTM1 expression level increased to considerably higher than basal levels (). Amino acid treatment also activated MTORC1, thus it is possible that MTORC1 non-specifically increased SQSTM1 protein synthesis either at a translational or transcriptional step. To rule out this possibility, we added Torin1 together with amino acids to suppress MTORC1. Even in this setting, the SQSTM1 protein levels increased following amino acid addition in Baf-treated cells (). Thus, MTORC1 reactivation does not have a role in amino acid-induced SQSTM1 protein synthesis. We also tested whether amino acid treatment could increase the SQSTM1 protein level in starved Rb1cc1 KO MEFs. However, exogenous amino acids did not result in an increase in the SQSTM1 level (Fig. S4). This may be because accumulation of SQSTM1 () masked an increase in SQSTM1.
Figure 5. Amino acid replenishment causes restoration of SQSTM1 in bafilomycinA1-treated cells. (A) Wild-type MEFs were cultured in starvation medium containing 0.1 µM bafilomycinA1 (Baf) for 1, 2, 4, 6, and 8 h. After 3 h of starvation treatment, the medium was replaced with regular DMEM containing amino acids with or without 250 nM Torin1. Densitometric quantification of SQSTM1 protein levels using ImageJ software is shown on the graph. Data represent the mean ± SEM of 3 independent experiments. (B) Wild-type MEFs were cultured in starvation medium containing 20 µM chloroquine (CQ) for 1, 2, 4, 6, and 8 h. After 3 h of starvation treatment, the medium was replaced with regular DMEM containing amino acids with or without 250 nM Torin1.
Collectively, the findings in wild-type cells demonstrated that exogenously added amino acids can compensate for the inhibitory effect of Baf on SQSTM1 protein synthesis, and suggest that amino acids that are generated by the autophagy–lysosomal pathway are used for SQSTM1 restoration during prolonged starvation.
Discussion
We have shown that SQSTM1, which is degraded by autophagy during short-term starvation, is restored to basal levels during prolonged (~4 to 8 h) starvation at least in MEFs and HepG2 cells. Furthermore, this depends on transcriptional upregulation of Sqstm1 mRNA and production of amino acids by autophagy, but not on MTORC1 reactivation. We propose the following model to explain these findings (). Under nutrient-rich conditions, SQSTM1 protein is synthesized using basal levels of Sqstm1 mRNA and an abundance of extracellular amino acids. When cells are subjected to short-term starvation, autophagy is induced, which degrades SQSTM1 protein as a selective substrate. At this time point, transcriptional upregulation of SQSTM1 has started, but only slightly. In addition, intracellular and extracellular amino acid pools should be limited. However, after prolonged starvation, high levels of Sqstm1 mRNA and sufficient amounts of intracellular amino acids, which are produced by autophagy, are now available. Thus, the SQSTM1 protein level is restored to basal levels by de novo protein synthesis.
Figure 6. A model of SQSTM1 restoration during prolonged starvation. Under nutrient-rich conditions, SQSTM1 protein is maintained at basal levels. During short-term starvation (~2 h), SQSTM1 is rapidly degraded by autophagy. As starvation-induced transcriptional upregulation of SQSTM1 is only modest, overall SQSTM1 protein levels decrease. Following prolonged starvation, high levels of Sqstm1 mRNA and sufficient amounts of intracellular amino acids derived from autophagy promote de novo synthesis of SQSTM1, which restores SQSTM1 protein expression to basal levels.
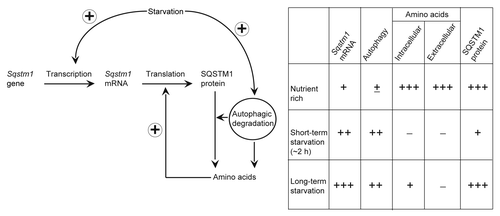
This restoration phenomenon of SQSTM1 was clearly observed in cells starved with both amino acids and serum, but not in cells starved only with either amino acids or serum (). Serum starvation alone does not induce transcriptional upregulation of SQSTM1. In contrast, amino acid starvation can induce Sqstm1 mRNA but the effect is modest. Moreover, amino acids sufficient for SQSTM1 synthesis may be produced only by full activation of autophagy induced by withdrawal of both amino acids and serum.
The promoter of the SQSTM1 gene is regulated by a variety of factors such as NFE2L2,Citation26,Citation27 SPDEF/PDEF (SAM pointed domain containing ETS transcription factor),Citation28 MAPK8/9/10 (mitogen-activated protein kinase 8/9/10, formerly also known as JNK1/2/3, respectively),Citation29 NFKB and XBP1,Citation30 but none of them seems to account for the starvation-induced upregulation of SQSTM1. Recently, it has been shown that TFEB (transcription factor EB), a transcriptional factor for lysosomal biogenesis, is activated upon starvation and induces expression of several autophagy-related proteins including SQSTM1.Citation31,Citation32 Thus, TFEB could be involved in SQSTM1 restoration. Another possible factor is CEBPB/C/EBPβ, [CCAAT/enhancer binding protein (C/EBP), β], which is one of the key transcription factors responsible for starvation,Citation33,Citation34 and is also implicated in circadian regulation of autophagy.Citation35 However, knockdown of CEBPB increased the level of Sqstm1 mRNA (unpublished observation). Furthermore, during revision of this paper, it was reported that amino acid starvation upregulates Sqstm1 transcription through the EIF2AK4/GCN2-EIF2A/eIFα-ATF4 and DDIT3/CHOP pathways.Citation36 These complex regulatory mechanisms may contribute to the different responses to prolonged starvation among cell lines. A more comprehensive study is needed to clarify the specific transcriptional factor responsible for SQSTM1 restoration during long-term starvation.
The findings of this study also illuminate the important role of autophagy as an amino acid generator. It is generally believed that autophagy-derived amino acids during starvation are used for protein synthesis, energy production, and gluconeogenesis.Citation4,Citation37 In yeast, amino acids produced by starvation-induced autophagy are important for synthesis of amino acid biosynthetic enzymes (e.g., Arg1/argininosuccinate synthetase), vacuolar enzymes (e.g., Ape1/aminopeptidase I and Prc1/carboxypeptidase Y),Citation38 respiratory chain proteins (e.g., cytochrome c oxidase components), and antioxidant enzymes (e.g., Cta1 and Ctt1).Citation39 To our knowledge, our study is the first to demonstrate in mammalian cells that autophagy-derived amino acids are used for upregulation of a specific protein during starvation.
What is the significance of SQSTM1 restoration during long-term starvation? It is notable that the restored level is quite similar to the original level in wild-type cells. Returning to this “default” level may be important for SQSTM1 to properly function in multiple pathways including selective autophagy.Citation40 This also seems to be the case for LC3. LC3-family proteins, which are required for autophagy, are paradoxically degraded by autophagy because they bind to the inner autophagosomal membrane. However, LC3A and LC3B are transcriptionally upregulated during starvation.Citation32 This could be to supplement the loss of LC3 and in order to adapt to persistent starvation. Another possibility is that restored SQSTM1 may be involved in reactivation of MTORC1 during prolonged amino acid starvation, because SQSTM1 interacts with MTORC1 to mediate amino acid signaling.Citation41 Indeed, the kinetics of SQSTM1 restoration are similar to those of MTORC1 reactivation ().
Finally, endogenous SQSTM1 should be used as an indicator of autophagy flux with caution, as its level is regulated by a number of factors. Generally, the rate of starvation-induced reduction in the SQSTM1 protein level is considered to correspond to the activity of autophagic degradation.Citation15,Citation16 However, our data showed that a clear reduction may be observed only during the first couple of hours of starvation, at least in MEFs and HepG2 cells. Prolonged starvation treatment causes SQSTM1 restoration, which may lead to underestimation of autophagic activity. Monitoring SQSTM1 under the control of an exogenous promoter would be better for this purpose, because GFP-SQSTM1 under the control of the CMV promoter can be degraded by autophagy, but is not restored even following prolonged starvation (). Artificial control of SQSTM1 expression using a tetracycline-regulated system would be an ideal method to measure autophagic flux.Citation42 We recommend that the level of Sqstm1 mRNA should be simultaneously determined along with the SQSTM1 protein level as an autophagy indicator.
Materials and Methods
Cell culture
Cells were maintained in Dulbecco’s modified Eagle’s medium (DMEM) (Sigma, D6546) supplemented with 10% fetal bovine serum (FBS), L-glutamine, and 50 µg/ml penicillin/streptomycin in a 5% CO2 chamber at 37 °C. rb1cc1–/– (Rb1cc1 knockout [KO])Citation43 and atg5–/– (Atg5 KO)Citation44 mouse embryonic fibroblasts (MEFs) were previously generated. MEFs stably expressing GFP-SQSTM1 were generated using pMXs-puro-GFP-SQSTM1 as previously described.Citation14 To induce starvation, cells were washed with phosphate-buffered saline (PBS) and incubated in amino acid-free DMEM without FBS (GIBCO, custom-made).
Western blotting
Cells were washed with ice-cold PBS, harvested in cold PBS and centrifuged at 3,000 rpm for 5 min. Cells were lysed in lysis buffer (50 mM TRIS-HCl, 150 mM NaCl, 1 mM EDTA, 1% Triton X-100, 10 mM NaF, 0.4 mM Na3VO4, 1 mM phenylmethylsulfonyl fluoride, and protease inhibitor cocktail [Complete EDTA-free protease inhibitor; Roche 05056489001]). The lysate was clarified by centrifugation at 15,000 rpm for 5 min at 4 °C and was mixed with 6× sample buffer. Samples were subsequently separated by SDS-PAGE and transferred to Immobilon-P Transfer Membrane (Millipore, IPVH00010). Immunoblot analysis was performed with the indicated antibodies and visualized with Immobilon Western (Millipore, WBKLS0500). Signal intensities were analyzed using an imaging analyzer (LAS-3000mini; Fujifilm) and Multi Gauge software (version 3.0; Fujifilm). Contrast and brightness adjustment was applied using Photoshop 7.0.1 (Adobe).
Antibodies and reagents
Rabbit polyclonal antibody against EIF4EBP1/4E-BP1 (9542), RPS6KB1 (2217), and RPS6KB1 pThr389 (9234) were purchased from Cell Signaling. Guinea pig polyclonal and rabbit polyclonal anti-SQSTM1/p62 antibodies were purchased from Progen (GP62-C) and MBL (PM 045), respectively. Anti-GFP (NACALAI TESQUE, 04404) and anti-Multi Ubiquitin (MBL, D058-3) antibodies were purchased. Anti-LC3BCitation45 antibody production was described previously. Horseradish peroxidase-conjugated anti-mouse (315035003) and anti-rabbit (111035144) IgG antibodies were purchased from Jackson ImmunoResearch, bafilomycin A1 (02911643) and α-amanitin (23109059) were purchased from Wako, cycloheximide (01810) and chloroquine (C6628) were purchased from Sigma, and Torin1 was purchased from Tocris Biosciences (4247). LysoTracker Red (L7528) was purchased from Molecular Probes.
Quantitative RT-PCR
Total RNA was extracted from MEFs using ISOGEN (Nippon Gene, 31102501) according to the manufacturer’s instructions. CDNA was generated with ReverTra Ace (Toyobo, TRT101) and was analyzed by quantitative real-time RT-PCR using the SYBR Green assay (Takara, RR018B). All data were normalized to Actb/β-actin or Gapdh expression. Primer sequences were:
Sqstm1: 5′-GCCAGAGGAACAGATGGAGT-3′
and 5′-TCCGATTCTG GCATCTGTAG-3′;
Actb: 5′-CTGGGTATGGAATCCTGTGG-3′
and 5′-GTACTTGCGCTCAGGAGGAG-3′.
GFP: 5′-ACAAGCAGAAGAACGGCATC-3′
and 5′-ACTGGGTGCTCAGGTAGTGG-3′
Gapdh: 5′-GCCAAGGTCATCCATGACAACT-3′
and 5′-GAGGGGCCATCCACAGTCT-3′.
Fluorescence microscopy
Cells were cultured on a glass-bottomed dish and directly observed. For examination by immunofluorescence microscopy, cells grown on gelatin-coated coverslips were fixed with 4% paraformaldehyde, permeabilized using 50 μg/ml digitonin, and then stained with specific antibodies. All images acquired using confocal laser microscope (FV1000D IX81; Olympus), using a 60× PlanApoN oil immersion lens (1.42 numerical aperture; Olympus).
Statistical analyses
All numerical data including error bars represent the mean ± SEM. Statistical comparisons were made using the unpaired Student t test.
| Abbreviations: | ||
| Baf | = | bafilomycin A1 |
| CHX | = | cycloheximide |
| CMV | = | cytomegalovirus |
| CQ | = | chloroquine |
| DMEM | = | Dulbecco’s modified Eagle’s medium |
| GFP | = | green fluorescent protein |
| KO | = | knockout |
| MAP1LC3 (LC3) | = | microtubule-associated protein 1 light chain 3 |
| LIR | = | LC3-interacting region |
| MEF | = | mouse embryonic fibroblast |
| MTORC1 | = | mechanistic target of rapamycin complex 1 |
| NFE2L2/Nrf2 | = | nuclear factor, erythroid 2-like 2 |
| NFKB/NFκB | = | nuclear factor of kappa light polypeptide gene enhancer in B-cells |
| PBS | = | phosphate-buffered saline |
| RB1CC1/FIP200 | = | RB1-inducible coiled-coil 1 |
| SQSTM1 | = | sequestosome 1 |
Additional material
Download Zip (762 KB)Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Acknowledgments
This work was supported in part by Grants-in-Aid for Scientific Research from the Ministry of Education, Culture, Sports, Science and Technology, Japan, by the Funding Program for Next Generation World-Leading Researchers and JSPS KAKENHI Grants-in-Aid for Scientific Research on Innovative Areas (Grant Number 25111005) (to N.M.). M.H.S. is supported by a scholarship from the Ministry of Education, Culture, Sports, Science and Technology (MEXT) in Japan.
References
- Rubinsztein DC. The roles of intracellular protein-degradation pathways in neurodegeneration. Nature 2006; 443:780 - 6; http://dx.doi.org/10.1038/nature05291; PMID: 17051204
- Levine B, Kroemer G. Autophagy in the pathogenesis of disease. Cell 2008; 132:27 - 42; http://dx.doi.org/10.1016/j.cell.2007.12.018; PMID: 18191218
- Rabinowitz JD, White E. Autophagy and metabolism. Science 2010; 330:1344 - 8; http://dx.doi.org/10.1126/science.1193497; PMID: 21127245
- Mizushima N, Komatsu M. Autophagy: renovation of cells and tissues. Cell 2011; 147:728 - 41; http://dx.doi.org/10.1016/j.cell.2011.10.026; PMID: 22078875
- Noda NN, Ohsumi Y, Inagaki F. Atg8-family interacting motif crucial for selective autophagy. FEBS Lett 2010; 584:1379 - 85; http://dx.doi.org/10.1016/j.febslet.2010.01.018; PMID: 20083108
- Johansen T, Lamark T. Selective autophagy mediated by autophagic adapter proteins. Autophagy 2011; 7:279 - 96; http://dx.doi.org/10.4161/auto.7.3.14487; PMID: 21189453
- Weidberg H, Shvets E, Elazar Z. Biogenesis and cargo selectivity of autophagosomes. Annu Rev Biochem 2011; 80:125 - 56; http://dx.doi.org/10.1146/annurev-biochem-052709-094552; PMID: 21548784
- Bjørkøy G, Lamark T, Brech A, Outzen H, Perander M, Øvervatn A, Stenmark H, Johansen T. p62/SQSTM1 forms protein aggregates degraded by autophagy and has a protective effect on huntingtin-induced cell death. J Cell Biol 2005; 171:603 - 14; http://dx.doi.org/10.1083/jcb.200507002; PMID: 16286508
- Pankiv S, Clausen TH, Lamark T, Brech A, Bruun JA, Outzen H, Øvervatn A, Bjørkøy G, Johansen T. p62/SQSTM1 binds directly to Atg8/LC3 to facilitate degradation of ubiquitinated protein aggregates by autophagy. J Biol Chem 2007; 282:24131 - 45; http://dx.doi.org/10.1074/jbc.M702824200; PMID: 17580304
- Wooten MW, Hu X, Babu JR, Seibenhener ML, Geetha T, Paine MG, Wooten MC. Signaling, polyubiquitination, trafficking, and inclusions: sequestosome 1/p62’s role in neurodegenerative disease. J Biomed Biotechnol 2006; 2006:62079; http://dx.doi.org/10.1155/JBB/2006/62079; PMID: 17047309
- Seibenhener ML, Geetha T, Wooten MW. Sequestosome 1/p62--more than just a scaffold. FEBS Lett 2007; 581:175 - 9; http://dx.doi.org/10.1016/j.febslet.2006.12.027; PMID: 17188686
- Moscat J, Diaz-Meco MT. p62 at the crossroads of autophagy, apoptosis, and cancer. Cell 2009; 137:1001 - 4; http://dx.doi.org/10.1016/j.cell.2009.05.023; PMID: 19524504
- Komatsu M, Kageyama S, Ichimura Y. p62/SQSTM1/A170: physiology and pathology. Pharmacol Res 2012; 66:457 - 62; http://dx.doi.org/10.1016/j.phrs.2012.07.004; PMID: 22841931
- Itakura E, Mizushima N. p62 Targeting to the autophagosome formation site requires self-oligomerization but not LC3 binding. J Cell Biol 2011; 192:17 - 27; http://dx.doi.org/10.1083/jcb.201009067; PMID: 21220506
- Klionsky DJ, Abdalla FC, Abeliovich H, Abraham RT, Acevedo-Arozena A, Adeli K, Agholme L, Agnello M, Agostinis P, Aguirre-Ghiso JA, et al. Guidelines for the use and interpretation of assays for monitoring autophagy. Autophagy 2012; 8:445 - 544; http://dx.doi.org/10.4161/auto.19496; PMID: 22966490
- Mizushima N, Yoshimori T, Levine B. Methods in mammalian autophagy research. Cell 2010; 140:313 - 26; http://dx.doi.org/10.1016/j.cell.2010.01.028; PMID: 20144757
- Komatsu M, Waguri S, Koike M, Sou YS, Ueno T, Hara T, Mizushima N, Iwata J, Ezaki J, Murata S, et al. Homeostatic levels of p62 control cytoplasmic inclusion body formation in autophagy-deficient mice. Cell 2007; 131:1149 - 63; http://dx.doi.org/10.1016/j.cell.2007.10.035; PMID: 18083104
- Komatsu M, Kurokawa H, Waguri S, Taguchi K, Kobayashi A, Ichimura Y, Sou YS, Ueno I, Sakamoto A, Tong KI, et al. The selective autophagy substrate p62 activates the stress responsive transcription factor Nrf2 through inactivation of Keap1. Nat Cell Biol 2010; 12:213 - 23; PMID: 20173742
- Lau A, Wang XJ, Zhao F, Villeneuve NF, Wu T, Jiang T, Sun Z, White E, Zhang DD. A noncanonical mechanism of Nrf2 activation by autophagy deficiency: direct interaction between Keap1 and p62. Mol Cell Biol 2010; 30:3275 - 85; http://dx.doi.org/10.1128/MCB.00248-10; PMID: 20421418
- Ichimura Y, Kumanomidou T, Sou YS, Mizushima T, Ezaki J, Ueno T, Kominami E, Yamane T, Tanaka K, Komatsu M. Structural basis for sorting mechanism of p62 in selective autophagy. J Biol Chem 2008; 283:22847 - 57; http://dx.doi.org/10.1074/jbc.M802182200; PMID: 18524774
- Mizushima N, Yoshimori T. How to interpret LC3 immunoblotting. Autophagy 2007; 3:542 - 5; PMID: 17611390
- Itakura E, Kishi-Itakura C, Mizushima N. The hairpin-type tail-anchored SNARE syntaxin 17 targets to autophagosomes for fusion with endosomes/lysosomes. Cell 2012; 151:1256 - 69; http://dx.doi.org/10.1016/j.cell.2012.11.001; PMID: 23217709
- Yu L, McPhee CK, Zheng L, Mardones GA, Rong Y, Peng J, Mi N, Zhao Y, Liu Z, Wan F, et al. Termination of autophagy and reformation of lysosomes regulated by mTOR. Nature 2010; 465:942 - 6; http://dx.doi.org/10.1038/nature09076; PMID: 20526321
- Thoreen CC, Kang SA, Chang JW, Liu Q, Zhang J, Gao Y, Reichling LJ, Sim T, Sabatini DM, Gray NS. An ATP-competitive mammalian target of rapamycin inhibitor reveals rapamycin-resistant functions of mTORC1. J Biol Chem 2009; 284:8023 - 32; http://dx.doi.org/10.1074/jbc.M900301200; PMID: 19150980
- Ni HM, Bockus A, Wozniak AL, Jones K, Weinman S, Yin XM, Ding WX. Dissecting the dynamic turnover of GFP-LC3 in the autolysosome. Autophagy 2011; 7:188 - 204; http://dx.doi.org/10.4161/auto.7.2.14181; PMID: 21107021
- Ishii T, Itoh K, Takahashi S, Sato H, Yanagawa T, Katoh Y, Bannai S, Yamamoto M. Transcription factor Nrf2 coordinately regulates a group of oxidative stress-inducible genes in macrophages. J Biol Chem 2000; 275:16023 - 9; http://dx.doi.org/10.1074/jbc.275.21.16023; PMID: 10821856
- Jain A, Lamark T, Sjøttem E, Larsen KB, Awuh JA, Øvervatn A, McMahon M, Hayes JD, Johansen T. p62/SQSTM1 is a target gene for transcription factor NRF2 and creates a positive feedback loop by inducing antioxidant response element-driven gene transcription. J Biol Chem 2010; 285:22576 - 91; http://dx.doi.org/10.1074/jbc.M110.118976; PMID: 20452972
- Thompson HG, Harris JW, Wold BJ, Lin F, Brody JP. p62 overexpression in breast tumors and regulation by prostate-derived Ets factor in breast cancer cells. Oncogene 2003; 22:2322 - 33; http://dx.doi.org/10.1038/sj.onc.1206325; PMID: 12700667
- Zou X, Feng Z, Li Y, Wang Y, Wertz K, Weber P, Fu Y, Liu J. Stimulation of GSH synthesis to prevent oxidative stress-induced apoptosis by hydroxytyrosol in human retinal pigment epithelial cells: activation of Nrf2 and JNK-p62/SQSTM1 pathways. J Nutr Biochem 2012; 23:994 - 1006; http://dx.doi.org/10.1016/j.jnutbio.2011.05.006; PMID: 21937211
- Liu XD, Ko S, Xu Y, Fattah EA, Xiang Q, Jagannath C, Ishii T, Komatsu M, Eissa NT. Transient aggregation of ubiquitinated proteins is a cytosolic unfolded protein response to inflammation and endoplasmic reticulum stress. J Biol Chem 2012; 287:19687 - 98; http://dx.doi.org/10.1074/jbc.M112.350934; PMID: 22518844
- Settembre C, Zoncu R, Medina DL, Vetrini F, Erdin S, Erdin S, Huynh T, Ferron M, Karsenty G, Vellard MC, et al. A lysosome-to-nucleus signalling mechanism senses and regulates the lysosome via mTOR and TFEB. EMBO J 2012; 31:1095 - 108; http://dx.doi.org/10.1038/emboj.2012.32; PMID: 22343943
- Settembre C, Di Malta C, Polito VA, Garcia Arencibia M, Vetrini F, Erdin S, Erdin SU, Huynh T, Medina D, Colella P, et al. TFEB links autophagy to lysosomal biogenesis. Science 2011; 332:1429 - 33; http://dx.doi.org/10.1126/science.1204592; PMID: 21617040
- Johnson PF. Molecular stop signs: regulation of cell-cycle arrest by C/EBP transcription factors. J Cell Sci 2005; 118:2545 - 55; http://dx.doi.org/10.1242/jcs.02459; PMID: 15944395
- Tsukada J, Yoshida Y, Kominato Y, Auron PE. The CCAAT/enhancer (C/EBP) family of basic-leucine zipper (bZIP) transcription factors is a multifaceted highly-regulated system for gene regulation. Cytokine 2011; 54:6 - 19; http://dx.doi.org/10.1016/j.cyto.2010.12.019; PMID: 21257317
- Ma D, Panda S, Lin JD. Temporal orchestration of circadian autophagy rhythm by C/EBPβ. EMBO J 2011; 30:4642 - 51; http://dx.doi.org/10.1038/emboj.2011.322; PMID: 21897364
- B’chir W, Maurin AC, Carraro V, Averous J, Jousse C, Muranishi Y, Parry L, Stepien G, Fafournoux P, Bruhat A. The eIF2α/ATF4 pathway is essential for stress-induced autophagy gene expression. Nucleic Acids Res 2013; 41:7683 - 99; http://dx.doi.org/10.1093/nar/gkt563; PMID: 23804767
- Kuma A, Mizushima N. Physiological role of autophagy as an intracellular recycling system: with an emphasis on nutrient metabolism. Semin Cell Dev Biol 2010; 21:683 - 90; http://dx.doi.org/10.1016/j.semcdb.2010.03.002; PMID: 20223289
- Onodera J, Ohsumi Y. Autophagy is required for maintenance of amino acid levels and protein synthesis under nitrogen starvation. J Biol Chem 2005; 280:31582 - 6; http://dx.doi.org/10.1074/jbc.M506736200; PMID: 16027116
- Suzuki SW, Onodera J, Ohsumi Y. Starvation induced cell death in autophagy-defective yeast mutants is caused by mitochondria dysfunction. PLoS One 2011; 6:e17412; http://dx.doi.org/10.1371/journal.pone.0017412; PMID: 21364763
- Moscat J, Diaz-Meco MT. p62: a versatile multitasker takes on cancer. Trends Biochem Sci 2012; 37:230 - 6; http://dx.doi.org/10.1016/j.tibs.2012.02.008; PMID: 22424619
- Duran A, Amanchy R, Linares JF, Joshi J, Abu-Baker S, Porollo A, Hansen M, Moscat J, Diaz-Meco MT. p62 is a key regulator of nutrient sensing in the mTORC1 pathway. Mol Cell 2011; 44:134 - 46; http://dx.doi.org/10.1016/j.molcel.2011.06.038; PMID: 21981924
- Larsen KB, Lamark T, Øvervatn A, Harneshaug I, Johansen T, Bjørkøy G. A reporter cell system to monitor autophagy based on p62/SQSTM1. Autophagy 2010; 6:784 - 93; http://dx.doi.org/10.4161/auto.6.6.12510; PMID: 20574168
- Gan B, Peng X, Nagy T, Alcaraz A, Gu H, Guan JL. Role of FIP200 in cardiac and liver development and its regulation of TNFalpha and TSC-mTOR signaling pathways. J Cell Biol 2006; 175:121 - 33; http://dx.doi.org/10.1083/jcb.200604129; PMID: 17015619
- Kuma A, Hatano M, Matsui M, Yamamoto A, Nakaya H, Yoshimori T, Ohsumi Y, Tokuhisa T, Mizushima N. The role of autophagy during the early neonatal starvation period. Nature 2004; 432:1032 - 6; http://dx.doi.org/10.1038/nature03029; PMID: 15525940
- Hosokawa N, Hara Y, Mizushima N. Generation of cell lines with tetracycline-regulated autophagy and a role for autophagy in controlling cell size. FEBS Lett 2006; 580:2623 - 9; http://dx.doi.org/10.1016/j.febslet.2006.04.008; PMID: 16647067
