Abstract
The members of the LC3/Atg8 family of proteins are covalently attached to phagophore and autophagosomal membranes. At the last step of the LC3 lipidation cascade, LC3 is transferred from the E2 enzyme ATG3 to phosphatidylethanolamine (PE). This transfer is stimulated by the ATG12–ATG5-ATG16L1 E3 complex, but the mechanism is not fully understood. We recently found that ATG12 of the E3 binds to a short sequence in the flexible region (FR) of ATG3 with high affinity, and that this interaction is critical for E2–E3 complex formation. These findings, together with detailed structural analyses of this interaction, define the properties of ATG12 and provide new insights of how LC3 transfer begins with ATG3 recruitment by ATG12.
Lipid conjugation of LC3 is essential for autophagosome formation and therefore extensively used to examine autophagic activity. Conjugation of LC3 to a lipid molecule, rather than a protein, is unusual for a ubiquitin-like protein. The reaction may target PE molecules specifically at the autophagosome-forming sites, even though PE is distributed throughout cellular membranes. These features are made possible by the unique E2–E3 pair of this ubiquitination-like reaction cascade. The E2 ATG3 has a canonical E2 conjugating domain that recognizes the lipid molecule. It also has a unique ~100 amino acid long flexible region, which is involved in E1 binding. The E3 factor, the ATG12–ATG5-ATG16L1 complex, localizes to phagophore membranes and stimulates the transfer of LC3 from ATG3 to PE. Thus, E3 localization determines the site of LC3 conjugation. However, it was not known how the E3 recruits ATG3.
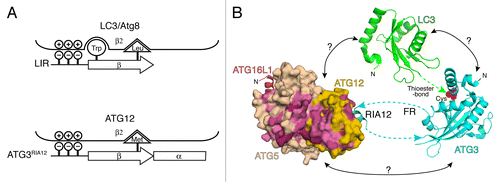
Through biophysical analyses on human proteins to dissect the E2–E3 interaction we found a ~30 residue-long peptide region within the FR of ATG3 that binds to E3. Subsequent co-crystallization of E3 with the binding peptide of ATG3 revealed the binding region to be a ~13 residue-long sequence within ATG3FR that binds exclusively to ATG12. Therefore, we referred to this peptide as the region interacting with ATG12 (RIA12) to specify the binding region accurately within the entire FR. At the N terminus of RIA12 is a 4 residue-long β-strand, which forms an intermolecular β-sheet with ATG12’s β2, followed by a short α-helix. Because LC3 and the LC3-interacting-region (LIR, also called LRS/AIM) interact with each other through a similar intermolecular β-sheet organization, we compared the RIA12-ATG12 and LIR-LC3 structures and found that the N-terminal β-strand of RIA12 and LIR superimpose well on the aligned ubiquitin-like folds (). A typical sequence of LIR is Trp-Xaa-Xaa-Leu and the side-chains of the Trp and Leu residues fit in respective hydrophobic pockets on the surface of LC3. The comparison revealed that Met157 of RIA12 superimposes on the Leu of the LIR, and correspondingly ATG12 has a hydrophobic pocket that accommodates Met157’s side-chain. Unlike LIR, RIA12 does not have a Trp residue at its N terminus, but Met157 is followed by a unique α-helix, which makes electrostatic and hydrophobic contacts with surface residues of ATG12. Highly acidic residues not resolved in our crystal structure, but immediately preceding the 13 visible residues of the RIA12, were found to contribute to the overall affinity. These electrostatic interactions at the N-terminal position with the β-sheet contacts described above are also found in the LIR-LC3 interaction. In summary, our data show that ATG12 uniquely binds to the peptide sequence in the RIA12 using a mechanism that is in part similar to the LIR-LC3 recognition. These findings, together with the fact that ATG12 and LC3 share the highest sequence homology with each other among all the ubiquitin-like proteins, suggest a common evolutionary root for these proteins.
Figure 1. Summary of structural information of the E2–E3 interaction. (A) A schematic presentation of the LC3-LIR (top) and ATG12-ATG3RIA12 (bottom) interactions. (B) The structures of the human ATG3RIA12-ATG12–ATG5-ATG16N (the N-terminal fragment of ATG16L1) (PDB ID: 4NAW) complex, Arabidopsis thaliana ATG3 (3VX8) and human LC3 (1UGM) are shown in ribbon models, while ATG12–ATG5 is shown in a surface model. The residues with the highest and high conservation are colored with magenta and pink, respectively. The cyan and green dotted lines indicate the missing connections between the N- or C-termini of the RIA12 and the E2 core of ATG3, and the C terminus of LC3 and the catalytic cysteine residue (red) of ATG3, respectively. The arrows at the ends of the dotted lines indicate the directions of protein chains. Possible but uncharacterized interactions are indicated with question marks.
In light of the similarity described above, we speculate that there may be more proteins that contain a RIA12-like sequence to bind ATG12. Because the sequence conservation of the RIA12 of ATG3 is clear only among higher eukaryotes, but not among fungi, and the fungal Atg3FR sequences are more diverse, any possible interactions involving a RIA12-like sequence might be specific to higher eukaryotes. In this regard, it is of note that such binding need not be related directly to autophagosome formation, as ATG12 appears to function for nonautophagic events as well as in higher eukaryotes. In our view, the poor conservation does not suggest that the E2–E3 interaction is fundamentally different in fungi. Rather, similar structural mechanisms would likely be observed with fungal Atg3 because their FRs are generally acidic and contain some amino acids with aliphatic side-chains, such as methionine and isoleucine. Indeed, the structure of S. cerevisiae Atg12–Atg5 shows the Met-binding pocket on its surface. Experimental investigation would clarify structural conservation in the E2–E3 interaction in fungi.
While our work explains how E3 recruits E2, it still remains unclear exactly how E3 stimulates LC3 transfer. Past structural studies on the SUMO and ubiquitin systems have shown that E3-ligases for these modifiers simultaneously bind to both E2 and SUMO/ubiquitin thioester-bonded to the E2, resulting in stabilization of an orientation and position of SUMO/ubiquitin on E2 optimal for the attack of the thioester-bond by the substrate lysine side-chains. If a similar mechanism is employed for LC3 transfer, we expect surfaces on LC3, ATG3, and ATG12–ATG5 to mediate new contacts among these proteins. Unlike E3-ligases for SUMO and ubiquitin, however, ATG12–ATG5 does not stably interact with either LC3 or the isolated E2 core of ATG3. Thus, at this point the mechanism for stimulation of LC3 transfer by ATG12–ATG5 remains unclear. We previously showed that the conjugation of ATG12 and ATG5 makes a composite surface across these 2 molecules that is important for LC3 lipidation. We are currently focusing on elucidating how this surface contributes to LC3 transfer (). Questions in this regard are: 1) What does the composite surface interact with? 2) Are there conformational changes in ATG12–ATG5 when it is bound to LC3-charged ATG3? 3) How does the active conformation of the E2 core of ATG3 look when it is poised for LC3 transfer? 4) How do LC3, ATG3, and ATG12–ATG5-ATG16L1 bind to membranes during the transfer? Clearly, more work is needed for a complete description of the mechanisms for LC3 lipidation.
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Acknowledgments
This work was supported by NIH R01GM092740 to TO.
