Abstract
Mitophagy is a degradative process that adapts the quantity and quality of mitochondria to the cellular needs. Mitochondria destined for degradation are marked by specific receptors that recruit the core autophagic machinery to the organellar surface. The organelle is then enclosed by a phagophore (PG) which fuses with the lysosome or vacuole where the mitochondrion is degraded. In spite of significant progress in recent years, several parts of the molecular machinery of mitophagy remain unknown. We used yeast as a model organism to screen for novel components and identified the mitochondria-ER tether ERMES (ER-mitochondria encounter structure) as a major player contributing to mitophagy and formation of mitophagosomes. Tethering of mitochondria to the ER appears to be important to supply the growing PG with lipids synthesized in the ER.
Mitochondria are involved in various cellular processes including oxidative phosphorylation, Fe/S-cluster biosynthesis, apoptosis, and many more. Because of their central role in cell physiology they are targets of several quality control mechanisms. While various proteases act at the molecular level to degrade misfolded or damaged proteins, mitophagy acts at the organellar level. Dysfunctional mitochondrial quality control is associated with neurodegenerative diseases, such as Parkinson disease, and thus, much effort has been made to gain insights into the molecular mechanisms governing mitophagy.
Due to its genetic amenability baker's yeast Saccharomyces cerevisiae is widely used as a model organism to study mitophagy. To induce mitophagy yeast cells are usually grown on nonfermentable carbon sources, which results in the substantial proliferation of mitochondria. Upon entry into post-log phase or starvation, surplus mitochondria are degraded by mitophagy to fuel cell metabolism. Previously reported screens of the yeast deletion collection (comprising mutants of all ca. 4800 nonessential yeast genes) identified the mitophagy receptor Atg32 under these growth conditions. However, all respiratory-deficient mutants were missed in these screens, as they cannot grow on nonfermentable carbon sources. We reasoned that these mutants could be particularly interesting because of their potential involvement in mitochondrial function. Thus, we developed a protocol that allows starvation of respiratory-deficient mutants grown on fermentable carbon sources and assayed mitophagy using a fluorescent reporter protein. Screening of roughly 380 mutants revealed many mutants with significantly increased or reduced mitophagy. Intriguingly, all mutants lacking genes encoding the 4 ERMES subunits showed strong mitophagy defects. The ERMES complex consists of the ER membrane protein Mmm1, the mitochondrial outer membrane proteins Mdm10 and Mdm34, and the soluble factor Mdm12. ERMES connects the ER and mitochondria and is involved in mitochondrial division, mitochondrial DNA segregation, and exchange of lipids between the ER and mitochondria. ERMES mutants are not defective in bulk autophagy, which suggests that the effect is specific for mitochondrial autophagy.
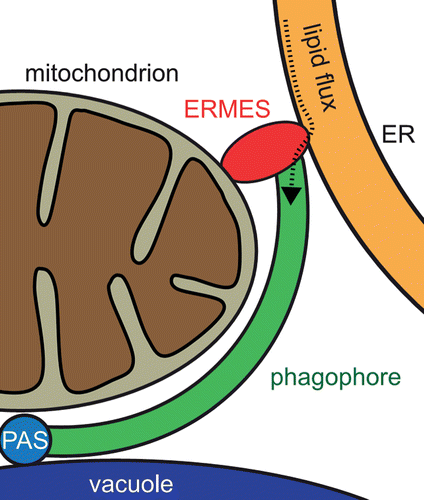
Atg8, a conserved component of the core machinery of autophagy, is involved in PG expansion. Split-YFP experiments showed that Mmm1 interacts with Atg8 in vivo, corroborating a role of ERMES in mitophagy. Fluorescence microscopy revealed that ERMES is present at the PG edge distal to the vacuole which likely corresponds to its growing end. As ERMES has been suggested to facilitate the exchange of lipids between membranes, we reasoned that ERMES could promote lipid flux from the ER to the growing PG. Consistently, Atg5-GFP, a marker of immature PGs, labels multiple PG structures accumulating at mitochondria in mmm1∆ yeast cells. Presumably these structures correspond to nascent mitophagosomes that cannot complete maturation in the absence of ERMES. Strikingly, expression of chiMERA, a protein artificially tethering ER and mitochondria, rescues mitophagy defects and the Atg5-GFP phenotype, which indicates that ER-mitochondria contacts per se are required for autophagic removal of mitochondria.
Since Atg8 and Atg32 colocalization is not affected in ERMES mutants, our data suggest a model in which ERMES acts at the step of PG expansion after Atg8-Atg32 interaction (). Presumably, growth of the PG depends on supply with lipids from the ER. In this scenario, ERMES mediates spatial proximity between the mitochondrion as autophagic cargo, the ER as membrane lipid source, and Atg8 as a PG expansion factor. Autophagic structures marked by Atg5-GFP accumulate in ERMES mutants, because PG generation is stalled due to insufficient lipid supply, and cells try to compensate this by induction of PG biogenesis at several sites.
Figure 1. Model of ERMES-mediated mitophagophore expansion. Phagophore (PG) generation is initiated at the phagophore assembly site (PAS) where Atg32 interacts with the PAS components Atg8 and Atg11 (not depicted) in juxtaposition to the vacuole. ERMES is localized at the distal end of the PG edge where it connects the mitochondrion destined for degradation, the PG, and the ER as lipid source. The putative lipid flux from the ER to the PG is depicted by a dotted line.
Several interesting questions remain. What happens to the ERMES components during mitophagy? Is ERMES degraded together with the mitochondrion, or does it remain associated with the mitochondrial tip outside the PG where another mitochondrial piece can be isolated for mitophagy? At least the physical link to the ER has to be resolved before the PG is closed. It is presently unknown how this is achieved. The fact that ERMES is involved in mitochondrial DNA segregation during mitochondrial fission points to another question: What happens to mitochondrial DNA during mitophagy? Is there a selection mechanism sorting out defective mitochondrial genomes? Recently, it was shown by the Klionsky group that pexophagy, the selective degradation of peroxisomes, depends on peroxisomal fission. Remarkably, peroxisomes marked for degradation are often in close vicinity to mitochondria. It would be interesting to see whether pexophagy is initiated at mitochondrial ER contacts and whether degradation of the 2 organelles is coupled.
Our screen revealed not only strains with reduced mitophagy, such as ERMES mutants, but also several candidates that show massively increased mitophagy. These strains are of particular interest because augmented mitophagy has been rarely observed until now. These mutants might accumulate mitochondrial defects rendering them more prone to mitophagy which could substantiate the idea of mitophagy as a quality control mechanism. We hope that in-depth characterization of candidate genes from this and other screens will provide further insights into the molecular mechanism and the physiological relevance of mitophagy.
| Abbreviations: | ||
| ERMES | = | ER-mitochondria encounter structure |
| PG | = | phagophore |
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
