Abstract
The conventional and most accepted method of measuring the lytic activity of a phage against its bacterial host is the plaque assay. This method is laborious, time consuming and expensive, especially in high throughput analyses where multiple phage-bacterial interactions are required to be monitored simultaneously. It can also vary considerably with the experimenter and by the growth and plating conditions. Alternatively, the lytic activity can be measured indirectly by following the decrease in optical density of the bacterial cultures owing to lysis. Here we describe an automated, high throughput, indirect liquid lysis assay to evaluate phage growth using the OmniLogTM system. The OmniLogTM system uses redox chemistry, employing cell respiration as a universal reporter. During active growth of bacteria, cellular respiration reduces a tetrazolium dye and produces a color change that is measured in an automated fashion. On the other hand, successful phage infection and subsequent growth of the phage in its host bacterium results in reduced bacterial growth and respiration and a concomitant reduction in color. Here we show that microtiter plate wells inoculated with Bacillus anthracis and phage show decreased or no growth, compared with the wells containing bacteria only or phage resistant bacteria plus phage. Also, we show differences in the kinetics of bacterial growth and the timing of appearance of phage resistant bacteria in the presence of individual phages or a cocktail of B. anthracis specific phages. The results of these experiments indicate that the OmniLogTM system could be used reliably for indirectly measuring phage growth in high throughput host range and phage and antibiotics combination studies.
Introduction
Bacteriophages are obligate intracellular parasites of bacteria, and in general they are highly host (bacteria) specific at species and often even at strain level.Citation1 There are four basic steps in the life cycle of a bacteriophage: (1) specific attachment of the phage to the bacterial surface, (2) injection of phage DNA into the host cytoplasm, (3) growth and multiplication of phage inside the bacterium and (4) eventual lysis of the host releasing progeny phage particles.Citation2 In nature, there is a plethora of phage-specific variations in each of these steps. Each of these steps can be monitored and measured by a variety of methodsCitation3 that can be broadly grouped into two categories: semi-solid medium and liquid assays.
An example of semi-solid medium assay is the plaque assay, which is somewhat more quantitative as compared with spot or cross-streak test.Citation4,Citation5 The plaque assay was originally described by one of the discoverers of phages, Felix d’Herelle,Citation6 and later modified by a number of other phage biologists.Citation7-Citation11 The plaque assay measures the formation of a clear zone in a bacterial lawn, resulting from amplification of a single phage particle. Although the plaque assay is simple, plaque morphologies and sizes can vary with the experimenter, media and other conditions. Plaque assay based enumeration gives the efficiency of plating (e.o.p.) of a given phage preparation in the bacterial strain in which the phage was prepared as compared with other susceptible strains. The presence of restriction modification systems in bacteria can heavily influence the efficiency of plating in a given host.Citation12
Liquid lysis assays generally monitor the reduction in the optical density of a bacterial culture upon infection, growth and lysis by phage. Recently, an automated lytic assay based on Bioscreen C analyzer was reported.Citation13 The drawback of this method is that bacterial cell debris resulting from lysis may contribute to the optical density values recorded and therefore might obscure or underestimate the real phage lytic activity. In contrast to methods described above, single step growth experiments offer a more precise measurement of phage growth in a given bacterial strain, giving data on each of the steps in the phage life cycle, such as adsorption efficiency, latent period, eclipse period, burst size, etc.Citation3,Citation8 However, single step growth experiments are not only resource and reagent intensive, for example, requiring a large number of plates, they are also labor intensive and therefore impractical in high-throughput analysis.
Automation is one potential solution for the inconsistencies and inconveniences inherent in many of the existing phage assays. Therefore, we tested the utility of the OmniLog instrument (Biolog Inc.) for automated assay of phage growth. The OmniLogTM instrument is a specialized plate incubator (with a capacity of 50 microtiter plates) coupled to a camera and a computer and is marketed for use with specialized phenotypic microarray plates (PM) or specialized plates for bacterial identification assays. In the case of the PM assays, the wells of each plate contain different carbon sources, secondary metabolites and even selective or inhibitory compounds/agents like antibiotics, and redox chemistry is employed to measure respiration of input microorganisms under the various conditions present in each well.Citation14 The PM technology has been successfully used in a variety of comparative analyses of phenotypes of organismsCitation15,Citation16 as well as to assess the accuracy of genome annotations.Citation17
In the current study, we have utilized the OmniLogTM system to monitor lack or reduction of bacterial growth in the presence of phage as compared with untreated conditions. The OmniLogTM system is designed to rely on redox chemistry, employing cell respiration as a universal reporter. If the growth of bacteria is strongly positive, the cells respire actively, reduce a tetrazolium dye and form a strong color. In the case of bacterial lysis by phage infection, respiration is slowed or stopped, and less color or no color is formed. In the OmniLogTM system, as many as 4,800 bacterial growth assays in the presence of phage can be performed simultaneously, allowing kinetics of bacterial growth and development of resistance to be monitored continuously over the course of the experiment. Here, we demonstrate the utility of the OmniLogTM system for indirect measuring of phage lytic activity using six individual Bacillus anthracis-specific phages as well as a cocktail of phages in combination with different B. anthracis strains. The results of these experiments indicated that the OmniLogTM system could be used reliably in high-throughput host range analysis and for evaluating additive or antagonistic interactions of phages and antibiotics. As compared with more traditional phage assays, the OmniLogTM system has the distinct advantage of automatic, real-time monitoring of phage infection and bacterial growth over the entire duration of the experiment, spanning several days if desired.
Results
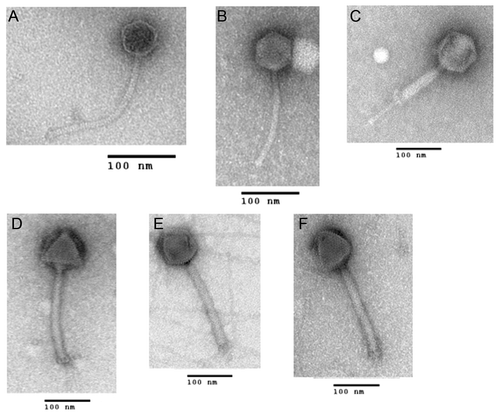
Morphology of B. anthracis phages used in this study
Six phages were used individually or as a cocktail for the in vitro indirect phage lytic assays using the OmniLogTM system. Phage Gamma is a CDC recommended diagnostic phage for B. anthracis. Giraffe was isolated from giraffe feces from a zoo on Long Island, New York and characterization of this phage will be published elsewhere (Ray Schuch, personal communication). The other four phages were isolated in this study from sewage collected from Great Seneca sewage treatment plant in Germantown, Maryland. Electron micrographs of the six phages used in this study are shown in . As published, Gamma () exhibits a typical morphology of a phage belonging to the family SiphoviridaeCitation18 with an icosahedral head and a long non-contractile tail and phage Giraffe exhibits a similar morphology (). Phage BA39 () appears to belong to the Myoviridae family (icosahedral head and contractile tail). Phages BA21, BA28, BA51 also appear to belong to the family Myoviridae based on tail width and bacterial growth inhibition profiles (see later) but this assignment is tentative.
Monitoring the kinetics of bacterial growth using OmniLogTM system upon infection with Giraffe phage
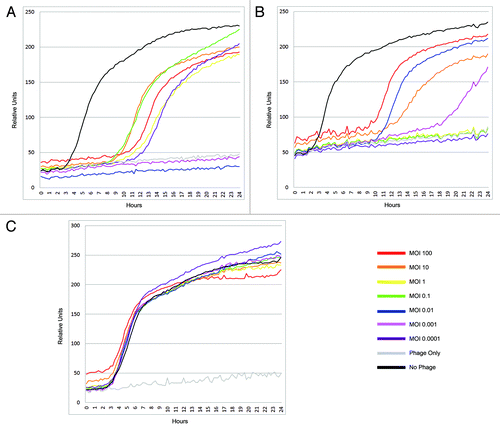
In order to assess the utility of the OmniLogTM system for indirect assay of phage lysis, we tested individual phages against their host, vegetative cells and spores of B. anthracis Sterne 7702. The kinetics of growth in wells seeded with B. anthracis Sterne strain 7702 vegetative cells or spores with and without infection by Giraffe phage are shown in , panels A and B respectively. The growth of 7702 without phage infection followed a typical sigmoidal growth curve with a lag period of ~3 h. Addition of phage at concentrations ranging from 0.0001to 100 MOIs completely suppressed bacterial growth for ~9 h after which growth resumed in majority of cases. There are some cases in which bacterial growth was completely suppressed for up to 24 h. However, there was no correlation with the concentration of phage and the appearance of resistance; for example, with only 0.001 or 0.01 MOI of phage, growth was completely suppressed, whereas at concentrations of 0.1 to 100 and 0.0001 MOI, growth was evident after 9 h. A similar profile was seen when spores were used as the inocula instead of vegetative cells. In this case, growth suppression was evident up to ~9 h after which growth resumed. However, complete suppression was observed up to 24 h in cases where phages were added at MOIs of 0.0001, 0.1 and 1. We hypothesize that the resurgence of growth after 9 h is the result of the presence of truly phage resistant or “persister” bacteria in the population.Citation19 The absence of a correlation with the concentration of the phage inocula could indicate that the appearance of phage resistant/persister bacteria followed a random fluctuation model.Citation20 Experiments where the cultures were monitored for up to 48 h showed a similar trend (data not shown). In , we presume that infection is occurring post germination of the spores, as all the phages used in this study have been shown to specifically infect vegetative B. anthracis and not spores (unpublished data). There is rapid germination and outgrowth of spores and subsequent infection without a further lag period since this time is comparable to the lag period seen in uninfected vegetative cell controls. We also tested the kinetics of growth of Giraffe phage-resistant mutant upon exposure to Giraffe (). This strain continued to grow even in the presence of very high concentrations of phage (100 MOI), similar to the control without any added phage. There was no evidence of “lysis from without,” even at these high MOIs.Citation21
Figure 2. Kinetics of bacterial growth in the presence of phage Giraffe. B. anthracis strain 7702 was grown at 37°C in tryptic soy media and 1% (v/v) of tetrazolium dye. The wells designed to receive bacteria were seeded with 106 cells. Microtiter plates for the phage assay were prepared as described in materials and methods. The tests were done with (A) vegetative cells, (B) spores and (C) Giraffe-resistant B. anthracis Sterne strain 8009, a derivative of the 7702 strain.
Monitoring the kinetics of bacterial growth using OmniLogTM system upon infection with Gamma phage
A slightly different profile was observed when Gamma phage was used in the kinetic experiments. Complete suppression of growth was observed in a few cases (0.01, 1 and 100 MOI) whereas in other cases (0.0001, 0.001 and 0.1 MOI) bacterial growth continued for a few hours (up to 5.25 h) and then reached a plateau. In some cases (0.1 and 10 MOI) suppression was prolonged a little longer, up to ~10 h, before growth resumed again (). Similar trends to those observed using vegetative cells were seen in wells seeded with spores except that the effects of various phage concentrations (i.e., the timing of resumption of growth) were more variable (). Notably, a striking difference was observed between Giraffe and Gamma; Giraffe completely suppressed bacterial growth for up to 9 h in all concentrations of phage before growth resumed, whereas in the case of Gamma infection, bacterial growth continued to occur in low phage concentrations. Results from experiments with shake flask cultures suggest that Giraffe exhibits a rapid lysis phenotype and hence the residual resistant cells present in the population are better able to rapidly grow and populate the culture. The rapid lysis phenotype is supported by the observation in shake flask culture experiments that Giraffe has shorter latent and eclipse periods (~5 min each) and a burst size of ~45 (unpublished data).
Figure 3. Kinetics of bacterial growth in the presence of phage Gamma. B. anthracis strain 7702 was grown at 37°C in tryptic soy media and 1% (v/v) of tetrazolium dye. The wells designed to receive bacteria were seeded with 106 cells. Microtiter plates for the phage assay were prepared as described in materials and methods. The tests were done with (A) vegetative cells, (B) spores and (C) gamma-resistant B. anthracis Sterne strain 8030, a derivative of the 7702 strain.
We also tested the kinetics of growth of Gamma phage-resistant mutant upon exposure to Gamma phage (). This strain continued to grow even in the presence of very high concentrations of phage (100 MOI), similar to the control without any added phage. There was no evidence of ‘lysis from without’ even at these high MOIs.Citation21
Monitoring the kinetics of bacterial growth using OmniLogTM system upon infection with T4-like phages
As compared with Giraffe and Gamma phages, the T4-like phages BA21, BA28, BA39 and BA51 exhibited a very different bacterial growth profile. With these four phages, bacterial growth inhibition was observed in almost all concentrations except for BA51 at 0.0001 and 0.001 MOI (). Also, wells that received spore inocula produced similar results as vegetative cells in that only low concentrations of phage, such as an MOI of 0.0001 to 0.001, exhibited any hint of bacterial growth (data not shown).
Figure 4. Kinetics of bacterial growth in the presence of T4 like phages. B. anthracis strain 7702 was grown at 37°C in tryptic soy media and 1% (v/v) of tetrazolium dye. The wells designed to receive bacteria were seeded with 106 cells. Microtiter plates for the phage assay were prepared as described in materials and methods. These experiments were performed with vegetative cells and using the phages: (A) BA21, (B) BA28, (C) BA39, (D) BA51 and (E) a mixture of Gamma, Giraffe, BA21, BA28, BA39 and BA51.
Monitoring the kinetics of bacterial growth using OmniLogTM system upon infection with a phage cocktail
Next we wished to test what effect a phage cocktail comprised of all six phages would have on growth of B. anthracis strain Sterne 7702. The results are shown in . In this experiment growth inhibition was observed at all phage concentrations for the entire duration of the experiment, indicating that the very few bacteria that are resistant to any given phage are still susceptible to infection by the other phages present in the cocktail indicating possibly different bacterial receptors are used by these phages. Experimentally, we have isolated resistant bacteria against each of the individual phages upon single phage treatment. Although these resistant mutants appear at very low frequencies (results not shown) their emergence after single phage treatment would still cause concern. Therefore, based on the apparent lack of resistant mutants post phage cocktail treatment, this experiment supports the idea of using a mixture of different phages for phage therapy experiment. The minimum number and combination of phages needed to completely arrest the growth of the bacteria were not evaluated in this study.
Monitoring the kinetics of Ciprofloxacin resistant bacteria using OmniLogTM system upon infection with Giraffe phage
In order to further evaluate the utility of the OmniLogTM system for indirect phage assays, phage infection in the presence and absence of antibiotic treatment was also examined. Wild type B. anthracis Sterne, in the absence of phage, was exposed to varying concentrations of ciprofloxacin ranging from 0.5 ug/ml up to 32 ug/ml. As expected, bacterial growth was inhibited at all concentrations () whereas the control, without any antibiotic, exhibited a typical sigmoidal growth curve. On the other hand, a medium level ciprofloxacin resistant mutant (HS2–7),Citation22 exhibited resistance to ciprofloxacin concentrations up to 4 ug/ml and sensitivity at concentrations above 4 ug/ml up to the highest concentration tested; i.e., 32 ug/ml (). When the additive effect of phage plus antibiotic was tested, it was found that the bacterial growth was completely inhibited in all cases (5C). Surprisingly, the Giraffe phage effectively suppressed the growth of HS2–7 even at very low multiplicity of infection and in the absence of ciprofloxacin treatment (). In this case, appearance of resistant bacteria was not seen as was observed with the strain 7702 (). We speculate that this difference is due to strain specific variations in the infectivity and growth of Giraffe. HS2–7 is a derivative of ΔANR which is a B. anthracis strain lacking both pXO1 and pXO2 plasmids, whereas 7702 is a Sterne strain lacking only pXO2. It is not clear at this time how the absence of the two plasmids prevents the emergence of resistance. It is possible that the absence of pXO1 and pXO2 plasmids may allow for efficient infection and killing of the host and possibly suppression of resistance.
Figure 5. Kinetics of bacterial growth in the presence of phage Giraffe or ciprofloxacin. B. anthracis HS2–7, a ciprofloxacin resistant strain, was grown at 37°C in tryptic soy media and 1% (v/v) of tetrazolium dye. The wells designed to receive bacteria were seeded with 106 cells. The Giraffe phage PFU ranged from 108 PFU to 102 PFU and ciprofloxacin concentrations ranged from 32 µg/ml to 0.5 µg/ml. One well was used as a phage only control and another as a bacterial positive control. The experiments were performed with (A) B. anthracis Sterne 7702 plus ciprofloxacin, (B) HS2–7 plus ciprofloxacin. (C) HS2–7 plus ciprofloxacin and Giraffe phage and (D) HS2–7 plus Giraffe.
Discussion
Although originally designed to monitor bacterial growth under a range of varying growth conditions, we have successfully adapted the OmniLogTM system for indirectly monitoring phage growth. Conventionally, one can monitor bacterial growth by following the increase in the optical density of the culture and a lack thereof; i.e., decrease in optical density due to lysis by phage. The OmniLogTM system, however, circumvents this by monitoring the respiration rates in the well, which is directly proportional to the number of actively growing cells. Measuring respiration, instead of optical density, allows us to measure the living cells while excluding the dead cells and other debris that may contribute to increasing the optical density in a well.Citation12 Also, the OmniLogTM system provides an unparalleled high-throughput capability (4800 phage assays), with continuous and automated monitoring for the entire duration of the experiment.
The advantage of real time, continuous monitoring is evident in infections with the phage Giraffe. Conventional endpoint analysis of an overnight phage growth experiment would give false impression of no lysis because of the turbidity of the culture upon overnight incubation whereas in reality Giraffe caused full lysis prior to the establishment of bacterial growth from founder persister or resistant bacteria present in the population. Continuous monitoring reveals the real kinetics of the bacterial growth upon infection. Also, phage specific variations can be uncovered in long-term continuous monitoring of indirect phage assays. The ‘rapid lysis’ phenotype of Giraffe is distinct from that of Gamma in which there is initial growth of bacteria before phage induced killing can happen (this may be due to shorter eclipse time in Giraffe as compared with Gamma). Similarly, the T4-like phages that are obligatorily lytic have different profiles and are efficient bacterial killers even at low MOI.
Lastly, the additive, synergistic or antagonistic effect of a phage-antibiotic combination has been tested in this study, using the Giraffe phage in combination with ciprofloxacin against a ciprofloxacin-resistant strain of B. anthracis. The strain HS2–7 is known to exhibit medium level resistance (up to 4 μg/ml) but is completely susceptible to Giraffe phage.Citation22 When combined in the same wells, the phage and antibiotic seem to work additively to produce virtually no growth in the course of 48 h. This suggests that this phage could be used therapeutically in the event of an infection from an antibiotic resistant strain of B. anthracis.
It is apparent that OmniLogTM cannot determine the kinetics of phage growth in each well, thereby requiring one to extract the contents of the wells and enumerate the phage present. Also, the OmniLogTM system may have to be modified for phage assays of anaerobic bacteria. Despite these caveats, we demonstrate that the OmniLogTM system can be successfully used for monitoring phage growth in high throughput experiments such as host range analysis and combination studies involving antibiotics and phages.
Materials and Methods
The bacterial strains and phages used in this study are listed in . Additionally, both vegetative cells and spores of B. anthracis Sterne 7702 and vegetative cells of phage resistant mutants of 7702 were used in this study. All the phage methods were followed using standard protocols.Citation4 CsCl purification of phage was done according to published protocol.Citation23 CsCl purified phage lysate was used for electron microscopy by negative staining with 2% uranyl acetate following standard procedures.Citation4 Preparation of spores was done according to published protocol with some modifications.Citation24
Table 1. List of bacterial strains and phages used in this study
Phage strains: Four of the six phages used in this study were obtained from sewage water. These phages are designated as B. anthracis BA21, BA51, BA28 and BA39. B. anthracis specific Gamma phage was obtained from NMRC phage stock. Giraffe phage was from the collection of Dr. Raymond Schuch.
Isolation of phage from raw sewage using polyethylene glycol
Sewage samples (2 gallons total) were obtained from the Great Seneca sewage treatment plant in Germantown, MD 20874. The sewage samples were placed in six Beckman plastic bottles (380mL volume), followed by addition of 22 g of NaCl and then the mixture was allowed to stand on ice for one hour. The samples were then centrifuged at 10,000 x g for 10 min. The supernatant was transferred to a separate container and mixed with polyethylene glycol (PEG, MW 8,000; Fisher Scientific) to provide a final concentration of 10% (w/v). The mixture was then allowed to sit for one hour on ice and then centrifuged at 12,000 x g for 20 min. The supernatant was discarded and the resulting precipitate was suspended in 5ml of phage dilution buffer (10mM NaCl, 50mM Tris-Cl pH 8, 10mM MgCl2) or SM buffer (100mM NaCl, 8mM MgSO4 · 7H2O, 50mM Tris-Cl, 0.002% gelatin (w/v)). This phage fraction was used as seed stock for isolation of phage against any given bacterium.
Amplification of phages in the sewage sample
In some experiments an alternative approach was used for phage isolation using the sewage directly as a source without purifying the viral fraction by ultra centrifugation. 100 ml of sewage was added to a 250 ml flask containing 3 g of Tryptic soy (TS) media and mixed thoroughly to dissolve the media with sewage water. 0.2 ml of overnight bacterial culture was added to the flask and then incubated with vigorous shaking (225 rpm) at 37⁰C. After 6 h incubation, a 5 ml aliquot from each flask was filtered through 0.22 µm Millex GV sterile filters (Millipore). Another 1 ml aliquot from each flask was treated with 50 µl of chloroform for 15 min at room temperature. Chloroform treated sample was centrifuged at 5,000 × g for 1 min. The aqueous phase of this processed sample was transferred to a new 1.5 ml eppendorf tube and used for further analysis. 50 µl aliquots of the filtered or chloroform treated sample were mixed with 100 µl-of bacterial isolate and incubated at 37⁰C for 20 min prior to plating by soft agar overlay technique. Plates were incubated at 37⁰C overnight and the next day plates were carefully examined for appearance of phage plaques.
Preparation of microtiter plates for Biolog assay
Bacterial cultures for the microtiter plate assay were prepared from colonies grown from an overnight culture on Tryptic Soy Agar (TSA) plates and suspended in phosphate buffered saline (pH 7.4) to an optical density of 0.1, which corresponded to roughly 108 colony forming units/ml. After further dilution, 106 CFUs were deposited in each well. Individual phages or a cocktail of Giraffe, BA21, BA28, BA39, BA51 and Gamma were mixed in equal proportions and used to infect the B. anthracis host. The phage titers used ranged from 108 PFUs to 102 PFUs per well, corresponding to a multiplicity of infection (MOI) of 100 to 0.0001.
Microtiter plates (96 well) plates were prepared as follows. In each well, 90 µl of TS broth mixed with 1% v/v tetrazolium dye was added as growth medium followed by the addition of 10 µl of (109 PFU) phage per well in one column, making a 108 PFU/well dilution. 10-fold dilutions were made in the subsequent columns down to 102 PFU per well, leaving 90 µl left in each well. The last column received media and phage alone, with a total volume of 100 µl. Following dilution of phage, 10 µl of 108 CFU/ml of B. anthracis cells or spores were added to each well designed to have bacteria, increasing the volume to a total of 100 µl and giving a final concentration of 106 CFU per well. The 96 well plates were then incubated in the OmniLogTM system at 37°C for 48 or 72 h. All experiments represent biological replicates of three except the ciprofloxacin experiment which was done once.
Acknowledgments
This work was supported by the Transformational Medical Technologies Program, under the contract TMTI_IB06RSQ002 through the Defense Threat Reduction Agency to SS. The views expressed in this article are those of the authors and do not necessarily reflect the official policy or position of the Department of the Navy, Department of Defense, nor the US. Government. We would like to thank Mr. Michael Delannoy (The Johns Hopkins University Electron Microscopy facility) for preparing the electron micrographs of the bacteriophages.
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
References
- Ackermann HW, Audurier A, Berthiaume L, Jones LA, Mayo JA, Vidaver AK. Guidelines for bacteriophage characterization. Adv Virus Res 1978; 23:1 - 24; http://dx.doi.org/10.1016/S0065-3527(08)60096-2; PMID: 34986
- Stent GS. Molecular Biology of bacterial viruses. San Francisco: W. H. Freeman, 1963.
- Hyman P, Abedon ST. Practical methods for determining phage growth parameters. Methods Mol Biol 2009; 501:175 - 202; http://dx.doi.org/10.1007/978-1-60327-164-6_18; PMID: 19066822
- Carlson K. Working with bacteriophages:common techniques and methodological approaches. In: Kutter E, and Sulakvelidze, A, ed. Bacteriophages: Biology and Application. Boca Raton, FL: CRC Press, 2004:437-94.
- Merabishvili M, Pirnay JP, Verbeken G, Chanishvili N, Tediashvili M, Lashkhi N, et al. Quality-controlled small-scale production of a well-defined bacteriophage cocktail for use in human clinical trials. PLoS One 2009; 4:e4944; http://dx.doi.org/10.1371/journal.pone.0004944; PMID: 19300511
- d'Herelle F. Sur un microbe invisible antagoniste des bacilles dysenterique. C R Acad Sci Paris 1917; 165:373 - 5
- Dreyer G, Campbell-Renton ML. The quantitative determination of bacteriophage activity and its application to the study of the Twort-d'Herellen phenomenon. J Path And Bact 1933; 36:399 - 423; http://dx.doi.org/10.1002/path.1700360305
- Ellis EL, Delbrück M. The Growth of Bacteriophage. J Gen Physiol 1939; 22:365 - 84; http://dx.doi.org/10.1085/jgp.22.3.365; PMID: 19873108
- Gratia A. Numerical relations between lysogenic bacteria and particles of bacteriophage. Ann Inst Pasteur (Paris) 1936; 57:652
- Hershey AD, Bronfenbrenner J. Stepwise Liberation of Poorly Sorbed Bacteriophages. J Bacteriol 1943; 45:211 - 8; PMID: 16560627
- Adams MH. Enumeration of bacteriophage particles. Bacteriophages. London: Interscience publishers, Ltd, 1959:27-34.
- Arber W, Dussoix D. Host specificity of DNA produced by Escherichia coli. I. Host controlled modification of bacteriophage lambda. J Mol Biol 1962; 5:18 - 36; http://dx.doi.org/10.1016/S0022-2836(62)80058-8; PMID: 13862047
- Cooper CJ, Denyer SP, Maillard JY. Rapid and quantitative automated measurement of bacteriophage activity against cystic fibrosis isolates of Pseudomonas aeruginosa. J Appl Microbiol 2011; 110:631 - 40; http://dx.doi.org/10.1111/j.1365-2672.2010.04928.x; PMID: 21205097
- Bochner BR. Sleuthing out bacterial identities. Nature 1989; 339:157 - 8; http://dx.doi.org/10.1038/339157a0; PMID: 2654644
- Bochner BR. New technologies to assess genotype-phenotype relationships. Nat Rev Genet 2003; 4:309 - 14; http://dx.doi.org/10.1038/nrg1046; PMID: 12671661
- Bochner BR. Global phenotypic characterization of bacteria. FEMS Microbiol Rev 2009; 33:191 - 205; http://dx.doi.org/10.1111/j.1574-6976.2008.00149.x; PMID: 19054113
- Johnson DA, Tetu SG, Phillippy K, Chen J, Ren Q, Paulsen IT. High-throughput phenotypic characterization of Pseudomonas aeruginosa membrane transport genes. PLoS Genet 2008; 4:e1000211; http://dx.doi.org/10.1371/journal.pgen.1000211; PMID: 18833300
- Watanabe T, Morimoto A, Shiomi T. The fine structure and the protein composition of gamma phage of Bacillus anthracis. Can J Microbiol 1975; 21:1889 - 92; http://dx.doi.org/10.1139/m75-275; PMID: 811348
- Lewis K. Persister cells and the paradox of chronic infections. ASM Microbe 2010; 5:429 - 37
- Luria SE, Delbrück M. Mutations of Bacteria from Virus Sensitivity to Virus Resistance. Genetics 1943; 28:491 - 511; PMID: 17247100
- Abedon ST. Lysis from without. Bacteriophage 2011; 1:46 - 9; http://dx.doi.org/10.4161/bact.1.1.13980; PMID: 21687534
- Price LB, Vogler A, Pearson T, Busch JD, Schupp JM, Keim P. In vitro selection and characterization of Bacillus anthracis mutants with high-level resistance to ciprofloxacin. Antimicrob Agents Chemother 2003; 47:2362 - 5; http://dx.doi.org/10.1128/AAC.47.7.2362-2365.2003; PMID: 12821500
- Sambrook J, Fritsch EF, Maniatis T. Molecular cloning: a laboratory manual. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press, 1989.
- Mukhopadhyay S, Akmal A, Stewart AC, Hsia RC, Read TD. Identification of Bacillus anthracis spore component antigens conserved across diverse Bacillus cereus sensu lato strains. Mol Cell Proteomics 2009; 8:1174 - 91; http://dx.doi.org/10.1074/mcp.M800403-MCP200; PMID: 19208616