Abstract
Listeria monocytogenes is a significant food-borne pathogen and the causative agent of listeriosis, a disease which manifests as meningitis in immunocompromised adults or infection of the foetus and miscarriage in pregnant women. We have previously used Lactococcus lactis, a GRAS (Generally Regarded As Safe) organism, as a vaccine vector against listeriosis by engineering plasmid-mediated expression of the immunodominant antigen from L. monocytogenes, listeriolysin O (LLO). However, the environmental release of an engineered vaccine vector carrying a replicating plasmid during clinical usage may raise safety concerns. Here we describe the integration of the LLO gene (hly) into the L. lactis chromosome through homologous double crossover to allow stable expression, in order to avoid the use of antibiotic selection markers and to eliminate the requirement for a plasmid-based system. The approach was designed to simultaneously eliminate the pyrG gene encoding the CTP synthase which is responsible for converting UTP to CTP in a unique step in the de novo pyrimidine synthesis in L. lactis. This gene was targeted in order to restrict bacterial replication outside of the host (biological containment). The resulting cytidine auxotroph was able to secrete LLO constitutively and could elicit LLO91-99-specific CD8+ T lymphocytes in the murine infection model. Moreover, protection against lethal challenge with L. monocytogenes was accomplished after intraperitoneal (IP) vaccination with the constructed strain. The implications for the use of cytidine auxotropy in biological containment are discussed.
Introduction
Listeria monocytogenes is a food-borne pathogen that can lead to meningitis in immunocompromised adults or fetal infection and potentially miscarriage in pregnant women.Citation1 Listeriolysin O (LLO) is the major virulence factor of L. monocytogenes and enables the escape of the pathogen from the phagosome to the cytoplasm of infected host cells. This unique mechanism depends on the ability of LLO to interact with cholesterol in the phagosomal membrane where it oligomerizes and creates pores through which bacteria can escape to the cytosol.Citation2 Being an intracellular microorganism, L. monocytogenes evades host antibody-mediated immunity and protection against infection is mainly accomplished through cytotoxic cell-mediated immunity.Citation3,Citation4 The development of cytotoxic CD8+ immunity against major listerial antigens (including LLO) is particularly important for protection against listeriosis.Citation4
Lactococcus lactis is a Gram-positive bacterium that is widely used in the food industry. Due to its GRAS (Generally Regarded As Safe) status, L. lactis has been extensively investigated as a vaccine vector by expressing heterologous antigens of various pathogens.Citation5 Recently we investigated L. lactis as a potential vaccine vector against listeriosis by expressing LLO constitutively or inducibly in various cellular compartments.Citation6,Citation7 In the latter study the LLO gene (hly) was cloned in plasmid vectors and transformed into L. lactis. However, ultimately the clinical use of a plasmid-borne system will raise safety concerns about the release of vectors expressing antibiotic resistance markers which have a risk of transfer to pathogens in the environment. Since promising immunological outcomes were obtained using plasmid-mediated expression of LLO in L. lactis,Citation6 we endeavoured in the present study to stably integrate hly into the chromosome of L. lactis for constitutive expression of LLO. The strong constitutive lactococcal P23 promoterCitation8 was chosen to drive LLO expression and a construct was designed to replace the lactococcal pyrG gene using the pORI280/pVE6007 integration system.Citation9,Citation10 The pyrG gene was targeted because it encodes the CTP synthase responsible for the unique de novo pathway that converts UTP to CTP in L. lactis.Citation11 Consequently, when pyrG is knocked out, a requirement for cytidine is established and cytidine has to be added in culture media for bacterial survival.Citation11 It was previously reported that mutations in the thymidylate synthase (thyA) gene of bacteria led to strict thymidine or thymine auxotrophy and rapid cell death in thymidine-free media (thymine-less death).Citation12 The previous concept was utilized by Steidler et al.Citation13 to create a biologically contained L. lactis that secretes hIL-10 (human interleukin 10) to treat inflammatory bowel disease (IBD) by the oral route. When the hIL-10 expression cassette replaced thyA in L. lactis, a strict thymidine auxotroph was created which rapidly died upon thymidine starvation and is therefore ‘biologically contained’.Citation13 This biological containment property is proposed to prevent the dissemination of genetically modified bacteria in the environment where pyrimidine is limiting.
In brief, in the present work we created a cytidine auxotroph of L. lactis that constitutively secretes LLO from an integrated construct. In vivo vaccination using the created strain was investigated in mice where it resulted in an LLO-specific CD8+ response and protection upon challenge with wild type L. monocytogenes. This construct will provide a platform for the development of future vaccines and for delivery of further heterologous antigens.
Results
Success of replacement recombination in L. lactis MG1363 chromosome and production of LLO.
We constructed a pORI280 plasmid vector carrying the hly gene from L. monocytogenes under the influence of the P23 promoter and flanked by appropriate sites for integration in place of the pyrG gene of L. lactis ( and ). Integration of pORIP23:SEC-LLO (single crossover) in the L. lactis MG1363 chromosome followed by excision of pORI280 along with pyrG (including its native promoter), (i.e., double crossover), was confirmed by a number of PCR reactions () using primers outlined in . The resulting strain, L. lactis MG1363 ΔpyrG (P23:SEC-LLO), was examined for LLO secretion by TCA-precipitation of the supernatant followed by western blot using primary rabbit anti-LLO antibodies (Diatheva, Italy). A specific band of LLO was obtained at the expected protein size (about 57 KDa) (). Culture supernatant of L. lactis MG1363 ΔpyrG (P23:SEC-LLO) had complete hemolytic units (CHU) of 8 confirming the secretion of biologically active LLO.
Deletion of pyrG gene has a bacteriostatic rather than bactericidal effect on the cytidine auxotroph L. lactis MG1363 ΔpyrG (P23:SEC-LLO).
The pyrG gene is reported to encode the enzyme CTP synthase.Citation11 This enzyme is responsible for the conversion of UTP to CTP and it is the only pathway for the de novo synthesis of CTP. We examined if pyrG deletion would result in a strictly auxotrophic cytidine mutant and if this deletion would have a bacteriostatic or bactericidal effect on the resulting mutant in cytidine deprived culture medium. Upon growing L. lactis MG1363 ΔpyrG (P23:SEC-LLO) in CDM lacking cytidine, there was a slow decline of the bacteria from 6.5 log CFU/ml to about 5 log CFU/ml over a 15 day period (). This finding indicates that the absence of cytidine from the culture medium is generally bacteriostatic rather than bactericidal. When the same mutant was grown in CDM containing 20 µg/ml cytidine, a phase of initial growth occurred starting from 6.5 log CFU/ml up to 8 log CFU/ml in the first 24 h, followed by a slow decline until it reached about 6.5 log CFU/ml again on day 15. This suggests that upon consumption of the provided cytidine from the CDM culture medium, the resulting cytidine starvation condition is bacteriostatic ().
Survival of the cytidine auxotroph L. lactis MG1363 ΔpyrG (P23:SEC-LLO) was also examined in autoclaved soil as an attempted simulation of a natural environmental situation. No significant difference in survival was observed between the cytidine auxotroph and the wild type MG1363 strain (). This indicates that cytidine auxotrophy does not lead to a dramatic cell death in conditions of cytidine starvation and suggests that other approaches (such as deletion of the thyA locus) are necessary for full biological containment.Citation13
CD8+ T lymphocytes specific for the H2-Kd-restricted LLO91–99 epitope are elicited by L. lactis MG1363 ΔpyrG (P23:SEC-LLO) following IP immunization.
The ELISPOT assay was used to analyze the development of LLO91–99-specific CD8+ cells. The mouse mastocytoma cells P815-1-1 were used as antigen presenting cells (APC) as they express restricted H2-Kd MHC class I molecules, so any resulting spots are due to LLO-specific CD8+ cells.Citation22 The cellular immune response was examined 4 weeks after the last IP booster. Groups injected with L. lactis MG1363 ΔpyrG (P23:SEC-LLO) or the positive control L. monocytogenes EGDe showed significant LLO91–99-specific spots (p < 0.05) (). On the contrary, no specific spots were observed with groups treated with the wild type L. lactis MG1363 or the PBS-treated group ().
Protection against L. monocytogenes challenge following IP vaccination.
Mice were challenged intraperitoneally with L. monocytogenes EGDe 4 weeks after the final vaccine booster. Three days following challenge, mice were euthanized and the listerial count was determined in the spleens. Bacterial count results following the IP vaccination revealed significant protection (p < 0.05) as evidenced by the low listerial count in spleens of mice vaccinated with L. lactis MG1363 ΔpyrG (P23:SEC-LLO) (). In contrast, high listerial counts were observed in the organs of the PBS-treated groups or groups treated with the control strain L. lactis MG1363 (). These results reflect the previous ELISPOT results () where L. lactis MG1363 ΔpyrG (P23:SEC-LLO) could elicit LLO91–99-specific spots.
Discussion
L. lactis has been repeatedly examined as a safe and effective vaccine delivery platform for the delivery of various heterologous antigens to the immune system.Citation5 In our previous work, we successfully expressed listeriolysin O (LLO) of L. monocytogenes in L. lactis under inducible and constitutive conditions.Citation6 The hly gene of LLO was expressed on plasmid vectors and immunizations with the constructed strains elicited specific protective immune responses against listeriosis in the murine infection model.Citation6 Given these promising results, we have designed and constructed a vaccine delivery platform in which LLO is expressed in L. lactis through chromosomal integration rather than plasmid-mediated cloning. This ensures stability of expression, avoids the use of antibiotic selection markers and reduces the likelihood of gene transfer to other bacterial species in the natural environment. Moreover, we have chosen a strong lactococcal constitutive promoter (P23)Citation8 to drive the expression of LLO which allows the continuous production of LLO without the need for an inducer. The system was designed to replace the pyrG gene of L. lactis MG1363 with the LLO expression cassette using homologous recombination via the integrative pORI280 plasmid system ( and ). The integration process was confirmed by PCR reactions throughout the engineering of this stable delivery vector (). Primers specific for pyrG (30 and 31) and primers specific for hybrid products of the LLO expression cassette and PRE-pyrG or POST-pyrG (27/29 and 28/internal 3 primer pairs respectively) were used. This confirmed the loss of the pyrG gene and the correct chromosomal integration of the LLO expression cassette ( and ). The resulting strain was also confirmed for constitutive secretion of LLO ().
The L. lactis MG1363 ΔpyrG (P23:SEC-LLO) strain successfully elicited LLO91–99-specific CD8+ lymphocytes and provided significant protection against lethal L. monocytogenes challenge following IP vaccination in our model system. The LLO-expressing L. lactis most likely elicited this specific CD8+ immune following phagocytosis by peritoneal macrophages which are the main antigen presenting cells (APCs) in the peritoneum.Citation24 Subsequently these macrophages are known to process antigen (LLO) and present specific epitopes in the context of MHC class I complex to CD8+ cells. Although some CD8+ T cells may be residing in the peritoneum,Citation25 the main stages in antigen presentation are assumed to occur in peripheral lymph nodes upon drainage of the peritoneal macrophages, or drainage of unphagocytosed bacteria, from the peritoneal cavity through the lymphatics.Citation26 This leads ultimately to systemic activation and proliferation of specific CD8+ lymphocytes as seen in our study.
The current data support our previous work using replicating plasmid vectors to deliver LLO and indicate that integration of hly under a strong constitutive promoter does not adversely affect the efficacy of the strain for vaccine delivery. The delivery of LLO permeabilizes the phagosomal membrane in antigen presenting cellsCitation2 and we propose that the L. lactis MG1363 ΔpyrG (P23:SEC-LLO) vector provides a potential delivery platform for the delivery of other heterologous antigens to the cytoplasmic MHC class I pathway for the generation of a strong CD8+ T cell response. Future work in our laboratory will test this hypothesis using a range of test antigens.
In the present system deletion of pyrG was targeted as we postulated that this would lead to cytidine auxotropy and limited survival of the vaccine strain in the natural environment (biological containment). Although mutation of pyrG has previously been reported, those studies were mainly functional studies and it was not determined whether mutation resulted in a bacteriostatic or bactericidal effect on L. lactis during cytidine starvation.Citation11,Citation27 Here we found that cytidine auxotrophy was indeed achieved in the L. lactis MG1363 ΔpyrG (P23:SEC-LLO) strain. However, the strain did not demonstrate significant cell death upon cytidine starvation in CDM and the effect was found to be mainly bacteriostatic rather than bactericidal. Moreover, we attempted to simulate environmental conditions as closely as possible by inoculating the bacteria into autoclaved soil. However, no significant difference in survival was found between wild type L. lactis MG1363 and L. lactis MG1363 ΔpyrG (P23:SEC-LLO) in this environment. These findings are in contrast to the work of Steidler et al. who demonstrated pyrimidine auxotrophy in L. lactis through deletion of the thymidylate synthase gene (thyA) leading to a “thymine-less death” of the cells upon thymidine or thymine deprivation.Citation13 It was found that their L. lactis ΔthyA mutant was completely killed within 3 days in thymidine-free culture medium.Citation13 Collectively our work suggests that cytidine auxotropy in L. lactis is bacteriostatic whilst thymidine auxotropy is bactericidal in media depleted of pyrimidines. The mechanisms underpinning these different responses remain unknown. We propose that future studies could focus upon creation of a double mutant in both thyA and pyrG to determine whether the double mutation offers any benefits over the thyA mutation with respect to biological containment.
Overall, we demonstrate the feasibility of chromosomal integration of the LLO gene (hly) through homologous double crossover in L. lactis and the successful expression of LLO in this system. The subsequent pyrG deletion at the site of hly integration resulted in cytidine auxotrophy which mainly had a bacteriostatic effect on the vaccine strain. LLO-specific immune responses and protection against listeriosis were elicited by the vaccine strain after IP vaccination. We propose that the current vaccine strain may act as a carrier for other heterologous antigens of any pathogen where LLO would facilitate antigen escape to the cytoplasm and subsequent presentation through the MHC class I pathway. For strict biological containment of the vaccine strain, we recommend that the accompanying heterologous antigen be integrated in place of the thyA gene of L. lactis ΔpyrG (P23:SEC-LLO) thereby resulting in double pyrimidine auxotrophy.
Materials and Methods
Bacterial strains, plasmids and culture conditions.
A summary of bacterial strains and plasmids used in this study is shown in . Luria-Bertani (LB) broth was used for Escherichia coli cultures while M17 broth (Oxoid) supplemented with 0.5% glucose (i.e., GM17) was used for Lactococcus. For L. monocytogenes, brain heart infusion (BHI) broth (Oxoid) was used. Technical agar (Merck) was added (1.5% w/v) when solid media were required. Incubation temperatures were 30°C for L. lactis and 37°C for L. monocytogenes and E. coli respectively unless otherwise stated. When required, Erythromycin (Em) was used at a concentration of 200 µg/ml for E. coli and at 5 µg/ml for Lactococcus. Chloramphenicol (Cm) was used at a concentration of 10 µg/ml when required. All cell culture media and reagents were obtained from Gibco unless otherwise stated.
Cloning of hly in pORI280 integration vector.
Cloning was performed using the integration plasmid pORI280,Citation10 to allow replacement recombination of hly gene in place of the essential pyrG gene in the genome of L. lactis MG1363. The Splicing by Overlap Extension (SOE) techniqueCitation14 was used to create a construct composed of the following in order: 839 bp that naturally exist upstream from the native promoter of pyrG gene in L. lactis MG1363 genome (hereafter referred to as PRE-pyrG), the constitutive P23 promoter,Citation8 the secretion signal of the Usp45 protein (designated SEC in this work)Citation15 with downstream hly gene (without its native secretion signal),Citation16 and 1,000 bp that naturally exist downstream of the pyrG gene in L. lactis MG1363 genome (hereafter designated POST-pyrG). All PCR reactions were performed using the high fidelity KOD hot start DNA polymerase (Novagen) following manufacturer's instructions.
Primers used in the PCR procedures are outlined in . Briefly, to create the construct, primers 16 and 24 were used to amplify the PRE-pyrG region using a genomic preparation from L. lactis MG1363 as a template. Genomic preparation from Lactococcus Wg2 was used as a template to amplify the constitutive promoter P23 using primers 22 and 23. The previous two PCR products (i.e., PRE-pyrG and P23 promoter) were spliced by the SOE technique at 1:1 molar ratio using primers 16 and 23 resulting in the spliced product PRE-pyrG/P23. A Histagged listeriolysin O gene (hly) with upstream Usp45 secretion signal (SEC) was amplified using primers 25 and 15 with plasmid pNZPnisA:SEC-LLO (previously created in our workCitation6) as a template. This resulting SEC-LLO product was further spliced downstream of the PRE-pyrG/P23 product by a SOE-PCR reaction using primers 16 and 15 yielding the product PRE-pyrG/P23/SEC-LLO. Finally, using primers 16 and 26, this latter spliced product was further PCR-spliced to the POST-pyrG region (obtained by PCR amplification from L. lactis MG1363 genome using primers 18 and 26) to give the final construct PRE-pyrG/P23/SEC-LLO/POST-pyrG (). The final SOE construct was sequentially digested with BglII and XbaI respectively and then ligated into a similarly digested pORI280 vector. Ligation was performed using T4 DNA ligase (Roche) and the ligation reaction was transformed into electro-competent E. coli EC101 (RepA+) using Gene Pulser (Biorad) and cells were plated onto LB agar (containing 64 µg/ml X-gal and 200 µg/ml Em). Plates were incubated at 37°C for 24 h and blue colonies were checked by colony PCR. One Positive EC101 colony (containing plasmid pORIP23:SEC-LLO) was selected and stocked at −80°C.
Plasmid was extracted from E. coli EC101 (pORIP23:SEC-LLO) using the Qiagen Miniprep Kit (Qiagen). The integrity of the DNA sequence was confirmed by sequencing (Cogenics, UK).
Transformation of Lactococcus lactis MG1363 (pVE6007) with pORIP23:SEC-LLO and induction of replacement recombination.
Electrocompetent L. lactis MG1363 (pVE6007), prepared as previously described,Citation17 was transformed with pORIP23:SEC-LLO using Gene Pulser (Biorad) and plated onto GM17 agar containing Em (5 µg/ml) and X-gal (64 µg/ml). Plates were incubated at 30°C for 24–48 h. Blue colonies were checked by colony PCR for successful transformation. For induction of chromosomal integration (single crossover), a positive colony was subcultured once in GM17 (Em 5 µg/ml) at 30°C overnight, then subcultured overnight twice sequentially (0.1% v/v inoculum) in pre-warmed GM17 broth (Em 5 µg/ml) at 37°C. The latter high temperature (37°C) promotes the loss of the temperature-sensitive RepA+ pVE6007 vector inducing the chromosomal integration of the RepA− pORIP23:SEC-LLO. This integration is guided through the homology between the constructed integration plasmid and the chromosomal DNA flanking the pyrG gene (). After growth of the second 37°C culture, a loopful was taken from the broth and streaked onto pre-warmed GM17 agar (5 µg/ml Em, 64 µg/ml X-gal) and incubated at 37°C for 24–48 h. Colonies were checked by replica plating on GM17 agar containing 10 µg/ml Cm to ensure chloramphenicol sensitivity (i.e., loss of pVE6007). Single crossover integration was confirmed by PCR and also by the absence of any plasmid content upon using the Qiagen Miniprep kit for plasmid extraction.
Excision of the integrated pORI280 derivative from the chromosome was performed by passaging (continuous subculturing using 0.1% v/v overnight inocula) at 37°C in antibiotic-free prewarmed (37°C) GM17 broth (1 mg/ml cytidine) and spreading at intervals on GM17 agar containing 64 µg/ml X-gal and 1 mg/ml cytidine. White colonies were picked up and checked by PCR to differentiate between cells with successful double crossover (P23/SEC-LLO in place of pyrG gene) and wild type revertants. Since CTP synthase, encoded by the pyrG gene, is essential for L. lactis, cytidine was added to all media in the second crossover procedures to facilitate the creation of the cytidine auxotroph L. lactis MG1363 ΔpyrG (P23:SEC-LLO).
Investigation of LLO production and haemolytic activity.
The overnight culture supernatant of L. lactis MG1363 ΔpyrG (P23:SEC-LLO) was collected. Proteins from this supernatant were precipitated as previously describedCitation18 by adding cold trichloroacetic acid (TCA) solution (75% w/v) to attain a final TCA concentration of 15% w/v.
For SDS-PAGE, a 12% SDS-polyacrylamide separating gel and 4% stacking gel were used. For western blot analysis, gels were blotted against a nitrocellulose membrane (Hybond-ECL™, Amersham Biosciences UK, Ltd.) in a semi-dry western transfer apparatus. Membranes were blocked overnight at 4°C in 5% skimmed milk in TBS buffer (0.8% sodium chloride, 20 mM Tris-HCl, pH 7.6). Primary rabbit anti-LLO antibody (Diatheva, Italy) and secondary anti-rabbit antibody (ECL western blotting system) were used at 1/1,000 and 1/1,500 dilutions in 5% and 10% skimmed milk in TBS buffer respectively. Western blot detection was performed using the Amersham ECL western blotting system (Amersham Biosciences UK, Ltd.) according to the protocol recommended by the manufacturer.
Haemolytic activity of the secreted LLO was assayed to ensure biological activity using the method described by Kohda C et al.Citation19 with some modifications. Briefly, aliquots of 100 µl of 0.5% v/v sheep red blood cells (RBCs) suspended in PBS (pH 5.9) were distributed in 1.5 ml Eppendorf tubes. Twofold serial dilutions (in PBS pH 5.9) of dialyzed culture supernatant were added to each tube to a final volume of 1 ml. A positive control (distilled water) and negative controls (PBS, pH 5.9 and supernatant from wild type L. lactis) were also included. Tubes were incubated statically at 37°C for 45 min after which they were centrifuged and supernatants collected. Absorbance was measured at 415 nm and hemolytic activity was assessed. Hemolytic activity was expressed in terms of complete hemolytic units (CHU) defined as the reciprocal of the highest dilution of supernatant showing complete haemolysis.Citation20
Investigation of the biological containment property of L. lactis MG1363 ΔpyrG (P23:SEC-LLO).
The pyrG gene encodes the only CTP synthase in L. lactis MG1363 which is essential for the de novo synthesis of CTP from UTP as previously reported.Citation11 Growth experiments were carried out at time intervals, in the presence or absence of cytidine, to examine the effect of pyrG deletion on the survival of L. lactis MG1363 ΔpyrG (P23:SEC-LLO). Briefly, Lactococcus Chemically Defined Medium (CDM)Citation21 with or without cytidine (20 µg/ml) was used to grow L. lactis MG1363 ΔpyrG (P23:SEC-LLO). Bacteria were first inoculated into the corresponding media at time zero and this culture was split to small aliquots incubated at 30°C statically. The bacterial count was examined at different time intervals by plating onto GM17 agar containing 1 mg/ml cytidine to examine the growth or decline of the bacteria. To examine bacterial survival in nature, another experiment was also conducted by inoculating bacteria (suspended in 5 ml PBS) in 5 g autoclaved soil where proper mixing was performed by vigorous vortexing and inoculated soil was incubated at 30°C statically. The viable count was determined at intervals by resuspending duplicate samples of the inoculated soil in 10 ml PBS and plating onto GM17 agar containing 1 mg/ml cytidine and the count was calculated as CFU/ml of the resuspended soil.
Animals and immunization protocols.
Female BALB/c mice, 6–8 weeks in age, were used in all animal experiments. All animal procedures were carried out according to institutional ethical guidelines and performed in a dedicated facility. Animals were divided into groups of five mice each. Mice were given five intraperitoneal (IP) doses of L. lactis MG1363 ΔpyrG (P23:SEC-LLO) on days 1, 2, 8, 15 and 28. L. lactis MG1363-treated groups and phosphate-buffered saline (PBS)-treated groups were included as negative controls. The IP doses were five × 103 and 106 CFU/mouse on days 1 and 2 respectively followed by three booster doses of 2 × 107 CFU/mouse. Overnight cultures of L. lactis strains were inoculated into fresh GM17 broth and grown to an OD600 of 0.8 at which point cells were pelleted, washed twice with PBS and resuspended again in PBS for inoculation. Final dose volume per mouse was 200 µl. Groups treated with sub-lethal IP L. monocytogenes EGDe (2 × 103 CFU/mouse for only 4 doses on days 1, 8, 15 and 28) were also included as a positive control.
Detection of LLO-specific CD8+ T cells by the enzymelinked immunospot (ELISPOT) test.
Four weeks after the final vaccination booster, the ELISPOT test was performed as described previouslyCitation22 to detect cytotoxic CD8+ cells specific to the H2-Kd-restricted LLO epitope, LLO91–99, GYKDGNEYI (Peptide Protein Research, UK). Mouse mastocytoma line P815-1-1 cells (The European Collection of Cell Culture; ECACC) were used as antigen presenting cells (APCs). P815-1-1 cells pulsed with 10−6 M of LLO91–99 peptide or non-pulsed were used to stimulate splenocytes isolated from mice after the vaccination regimen. Numbers of LLO91–99-specific IFNγ-secreting cells were counted using a stereomicroscope.
Challenge test using IP L. monocytogenes EGDe.
Listerial challenge was performed as previously described.Citation23 Briefly, mice were challenged 4 weeks after the last vaccination booster with intraperitoneal (IP) injection of 200 µl of 2 × 106 CFU/ml (i.e., 2 × 105 CFU/mouse) L. monocytogenes EGDe. Mice were euthanized 3 days post-challenge and the listerial burden was determined in spleens through organ homogenization, serial dilutions and plating on BHI agar plates. Plates were incubated at 37°C for 2 days and Listeria counts were calculated per organ. The limit of detection (LOD) of Listeria was 50 CFU per organ.
Interpretation of data and statistical analysis.
Statistical analysis was performed using One Way ANOVA with post hoc analysis by the Holm-Sidak method or by using the Student's t-test. Results with p values of less than 0.05 were considered statistically significant.
Abbreviations
| LLO | = | listeriolysin O |
| CTP | = | cytidine triphosphate |
| UTP | = | uridine triphosphate |
| IP | = | intraperitoneal |
| Em | = | erythromycin |
| Cm | = | chloramphenicol |
| SOE | = | splicing by overlap extension |
| SEC | = | Usp45 secretion signal |
| SDS-PAGE | = | sodium dodecyl sulphate-polyacrylamide gel electrophoresis |
| TCA | = | trichloroacetic acid |
| ELISPOT | = | enzyme-linked immunospot test |
| APCs | = | antigen presenting cells |
| CDM | = | chemically defined medium |
Figures and Tables
Figure 1 Cloning of LLO expression cassette in the integrative RepA- plasmid pORI280. The uppermost construct was created using the splicing by overlap extension (SOE) technique as described in the material and methods section. Insertion in pORI280 was achieved through restriction digestion (XbaI and BgIII) followed by ligation resulting in the formation of pORIP23:SEC-LLO.
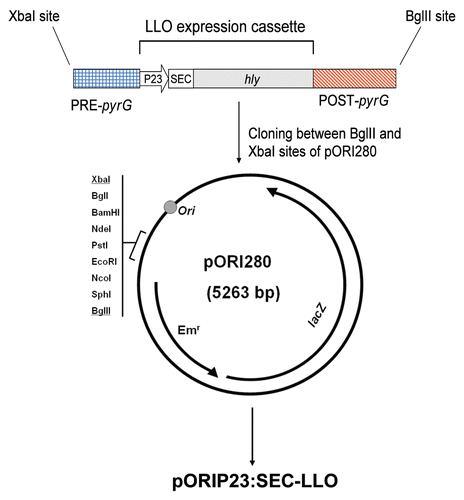
Figure 2 Diagrammatic representation of the replacement recombination (homologous double crossover) process. Primers used in PCR checking of the recombination are indicated by arrows with primer codes mentioned (see ).
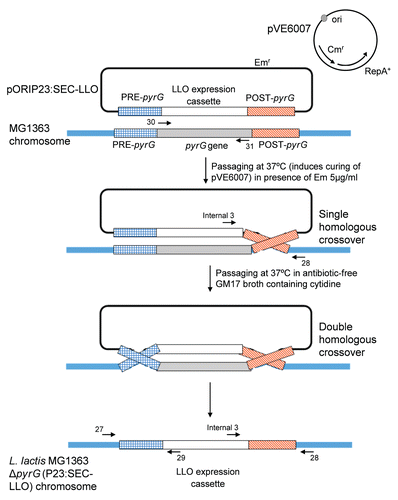
Figure 3 (A) Confirmatory PCR reactions to check for LLO expression cassette integration in L. lactis MG1363 chromosome and concomitant deletion of pyrG gene. Lanes 1–3: PCR reactions done on genome of L. lactis MG1363 with single crossover integration of pORIP23:SEC-LLO. Lanes 4–6: PCR reactions done on genome of L. lactis MG1363 with double crossover integration i.e., L. lactis MG1363 ΔpyrG (P23:SEC-LLO). Lanes 7–9: PCR reactions done on genome of wild type L. lactis MG1363. Primers 30 and 31, which are specific for pyrG gene, were used in PCRs of lanes 1, 4 and 7. Primers 27 and 29 were used in PCRs of lanes 2, 5 and 8 while primers 28 and internal 3 were used in lanes 3, 6 and 9. Primer sequences and names are summarized in . See for more details. (B) Western blot of precipitated LLO from the supernatant of L. lactis MG1363 ΔpyrG (P23:SEC-LLO) using anti-LLO antibodies.
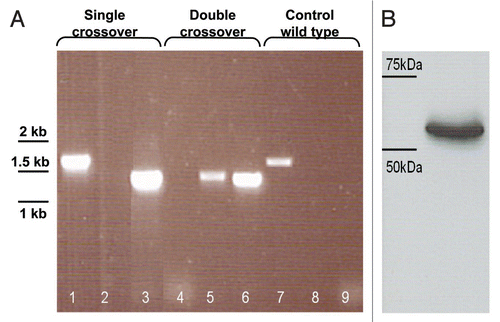
Figure 4 (A) Survival of L. lactis MG1363 ΔpyrG (P23:SEC-LLO) in chemically defined medium (CDM) without or with cytidine 20 µg/ml. (B) Examination of L. lactis MG1363 ΔpyrG (P23:SEC-LLO) survival in autoclaved soil.
Figure 5 (A) ELISPOT test results four weeks after IP vaccination regimen. Mouse groups (n = 5) were vaccinated by IP injection on days 1, 2, 8, 15 and 28 then examined by ELISPOT 4 weeks later. (B) Results of the challenge experiment four weeks after the IP immunization regimen. Mouse groups (n = 5) were IP-vaccinated on days 1, 2, 8, 15 and 28 then challenged IP with L. monocytogenes EGDe 4 weeks later. Mice were euthanized 3 days following challenge and Listeria count was determined in spleens. *p < 0.05 as compared to negative control groups. Error bars represent the mean ± SEM (Standard Error of the Mean). LOD: Limit of detection of the test.
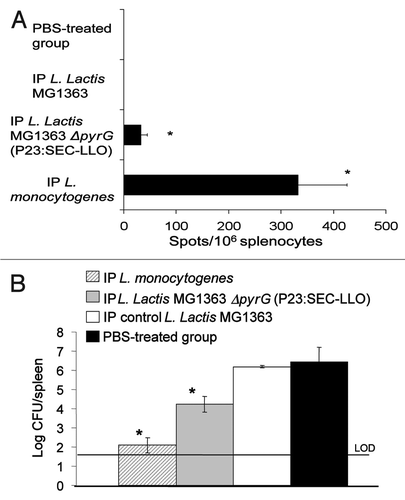
Table 1 Bacterial strains and plasmid vectors used in the present study
Table 2 Oligonucleotide primers used in this study
References
- Meng J, Doyle MP. Emerging issues in microbiological food safety. Annu Rev Nutr 1997; 17:255 - 275
- Vazquez-Boland JA, Kuhn M, Berche P, Chakraborty T, Dominguez-Bernal G, Goebel W, et al. Listeria pathogenesis and molecular virulence determinants. Clin Microbiol Rev 2001; 14:584 - 640
- Bahey-El-Din M, Griffin BT, Gahan CGM. Sleator R, Hill C. Attack and counter-attack: Targeted immunomodulation using bacterial virulence factors. Patho-Biotechnology 2008; Austin Landes Bioscience 163 - 172
- Pamer EG. Immune responses to Listeria monocytogenes. Nat Rev Immunol 2004; 4:812 - 823
- Nouaille S, Ribeiro LA, Miyoshi A, Pontes D, Le Loir Y, Oliveira SC, et al. Heterologous protein production and delivery systems for Lactococcus lactis. Genet Mol Res 2003; 2:102 - 111
- Bahey-El-Din M, Casey PG, Griffin BT, Gahan CG. Lactococcus lactis-expressing listeriolysin O (LLO) provides protection and specific CD8(+) T cells against Listeria monocytogenes in the murine infection model. Vaccine 2008; 26:5304 - 5314
- Bahey-El-Din M, Griffin BT, Gahan CG. Nisin inducible production of listeriolysin O in Lactococcus lactis NZ9000. Microb Cell Fact 2008; 7:24
- van der Vossen JM, van der Lelie D, Venema G. Isolation and characterization of Streptococcus cremoris Wg2-specific promoters. Appl Environ Microbiol 1987; 53:2452 - 2457
- Cotter PD, Hill C, Ross RP. A food-grade approach for functional analysis and modification of native plasmids in Lactococcus lactis. Appl Environ Microbiol 2003; 69:702 - 706
- Leenhouts K, Buist G, Bolhuis A, ten Berge A, Kiel J, Mierau I, et al. A general system for generating unlabelled gene replacements in bacterial chromosomes. Mol Gen Genet 1996; 253:217 - 224
- Wadskov-Hansen SL, Willemoes M, Martinussen J, Hammer K, Neuhard J, Larsen S. Cloning and verification of the Lactococcus lactis pyrG gene and characterization of the gene product, CTP synthase. J Biol Chem 2001; 276:38002 - 38009
- Ahmad SI, Kirk SH, Eisenstark A. Thymine metabolism and thymineless death in prokaryotes and eukaryotes. Annu Rev Microbiol 1998; 52:591 - 625
- Steidler L, Neirynck S, Huyghebaert N, Snoeck V, Vermeire A, Goddeeris B, et al. Biological containment of genetically modified Lactococcus lactis for intestinal delivery of human interleukin 10. Nat Biotechnol 2003; 21:785 - 789
- Horton RM, Cai ZL, Ho SN, Pease LR. Gene splicing by overlap extension: tailor-made genes using the polymerase chain reaction. Biotechniques 1990; 8:528 - 535
- van Asseldonk M, Rutten G, Oteman M, Siezen RJ, de Vos WM, Simons G. Cloning of usp45, a gene encoding a secreted protein from Lactococcus lactis subsp. lactis MG1363. Gene 1990; 95:155 - 160
- Glaser P, Frangeul L, Buchrieser C, Rusniok C, Amend A, Baquero F, et al. Comparative genomics of Listeria species. Science 2001; 294:849 - 852
- Holo H, Nes IF. High-frequency transformation, by electroporation, of Lactococcus lactis subsp. cremoris grown with glycine in osmotically stabilized media. Appl Environ Microbiol 1989; 55:3119 - 3123
- Piard JC, Hautefort I, Fischetti VA, Ehrlich SD, Fons M, Gruss A. Cell wall anchoring of the Streptococcus pyogenes M6 protein in various lactic acid bacteria. J Bacteriol 1997; 179:3068 - 3072
- Kohda C, Kawamura I, Baba H, Nomura T, Ito Y, Kimoto T, et al. Dissociated linkage of cytokine-inducing activity and cytotoxicity to different domains of listeriolysin O from Listeria monocytogenes. Infect Immun 2002; 70:1334 - 1341
- Hess J, Grode L, Gentschev I, Fensterle J, Dietrich G, Goebel W, et al. Secretion of different listeriolysin cognates by recombinant attenuated Salmonella typhimurium: superior efficacy of haemolytic over nonhaemolytic constructs after oral vaccination. Microbes Infect 2000; 2:1799 - 1806
- Poolman B, Konings WN. Relation of growth of Streptococcus lactis and Streptococcus cremoris to amino acid transport. J Bacteriol 1988; 170:700 - 707
- Carvalho LH, Hafalla JC, Zavala F. ELISPOT assay to measure antigen-specific murine CD8(+) T cell responses. J Immunol Methods 2001; 252:207 - 218
- Stack HM, Sleator RD, Bowers M, Hill C, Gahan CG. Role for HtrA in stress induction and virulence potential in Listeria monocytogenes. Appl Environ Microbiol 2005; 71:4241 - 4247
- Thatte J, Rath S, Bal V. Analysis of immunization route-related variation in the immune response to heat-killed Salmonella typhimurium in mice. Infect Immun 1995; 63:99 - 103
- Gutt CN, Hollander D, Brier CH, Kim ZG, Lorenz M. Influence of laparoscopy and laparotomy on systemic and peritoneal T lymphocytes in a rat model. Int J Colorectal Dis 2001; 16:216 - 220
- Pavlidis TE. Cellular changes in association with defense mechanisms in intra-abdominal sepsis. Minerva Chir 2003; 58:777 - 781
- Jorgensen CM, Hammer K, Martinussen J. CTP limitation increases expression of CTP synthase in Lactococcus lactis. J Bacteriol 2003; 185:6562 - 6574
- Law J, Buist G, Haandrikman A, Kok J, Venema G, Leenhouts K. A system to generate chromosomal mutations in Lactococcus lactis which allows fast analysis of targeted genes. J Bacteriol 1995; 177:7011 - 7018
- Gasson MJ. Plasmid complements of Streptococcus lactis NCDO 712 and other lactic streptococci after protoplast-induced curing. J Bacteriol 1983; 154:1 - 9
- Maguin E, Duwat P, Hege T, Ehrlich D, Gruss A. New thermosensitive plasmid for gram-positive bacteria. J Bacteriol 1992; 174:5633 - 5638