Abstract
The THO complex is a nuclear structure whose architecture is conserved among all kingdoms and plays an important role in mRNP biogenesis connecting transcription elongation with mRNA maturation and export. Recent data indicates that the THO complex is necessary for the proper expression of some genes, assurance of genetic stability by preventing transcription-associated recombination. Yeast THO has been described as a heterotetramer (Tho2, Hpr1, Mft1 and Thp2) that performs several functions through the interaction with other proteins like Tex1 or the mRNA export factors Sub2 and Yra1, with which it forms the TRanscription and EXport complex (TREX). In this article we review the cellular role of THO, which we show to be composed of five subunits with Tex1 being also an integral part of the complex. We also show a low-resolution structure of THO and localize some of its components. We discuss the consequences of THO interaction with nucleic acids through the unfolded C-terminal region of Tho2, highlighting the importance of unfolded regions in eukaryotic proteins. Finally, we comment on THO recruitment to active chromatin, a role that is linked to mRNA biogenesis.
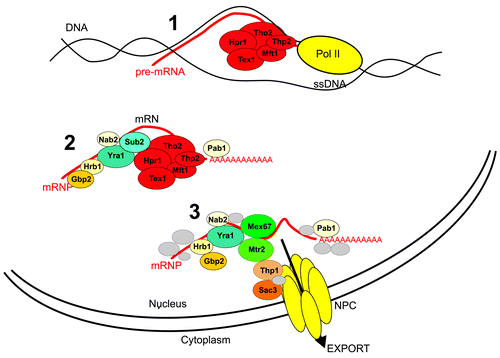
Transcription of pre-mRNA, its maturation and mRNA export to the cytoplasm are a chain of processes involving cooperation of many proteins and assemblies. Over the years these processes have been intensively, albeit independently investigated. However, the acquired knowledge has led to a new vision of these processes as working in a coordinated manner. One of the examples of the connection between all the steps involved in RNA biogenesis is the role of the THO complex, thought to be formed by Tho2 (180 kDa), Hpr1 (90 kDa), Mft1 (45 kDa), and Thp2 (30 kDa) proteins.Citation1 During transcription elongation, the THO complex is involved in packing pre-mRNA molecules into RNA–protein assemblies termed mRNPsCitation2 and is also essential for efficient co-transcriptional recruitment of mRNA export factors Yra1 and Sub2Citation3 (). Disruption of any of these tightly coupled steps of production of translation-competent mRNA in the cytoplasm leads to the activation of the RNA surveillance pathway and to the subsequent degradation of non-active mRNA molecules.Citation4 Lack of any of the THO subunits in yeast results in several molecular phenotypes like impairment of mRNP formation, that in turn leads to defects in transcription elongation and export. Lack of the Hpr1 subunit causes mRNA to remain trapped in the transcription sites giving rise to RNA/DNA hybrids (R-loops), which causes hyperrecombination and genomic instability.Citation5 Deletion of the Mtf1 subunit promotes the accumulation of transcribed but not matured and exported RNA, which, together with transcriptionally active chromatin, pieces of RNA export machinery and nuclear pore complexes (NPC), forms large aggregates called heavy chromatin.Citation6 Besides, the expression of long genes containing internal repeats is markedly reduced in tho2 mutants.Citation7
Figure 1. The THO complex couples RNA transcription elongation, mRNP formation and export. The THO binds active chromatin promoting association of Sub2 and Yra1 with the mRNP particle what facilitates mRNP export and prevents RNA/DNA hybrid formation (R-loops).
Not much is known about the structure of THO and even the composition is a matter of debate. Two of its constituent proteins, Tho2 and Hpr1, present high sequence similarity among yeasts, plants, insects and mammals. The rest of THO core subunits is less conserved but they have nevertheless been proposed to play similar roles.Citation8 Several proteins have been described to interact with the THO such as Mex67, Gbp2 and Hrb1 (the serine-arginine-rich (SR)-like proteins) and the Prp19 complex, with is involved in splicing and transcription elongation.Citation3,Citation9-Citation11 However, a more stable and stronger interaction has been described between THO and three other proteins, the RNA helicase Sub2/UAP56, the RNA binding protein Yra1/Aly, and Tex1. The complex formed by THO and all these proteins has been termed TREX (from TRanscription and Export),Citation3 although the real subunit composition of both THO and TREX is still a matter of speculation. Tex1 was first described as a TREX component of unknown function which co-purified with THO core subunits in yeast,Citation3 although later was found to co-purify in stoichiometric amounts with THO isolated from Drosophila and human.Citation3,Citation12,Citation13 This evidence led to postulate that Tex1 could be a bona fide THO component, while Sub2/UAP56 and Yra1/Aly may only interact transiently with it.Citation3,Citation12,Citation13 Determination of the three-dimensional structure of the THO complex should therefore shed additional light on the composition of the genuine THO complex.
The Architecture of the THO Complex
THO was purified using a TAP-tagged Tho2Citation14 and this consistently rendered a very pure and stable complex composed of stoichiometric amounts of the four proteins already described as THO components (Tho2, Hpr1, Mft1 and Thp2) and a fifth protein, Tex1. This indicates that THO is a hetero-pentameric complex. The three-dimensional reconstruction of THO,Citation15 performed by electron microscopy and image processing, revealed a long and conspicuous structure with two lateral protrusions (). The resolution of the reconstruction (17 Å) is not good enough to locate the five subunits within the envelope of the complex, so other approaches were used to map some of the components.
Figure 2. Three-dimensional structure of THO, generated by electron microscopy and image processing. The THO complex (EMD-2053) comprises Tho2, Hpr1, Mft1,Thp2 and Tex1 proteins. Different electron microscopy experiments have helped to localized Hpr1, Tex1 and the C-terminal region of Tho2, which is involved in nucleic acid binding [and which we show here in close relation with the RNA polymerase (EMD-1322)]. The localization of the rest of the subunits is speculative.
![Figure 2. Three-dimensional structure of THO, generated by electron microscopy and image processing. The THO complex (EMD-2053) comprises Tho2, Hpr1, Mft1,Thp2 and Tex1 proteins. Different electron microscopy experiments have helped to localized Hpr1, Tex1 and the C-terminal region of Tho2, which is involved in nucleic acid binding [and which we show here in close relation with the RNA polymerase (EMD-1322)]. The localization of the rest of the subunits is speculative.](/cms/asset/4fc23f17-a9ef-43af-a397-38d2d8f14962/kbia_a_10921181_f0002.gif)
Labeling of THO with an anti-Hpr1 antibody that recognizes its C-terminal part allowed us to locate this region to one of the ends of the THO structure (). Hpr1 is the most evolutionary conserved and best functionally characterized THO subunitCitation16-Citation19 and it has been found that its C-terminal region is ubiquitinated in a transcription-dependent manner and the ubiquitin moiety is recognized by the C-terminal UBA domain of the export receptor Mex67. Structural studies have indicated that ubiquitinated Hpr1 and the NPC subunits, FG nucleoporins (Nups), may bind to the Mex67 UBA in a mutually exclusive manner.Citation16 This suggests that the Mex67 export factor may be recruited to the nascent transcript via ubiquitinated Hpr1 and then transferred to nuclear pore FG Nups. The localization of the C-terminal region of Hpr1 in a exposed area of the THO structure strengthens the notion of this area of Hpr1 as a protein-interacting region.
An independent, three-dimensional reconstruction of the THO purified from a ΔTex1 strain which generated a THO complex without Tex1, allowed us to locate this protein in one of the protruding masses of the complex, and the atomic model of Tex1, a 7-blade β-propeller structure, docked very well in the corresponding mass of THO (). The identification of Tex1 as a core component of the THO complex is in accordance with some phenotypes described previously in plant orthologs. Since its discovery, the function of Tex1 has remained elusive. Deletion of Tex1 in yeast does not lead to any detectable growth phenotype and we only have observed a small decrease in production of exogenous β-galactosidase mRNA, which is encoded by a long GC-reach gene particularly sensitive to THO complex.Citation15 In contrast to yeast, the plant Tex1 seems to play a crucial role. The Arabidopsis ortholog of Tex1 (AtTEX1) is important for biosynthesis of a subset of endogenous and transgene siRNAsCitation20,Citation21 such us TAS1 and TAS2 tasiRNAs, IR71 siRNAs, and PSuc2:PDS.Citation20,Citation21 AtTEX1 is however not essential for biogenesis of all small RNAs since miRNAs and most cellular endogenous siRNAs, including those that are dependent on Pol IV, are not affected by mutations in the AtTEX1 locus.Citation3 Not surprisingly, in addition to AtTex1, mutations in other THO components such as AtTHO1 and AtTHO6, also affect tasiRNAs biogenesis. Proteins with WD40 folds are known to be involved in protein-protein interactionCitation22 so it is possible that AtTEX1 plays a role in the interaction of THO with other proteins involved in the siRNA pathway. In yeast, where the RNAi machinery does not exist, Tex1 seems to play a less crucial role, although probably one which involves protein-protein interaction.
The THO Complex is Recruited to Active Chromatin through a Basic Unfolded Tail with Can Interact Directly with Nucleic Acids
A different labeling experiment was used to locate the C-terminal region of Tho2 at the tip of the other, thinner and longer protrusionCitation15 (). One of the main questions regarding the mechanism of THO-mediated mRNP biogenesis is how this complex is recruited to active chromatin. Two mechanisms have been proposed. The first one involves the mediation of other proteins in the recruitment, such as RNA polymerase II, which was already described to interact with THO,Citation23 or Syf1, a component of the Prp19 splicing complex that has been proposed to be involved in THO complex recruitment.Citation9 The second hypothesis proposes that THO interacts directly with DNA and RNA in vitro.Citation3,Citation12,Citation13 We have found that THO interacts with both DNA and RNA by the unfolded C-terminal end of Tho2 protein.Citation15 Without this C-terminal region THO complex is not able to bind nucleic acids in vitro, which in turn decreases the β-galactosidase expression and slightly increases the level of recombination. When Syf 1 is mutated, only about 50% of TREX is bound to chromatin, but this amount is sufficient for proper mRNA export.Citation9 These two findings lead to the conclusion that binding of THO to nucleic acids relies on a combination of both mechanisms, direct and mediated.
The properties of the Tho2 C-terminal region are somehow similar to the serine/arginine-rich domains of SR splicing factors.Citation24 These act as alternative splicing factors by enhancing or inhibiting particular splicing reactions and the SR region is predicted to be intrinsically disordered, as is the C-terminal region of Tho2.Citation25 Nevertheless their mechanism of action is still a matter of debate. There is some evidence on direct interaction with RNA,Citation24,Citation26 but also for the involvement in protein-protein interactions partially through self-multimerization/aggregation.Citation24 In fact, actions through RNA/DNA-protein or protein-protein interactions for both THO and SR splicing factors are not mutually exclusive and it is very likely that both modes are required for the interaction. The unfolded tails are very elongated in comparison to folded globular domains and even a single molecule could accommodate interactions with both RNA and several proteins at the same time. It is tempting to speculate that these interactions are mostly driven by charge rather than specific recognition and that individual interactions have rather low overall affinity, but the attachment at multiple sites provides sufficient strength to perform physiological functions.
Acknowledgments
This work was supported by the Spanish Ministry of Science and Innovation Grant BFU2010-15703/BMC to J.M.V. and by the Foundation for Polish Science Team Programme co-financed by the EU European Regional Development Fund.
References
- Chávez S, Beilharz T, Rondón AG, Erdjument-Bromage H, Tempst P, Svejstrup JQ, et al. A protein complex containing Tho2, Hpr1, Mft1 and a novel protein, Thp2, connects transcription elongation with mitotic recombination in Saccharomyces cerevisiae. EMBO J 2000; 19:5824 - 34; http://dx.doi.org/10.1093/emboj/19.21.5824; PMID: 11060033
- Köhler A, Hurt E. Exporting RNA from the nucleus to the cytoplasm. Nat Rev Mol Cell Biol 2007; 8:761 - 73; http://dx.doi.org/10.1038/nrm2255; PMID: 17786152
- Strässer K, Masuda S, Mason P, Pfannstiel J, Oppizzi M, Rodriguez-Navarro S, et al. TREX is a conserved complex coupling transcription with messenger RNA export. Nature 2002; 417:304 - 8; http://dx.doi.org/10.1038/nature746; PMID: 11979277
- Houseley J, LaCava J, Tollervey D. RNA-quality control by the exosome. Nat Rev Mol Cell Biol 2006; 7:529 - 39; http://dx.doi.org/10.1038/nrm1964; PMID: 16829983
- Huertas P, Aguilera A. Cotranscriptionally formed DNA:RNA hybrids mediate transcription elongation impairment and transcription-associated recombination. Mol Cell 2003; 12:711 - 21; http://dx.doi.org/10.1016/j.molcel.2003.08.010; PMID: 14527416
- Rougemaille M, Dieppois G, Kisseleva-Romanova E, Gudipati RK, Lemoine S, Blugeon C, et al. THO/Sub2p functions to coordinate 3′-end processing with gene-nuclear pore association. Cell 2008; 135:308 - 21; http://dx.doi.org/10.1016/j.cell.2008.08.005; PMID: 18957205
- Chávez S, García-Rubio M, Prado F, Aguilera A. Hpr1 is preferentially required for transcription of either long or G+C-rich DNA sequences in Saccharomyces cerevisiae. Mol Cell Biol 2001; 21:7054 - 64; http://dx.doi.org/10.1128/MCB.21.20.7054-7064.2001; PMID: 11564888
- Luna R, Jimeno S, Marín M, Huertas P, García-Rubio M, Aguilera A. Interdependence between transcription and mRNP processing and export, and its impact on genetic stability. Mol Cell 2005; 18:711 - 22; http://dx.doi.org/10.1016/j.molcel.2005.05.001; PMID: 15949445
- Chanarat S, Seizl M, Strässer K. The Prp19 complex is a novel transcription elongation factor required for TREX occupancy at transcribed genes. Genes Dev 2011; 25:1147 - 58; http://dx.doi.org/10.1101/gad.623411; PMID: 21576257
- Zenklusen D, Vinciguerra P, Wyss JC, Stutz F. Stable mRNP formation and export require cotranscriptional recruitment of the mRNA export factors Yra1p and Sub2p by Hpr1p. Mol Cell Biol 2002; 22:8241 - 53; http://dx.doi.org/10.1128/MCB.22.23.8241-8253.2002; PMID: 12417727
- Hurt E, Luo MJ, Röther S, Reed R, Strässer K. Cotranscriptional recruitment of the serine-arginine-rich (SR)-like proteins Gbp2 and Hrb1 to nascent mRNA via the TREX complex. Proc Natl Acad Sci U S A 2004; 101:1858 - 62; http://dx.doi.org/10.1073/pnas.0308663100; PMID: 14769921
- Jimeno S, Rondón AG, Luna R, Aguilera A. The yeast THO complex and mRNA export factors link RNA metabolism with transcription and genome instability. EMBO J 2002; 21:3526 - 35; http://dx.doi.org/10.1093/emboj/cdf335; PMID: 12093753
- Rehwinkel J, Herold A, Gari K, Köcher T, Rode M, Ciccarelli FL, et al. Genome-wide analysis of mRNAs regulated by the THO complex in Drosophila melanogaster. Nat Struct Mol Biol 2004; 11:558 - 66; http://dx.doi.org/10.1038/nsmb759; PMID: 15133499
- Dziembowski A, Seraphin B. Sample preparation for protein complex analysis by the TAP method. In: vonHagen J, ed. Sample preparation for proteomics, Weinheim: VCH Wiley, 2008.
- Peña A, Gewartowski K, Mroczek S, Cuéllar J, Szykowska A, Prokop A, et al. Architecture and nucleic acids recognition mechanism of the THO complex, an mRNP assembly factor. EMBO J 2012; 31:1605 - 16; http://dx.doi.org/10.1038/emboj.2012.10; PMID: 22314234
- Iglesias N, Tutucci E, Gwizdek C, Vinciguerra P, Von Dach E, Corbett AH, et al. Ubiquitin-mediated mRNP dynamics and surveillance prior to budding yeast mRNA export. Genes Dev 2010; 24:1927 - 38; http://dx.doi.org/10.1101/gad.583310; PMID: 20810649
- Gwizdek C, Iglesias N, Rodriguez MS, Ossareh-Nazari B, Hobeika M, Divita G, et al. Ubiquitin-associated domain of Mex67 synchronizes recruitment of the mRNA export machinery with transcription. Proc Natl Acad Sci U S A 2006; 103:16376 - 81; http://dx.doi.org/10.1073/pnas.0607941103; PMID: 17056718
- Hobeika M, Brockmann C, Iglesias N, Gwizdek C, Neuhaus D, Stutz F, et al. Coordination of Hpr1 and ubiquitin binding by the UBA domain of the mRNA export factor Mex67. Mol Biol Cell 2007; 18:2561 - 8; http://dx.doi.org/10.1091/mbc.E07-02-0153; PMID: 17475778
- Hobeika M, Brockmann C, Gruessing F, Neuhaus D, Divita G, Stewart M, et al. Structural requirements for the ubiquitin-associated domain of the mRNA export factor Mex67 to bind its specific targets, the transcription elongation THO complex component Hpr1 and nucleoporin FXFG repeats. J Biol Chem 2009; 284:17575 - 83; http://dx.doi.org/10.1074/jbc.M109.004374; PMID: 19401465
- Yelina NE, Smith LM, Jones AM, Patel K, Kelly KA, Baulcombe DC. Putative Arabidopsis THO/TREX mRNA export complex is involved in transgene and endogenous siRNA biosynthesis. Proc Natl Acad Sci U S A 2010; 107:13948 - 53; http://dx.doi.org/10.1073/pnas.0911341107; PMID: 20634427
- Jauvion V, Elmayan T, Vaucheret H. The conserved RNA trafficking proteins HPR1 and TEX1 are involved in the production of endogenous and exogenous small interfering RNA in Arabidopsis. Plant Cell 2010; 22:2697 - 709; http://dx.doi.org/10.1105/tpc.110.076638; PMID: 20798330
- Smith TF, Gaitatzes C, Saxena K, Neer EJ. The WD repeat: a common architecture for diverse functions. Trends Biochem Sci 1999; 24:181 - 5; http://dx.doi.org/10.1016/S0968-0004(99)01384-5; PMID: 10322433
- Li Y, Wang X, Zhang X, Goodrich DW. Human hHpr1/p84/Thoc1 regulates transcriptional elongation and physically links RNA polymerase II and RNA processing factors. Mol Cell Biol 2005; 25:4023 - 33; http://dx.doi.org/10.1128/MCB.25.10.4023-4033.2005; PMID: 15870275
- Shen H, Green MR. RS domains contact splicing signals and promote splicing by a common mechanism in yeast through humans. Genes Dev 2006; 20:1755 - 65; http://dx.doi.org/10.1101/gad.1422106; PMID: 16766678
- Haynes C, Iakoucheva LM. Serine/arginine-rich splicing factors belong to a class of intrinsically disordered proteins. Nucleic Acids Res 2006; 34:305 - 12; http://dx.doi.org/10.1093/nar/gkj424; PMID: 16407336
- Shen H, Green MR. A pathway of sequential arginine-serine-rich domain-splicing signal interactions during mammalian spliceosome assembly. Mol Cell 2004; 16:363 - 73; http://dx.doi.org/10.1016/j.molcel.2004.10.021; PMID: 15525510