Abstract
Research has shown that a greater variety of enzymes, as well as variety of microorganisms producing enzymes, can have an overall synergistic effect on the decomposition of lignocellulosic biomass for the production of value-added bio-products. Here, 8 cellulase-degrading bacterial isolates were selected to develop co-, tri-, and tetra-cultures for the decomposition of lignocellulosic biomass. Glucose and xylose equivalents released from imitation biomass media containing 0.5% (w/v) beechwood xylan and 0.5% (w/v) Avicel was measured using di-nitrosalicylic acid for all consortia, along with cell growth and survival. Thereafter, 6 co- and 2 tri-cultures with greatest decomposition were examined for ability to degrade Agave americana fiber. Interestingly, when strains were paired up in co-culture, four pairs: G+5, G+A, C+A1, and G+A1 produced high reducing sugars in 24 h: 6 µM, 8 µM, 8 µM, and finally, 6 µM, respectively. From 4 co-cultures with highest reducing sugar equivalents, tri- and tetra-cultures were produced. The bacterial consortia which had the highest reducing sugars detected were 2 tri-cultures: G + A1 + A4 and G + A1 + 5, displaying levels as high as 9 µM and 5 µM in day 1, respectively. All co- and tri-cultures maintained high cell survival for 14 days with 0.5 g ground Agave. Upon evaluating Agave dry weight after treatment, it was evident that almost half the biomass could be decomposed in 14 days. Scanning electron microscopy of treated Agave supported decomposition when compared with the control. These bacterial consortia have potential for further study of value-added by-product production during metabolism of lignocellulosic biomasses.
Introduction
The rising cost of petroleum, concern for exhaustion of fossil fuel sources, and increasing global carbon dioxide emissions have driven the onward exploration in renewable fuels from lignocellulosic biomasses. Different lignocellulosic biomasses exist which are potential candidates for the bioconversion to bioethanol and other value-added bioproducts; this includes a variety of grasses, as well as municipal and agricultural wastes. One major limiting step to the development of economically viable second generation biofuels is the complete efficient degradation of lignocellulosic biomass using enzymes or microorganisms producing enzymes. This rate limiting step is caused by barriers such as end product inhibition, a lack in variety of enzymes for complete hydrolysis, and the harsh environmental conditions of the industrial process.
Bacteria offer several amenable traits for the decomposition of lignocelluloses and can help overcome costly hurtles in the biodegradation process. For example, bacteria have high growth rates compared with other microorganisms; they also have the ability to adapt to a wide variety of environmental conditions such as pH, salinity, and temperature changes. Moreover, bacteria can be genetically modified to cut back on feedback inhibition or increase catalytic activity of the lignocellulolytic enzymes.Citation1
For several years, researchers have studied pure cultures of bacteria and observed their ability to degrade simple substrates such as cellulose and xylan; pure cultures have displayed less than satisfactory activity toward lignocelluloses.Citation2,Citation3 In the environment bacteria decompose biomass such as leaf litter and decaying plant matter in concert with other microorganisms, particularly other bacteria. Thus, it is ideal to consider the use of bacterial consortia for lignocellulose decomposition. In fact recently, several researchers have begun to focus on the ability of bacterial consortia to efficiently degrade different lignocellulosic biomasses.Citation1,Citation2,Citation4,Citation5
Previously, we isolated and identified 20 cellulase-producing bacteria from municipal wastes and peat. These bacterial isolates were characterized for their ability to degrade or modify different components of lignocellulose, including celluloses, xylan, and lignin.Citation6 Here, we selected 8 strains with varying lignocellulose-degrading abilities to build synthetic co-, tri-, and tetra-cultures to optimize lignocellulose decomposition with synergy between microorganisms.
Results
Decomposition of imitation biomass by single cultures
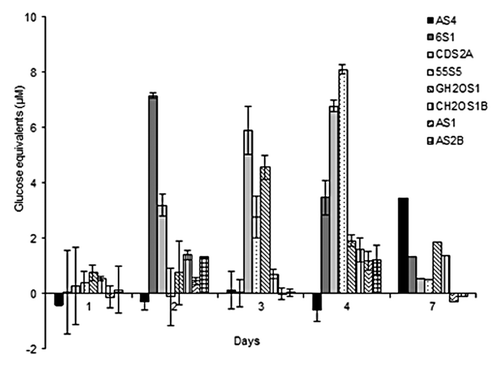
The reducing sugars released from imitation biomass containing crystalline cellulose and xylan were examined for 8 bacterial isolates selected based on previously characterized biomass-degrading capabilities (). As shown in , none of the isolates produced significantly detectable reducing sugars in 24 h (day 1), nor did they produce significantly detectable sugars after day 7 (data not shown). As well, there is a wide variation in reducing sugars production between individual bacterial isolates. For example, isolate 6S1 had a maximum detection of reducing sugars of 7.2 µM at day 2, while CDS2A had a peak at day 4 of 6.9 µM. Moreover, GH2OS1 peaked at day 4 producing 4.6 µM glucose equivalents, and 55S5 had the highest peak in reducing sugars of 8.1 µM at day 4. Finally, AS4 had a significantly lower peak in reducing sugars at day 7 of 3.1 µM. The remaining isolates, CH2OS1, AS1, and AS2B, did not produce significant amounts of reducing sugars when compared with the other isolates.
Table 1. Bacterial strains and their lignocellulose degrading characteristics for consortia development
Decomposition of imitation biomass by co-cultures
Similarly, the reducing sugars released from imitation biomass were examined for bacterial co-cultures prepared by pairing each of the 8 previously selected isolates with each other once. A total of 10 co-cultures were developed and were split into two groups of co-cultures containing 5 each for reducing sugar measurements (). In the first 5 co-cultures, all co-cultures produced significant reducing sugar equivalents at day 1 for nearly all of which was their peak reducing sugar production. For example, G + C, G + A4, C + A4, and G + 5 produced 8.2, 7.7, 6.6, and 5.9 µM of reducing sugar equivalents in 24 h, respectively. Meanwhile, the remaining co-culture, C + 5, peaked at day 7 producing 4.7 µM of reducing sugar equivalents (). From these results two co-cultures (G + 5 and G + A4) with the greatest potential toward lignocellulose were chosen from this group for the production of bacterial consortia, based on their ability to maintain approximately half of their activity up to 4 d.
Figure 2. Reducing sugar equivalents (µM) released from (A) first 5 groups of bacterial co-cultures and (B) an additional 5 bacterial co-cultures, during aerobic incubation over 7 d with imitation media: 0.5% (w/v) microcrystalline cellulose and 0.5% (w/v) beechwood xylan.
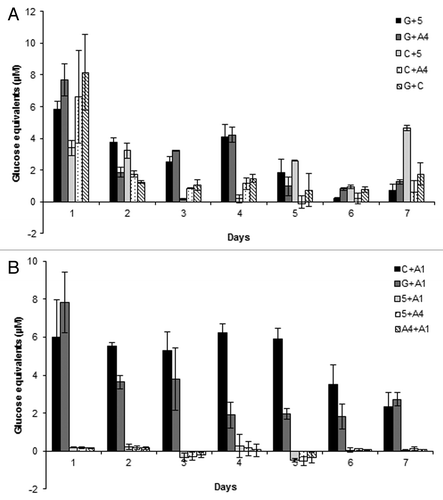
In regards to the second group of co-cultures, when looking at their reducing sugar production over 7 d in imitation media, it is readily apparent that only 2 co-cultures (C + A1 and G + A1) were capable of producing any detectable reducing sugars. G + A1 and C + A1 reducing sugars production peaked at day 1 at 7.8 and 6.0 µM, respectively. Specifically, co-culture C + A1 maintained an average reducing sugar production of 5.7 µM from day 2 to 5, while G + A1 maintained an average of 3.7 µM of reducing sugar equivalents from day 2 to 3. However, both maintained detectable reducing sugars at day 7. Both co-cultures C + A1 and G + A1 were chosen to be used in the creation of bacterial consortia.
Decomposition of imitation biomass and cell growth by tri- and tetra-cultures
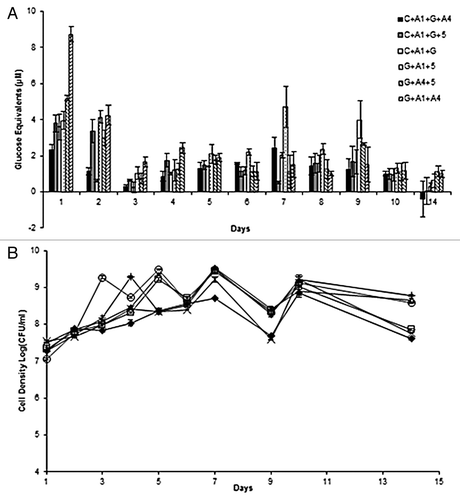
Based on the results from the reducing sugar production in co-cultures, co-cultures (C + A1, G + A1, G + A4, and G + 5) producing the greatest reducing sugars were selected to produce 2 tetra-cultures and 4 tri-cultures. That is the most combinations possible from 4 pairs of bacteria. The reducing sugars for tri- and tetra-cultures were examined on imitation media and it was found that all bacterial consortia produced reducing sugar equivalents on day 1. However the 2 tetra-cultures (C + A1 + G + A4 and C + A1 + G + 5) and one tri-culture (C + A1 + G) did not produce high or consistent amounts of reducing sugars with their highest peak of 2.3, 3.6, and 3.8 µM of reducing sugar equivalents at day 1. On the other hand, the remaining tri-cultures produced displayed good activity for reducing sugar production. For example, the tri-culture G + A1 + A4 peaked at day 1 producing 8.7 µM of reducing sugars and dropped to nearly half the amount of reducing sugar production (4.2 µM) on day 2. Moreover, tri-culture G + A1 + 5 produced more consistent activity by producing approximately 4 µM of reducing sugar equivalents on day 1 and 2, meanwhile peaked again to 4.7 µM at day 7 and 4 µM on day 9 (). These two tri-cultures were selected for treatment of exemplar lignocellulosic biomass: Agave. Additionally, the mixed cell density of tri- and tetra-cultures was determined through plate counting using the drop plate technique, and it was found that all bacterial consortia mixed growth could be detected using the drop plate counting technique ().
Figure 3. (A) Reducing sugar equivalents (µM) released by 2 tetra-culture and 4 tri-cultures of bacteria during aerobic incubation over 14 d with imitation media: 0.5% (w/v) microcrystalline cellulose and 0.5% (w/v) beechwood xylan. (B) Bacterial consortia mixed cell densities in log(CFU/ml) for bacterial consortia grown in imitation media for 14 d. Consortia: C + A1 + G + A4 (♦), C + A1 + G + 5 (□), G + A1 + A4 (Δ), C + A1 + G (x), G + A1 + 5 (+), G + A4 + 5 (○).
Decomposition of Agave by bacterial consortia
From the reducing sugar results of previous co-, tri-, and tetra-cultures, 6 co-cultures and 2 tri-cultures were selected as the greatest candidates for decomposition of lignocellulosic biomass, Agave fiber. In it is evident that all selected bacterial consortia could grow consistently on ground Agave for 14 d. On day 1, all cell densities were approximately 7.5 log(CFU/ml). Additionally, after day 3 all mixes plateaued growth to a constant average 8.3 log(CFU/ml) until day 14
Figure 4. (A) Bacterial consortia mixed cell growth in log(CFU/ml) after 14 d growth with ground Agave. Consortia: C + A1(♦), C + A4 (□), C + 5 (Δ), G + A1 (x), G + A4 (*), G + 5 (○), G + A1 + 5 (+), and G + A1 + A4 (‒). (B) Agave decomposition expressed as the dry weight percentage (%) compared with the control.
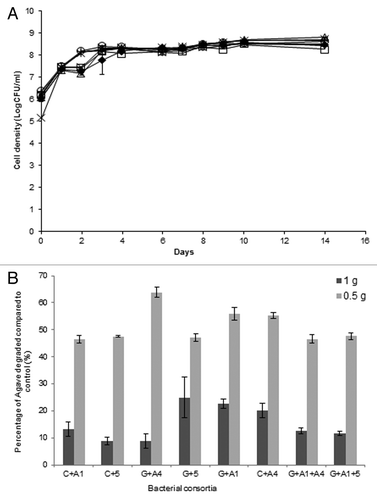
Upon evaluation of reducing sugars released from Agave by bacterial consortia, no significant reducing sugars were observed. However, it was evident that the bacteria were growing by utilizing something from the Agave, so the dry weight of the Agave was compared with the dry weight of the control and percentage of Agave degraded was calculated (). The bacterial consortia were grown in media containing 1% (w/v) Agave and 0.5% Agave, separately. It was found for consortia grown with 1% Agave the average decomposition of Agave dry weights was 15.32%. However, when the consortia were grown with 0.5% (w/v) Agave G + A4, G + A1, C + A4 decomposed 63.9, 55.9, and 55.3% Agave after 14 d, while the remaining consortia grown with 0.5% Agave decomposed an average 47.1%.
Agave morphology after decomposition (SEM)
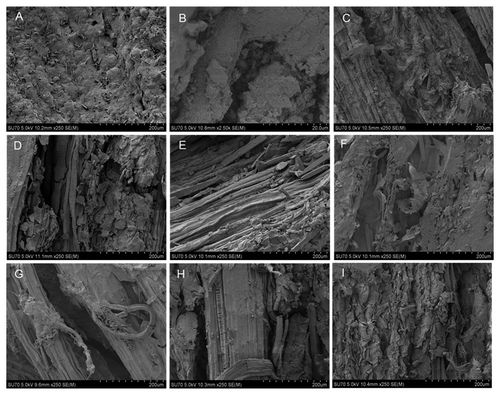
Images of the Agave control (no bacteria) and for each of 6 co-cultures and 2 tri-cultures after 14 d treatment were taken using the SEM. In control, it is evident that the agave biomass with no bacterial treatment is much smoother than the treated samples (), and no apparent decomposition has occurred. It is clearly evident in co-cultures (, C + A4; , G + A1; , G + 5) and tri-culture (, G + A1 + 5) that degradation of the Agave is occurring within 14 d. The fibers, which would be cellulose microtubules, have been exposed; similar fine fibers have been characterized in Agave in other studies.Citation7 Nonetheless, in co-cultures (, C + 5; , G + A4) and tri-culture (, G + A1 + A4), there is evidence of degradation by the disruption to the surface structure, leading to access of lower microtubules. Only the co-culture in , C + A1, showed the least decomposition and the smoothest surface comparable to control; however, evidence of degradation is represented by the crevasses.
Discussion
The use of bacterial consortia in the degradation of lignocelluloses has gained attention in bioproducts production considering consortia are recognized for their ability to perform more complicated tasks and more readily adapt to changes in the environment than mono-culture.Citation8 The synergy between enzymes produced by different microorganisms can help overcome the lack of efficient conversion by a single strain; this is because strains may produce high levels of some but not all enzymes required for efficient conversion. Researchers have commonly explored the potential for co-culture of microorganisms based on pre-hypothesized abilities for bacteria and/or fungi/yeast to co-exist and consolidate the degradation of cellulosic biomass with the fermentation of sugars to products such as ethanol.Citation9,Citation10,Citation11 In this study the exploration of co-, tri-, and tetra-cultures using new bacterial strains recently isolated for the degradation of lignocelluloses sets it apart from most other studies.
In 2005 a complex study was performed where researchers established a stable cellulose-degrading bacterial community consisting of 5 different bacterial species. The stability of the microbial community was dependent upon the network flow of substrates between species. The role of each species in the consortia was determined based on the cellulose degrading potential.Citation12 Then recently, similar to our work, a study explored the use of 5 bacterial isolates identified by 16rRNA gene sequence and their use in a bacterial consortium.Citation5 Similarly, using 8 bacterial strains previously isolated and characterized for their lignocellulolytic-degrading abilities, a variety of co-, tri-, and tetra-cultures were developed on imitation media containing crystalline cellulose and xylan.Citation6 Unlike studies using existing microflora, the bacteria used in this study were isolates identified by 16 rDNA sequencing similar to the previous study.
Interestingly, 6 co-cultures were among the greatest producers of reducing sugars from the imitation media. However, no tetra-cultures had significant reducing sugar production, possibly due to the complicated interactions between 4 strains in the consortia. Yet, 3 tri-cultures had significant reducing sugar production and were selected along with the 6 greatest co-cultures for the treatment of exemplar lignocellulosic biomass: Agave. When the Agave was treated with the selected co- and tri-cultures it was apparently degraded as was observed under the SEM. However, upon measuring of the reducing sugars released from the Agave, little reducing sugars could be detected. Due to the bacterial growth on Agave it is considered here that the lack of reducing sugars is a consequence of the consumption of sugars released. The Agave americana represents a more complex biomass and may take more energy to degrade because it may require production of different enzymes as it is comprised of cellulose (~68%), hemicellulose (~15%), moisture (~8%), lignin (~5%), and wax (~0.26%).Citation7 Thus, the predicted rate of degradation may be slower or accessibility to sugars more restricted than the imitation media. The dry weights were measured to provide further evidence to the hypothesis. From the dry weights, it is evident that a significant proportion of the Agave biomass is being assimilated. Using dry weights to determine biomass degradation has been done by researchers studying lignocellulose decomposition in bacteria and fungi.Citation13,Citation14 The fact that almost half the biomass was assimilated at 0.5 g/l Agave suggests significant assimilation may occur at a gradual rate. Perhaps testing the reducing sugars beyond 14 d may reveal more significant production of detectable reducing sugars, as the dry weight was taken after 21 d incubation.
Furthermore, considering the assimilation of Agave by the bacterial isolates suggests that in the process of metabolizing the sugars some products may also be produced which will be evaluated in future studies of these consortia. According to other researches, we may expect by-products such as ethanol, butanol, acetone, isopropanol, and xylitol.Citation15-Citation17 Agave was chosen as the biomass for this study as an example lignocellulosic biomass, and in the future the bacterial consortia developed here can be similarly exposed to other lignocellulosic biomasses.
The co-cultures and tri-cultures which were selected for degradation of Agave consisted of 5 of 8 different bacterial strains characterized with different lignocellulose degrading abilities: Bacillus sp. 55S5 (CMC, FP, Xyl, and Lig activities), Pseudomonas sp. AS1 (CMC, FP, Xyl, and Lig activities), Bacillus sp. CH2OS1 (with CMC, FP, and Xyl activities), and Pseudomonas sp. GH2OS1 (CMC and Xyl activities), and Microbacterium sp. AS4 (CMC and Lig activities). Genera of Gram positives and Gram negatives such as these have been implicated in the degradation of cellulose, some have of which have been found to produce value-added products from such hydrolysates. Although there is overlap in the characterized lignocellulolytic activities between these isolates, the specific activities of individually different enzymes have not been directly compared and seem to complement each other in the work presented here. The 6 co-cultures and 2 tri-cultures hold future potential for not only the decomposition of lignocellulosic biomass, such as agave but also the simultaneous production of value-added products.
Materials and Methods
Bacterial strains and media
The following eight bacterial isolates were used in this study: Bacillus sp. (55S5), Streptomyces sp. (AS1), Bacillus sp. (CH2OS1), Bacillus sp. (6S1), Microbacterium sp. (AS4), Exiguobacterium sp. (AS2B), Pseudomonas sp. (GH2OS1), and Chryseobacterium sp. (CDS2A). The strains and their previously characterized activities are described in . All strains were maintained on Luria Burtani (LB) agar plates at 4 °C or cultured overnight in 8 ml of LB broth prior to experiments. The base of all substrate media used contained salt media comprised of: NaNO3 1 g/l, K2HPO41 g/l, KCl 1 g/l, MgSO4 0.5 g/l, and yeast extract 0.3 g/l. The substrates crystalline cellulose and beechwood xylan were purchased from Sigma-Aldrich, and were of HPLC grade. As well, Agave americana leaf biomass (kindly provided by Jeffrey Phelps in Redding, California) was prepared as follows: dried at 70 °C in a drying oven (Thermo Fisher Scientific) for 24 h, ground in a Wiley mill, and then sieved through 2 mm pore size mesh. Fibers of the Agave americana biomass are composed of ~83% sugars, making it an excellent candidate for hydrolysis to sugar monomers.Citation18
Development of single, co- and tetra-cultures
For all experiments, all or some of the 8 bacterial isolates selected in the study were pre-grown in 6 ml of LB broth overnight at 30 °C, shaking at 200 rpm. For single cultures experiments 1 ml of overnight cultures was inoculated to individual flasks containing 100 ml of Dubois Salts Media supplemented with 0.5% (w/v) of Avicel and 0.5% (w/v) of beechwood xylan, referred to here as the imitation biomass medium. Co-cultures were designed using 5 of 8 bacterial isolates, (GH2OS1, CH2OS1, 55S5, AS1, and AS4), which presented the greatest potential in imitation media were paired with each other once; this was done by inoculating the imitation biomass medium with 500 µl of each strain in triplicate. Finally, all tri- and tetra-cultures were prepared by adding 500 µl of each selected strain (3 or 4 strains) and inoculated to the imitation biomass medium. All experiments were performed in triplicate. To simplify the strain identification in reference to co-, tri-, and tetra-cultures in the text, the following representative codes are used: GH2OS1 (G), CH2OS1 (C), 55S5 (5), AS1 (A1), and AS4 (A4), respectively.
Bacterial cell density in medium containing imitation biomass
The cell density of single, co-, tri-, and tetra-cultures was examined by the direct count drop plate technique. Cells were permitted to grow in the imitation biomass media for 21 d at 30 °C, shaking at 200 rpm. To determine cell density for each culture, 1 ml aliquots were removed from the flasks on the following days: 1, 2, 3, 4, 5, 6, 7, 10, 14, and 21. The 1 ml aliquots were then diluted to obtain countable colonies (30–300 colonies) and drop plated to LB agar in 6 µl aliquots. Then, drop plates were incubated for 18 h at 30 °C before being counted. All drop plates were done in triplicate and all experiments were repeated in triplicate. Cell densities were expressed in log (CFU/ml).
Reducing sugars
All cells in single, co-, tri-, and tetra-cultures were permitted to grow in imitation biomass media for 21 d at 30 °C, shaking at 200 rpm. To determine the reducing sugar equivalents released from the imitation biomass (salts media with 0.5% [w/v] of Avicel and 0.5% [w/v] of beechwood xylan), 1 ml aliquots of media were removed from each flask on days: 1, 2, 3, 4, 5, 6, 7, 10, 14, and 21. The 1 ml aliquots were centrifuged for 2 min at 1700 g to pellet cells, cell debris, and biomass. The clear supernatant was used to measure the reducing sugars using a modified microtiter plate assay.Citation19 Specifically, 60 µl of supernatant was loaded in triplicate to a 96-well microtiter plate. Following, 120 µl of di-nitro-salicylic acid solution (DNS) was added to each well. The plate was then incubated at 100 °C for 5 min. The plate was then allowed to cool before 36 µl was removed from each well and individually added to 160 µl of distilled water. The plate was then read using BioRad XMark Spectrophotometric plate reader at 540 nm with 15 s shaking prior to the reading. All experiments were performed in triplicate. The reducing sugars were expressed as nm glucose equivalents.
Decomposition of Agave with bacterial consortia
From the consortia previously tested, 8 that possessed the greatest potential in our imitation media were chosen to degrade Agave biomass, 6 co-cultures and 2 tri-cultures as follows: C + A1, C + A4, C + 5, G + A1, G + A4, G + 5, G + A1 + A4, and G + A1 + 5. All strains were grown for a 14 d period in 200 ml of salt media containing 0.5% and 1% Agave for separate experiments, incubated at 30 °C, and shaken at 200 rpm. Samples were collected and prepared by removing 1 ml aliquots into 1.5 ml microcentrifuge tubes, which were then centrifuged at 17 000 g for 2 min to obtain supernatant. Agave degradation was evaluated by measuring the reducing sugars in the supernatant using a microtiter plate method with DNS, as previously mentioned. Simultaneously, 0.5 ml of culture was removed and serially diluted as required to determine cell density by the drop plate counting technique, also previously described. All sampling was done from biological triplicates at days: 0–4, 6–10, and 14.
Agave dry weight after decomposition
Due to a lack of reducing sugars observed, to determine whether or not the Agave biomass was being degraded, the dry weight of the Agave was determined at 14 d treatment. To accomplish this, an additional biological replicate for the treatment of Agave with bacterial consortia as described previously was used which had not been sampled from; this was to ensure biomass was not lost. Identical growth conditions and concentrations of Agave were used. Briefly, each culture was filtered through pre-weighed Whatman No. 1 filter paper (M1); following that, the culture flasks were washed thrice with approximately 10 ml of distilled water to remove residual residue and flush through bacterial cells adhering to the biomass. The filter papers with biomass were dried in an oven at 70 °C for 72 h to obtain a constant weight and then weighed (M2). Dry weights were determined by subtracting M1 from M2, compared with the control.
Agave morphology after decomposition
A scanning electron microscope (SEM) was used to observe the agave morphology after decomposition with bacterial co-, tri-, and tetra-cultures previously selected. Dried Agave leaf was sliced using scalpels into approximately 1 cm2 pieces, and 1 g of Agave portions were placed in 100 ml of salts media, then autoclaved. Thereafter, the flasks were inoculated to prepare bacterial consortia as previously described before incubating at 30 °C, with shaking at 180 rpm. Samples were collected from the Agave control containing no bacteria and bacterial treated agave after 14 d incubation. The Agave biomass samples were filtered through grade 202 filter papers, washed twice with 5 ml sterile double-distilled water, and then dried in a glass petri dish at 70 °C for 48 h. Thereafter, each sample was coated with gold for 45 s in a DentonDeskII sputter coater (Denton Vacuum USA); then, samples were examined on an SEM (Hitachi SU-70) at 5 kV.
Disclosure of Potential Conflicts of Interest
No potential conflict of interest was disclosed.
Acknowledgments
The authors thank Elyssia Adamo of Lakehead University for her assistance toward the making of the scanning electron images.
References
- Pandey S, Singh S, Yadav AN, Nain L, Saxena AK. Phylogenetic diversity and characterization of novel and efficient cellulase producing bacterial isolates from various extreme environments. Biosci Biotechnol Biochem 2013; 77:1474 - 80; http://dx.doi.org/10.1271/bbb.130121; PMID: 23832366
- Hui W, Jiajia L, Yucai L, Peng G, Xiaofen W, Kazuhiro M, Zongjun C. Bioconversion of un-pretreated lignocellulosic materials by a microbial consortium XDC-2. Bioresour Technol 2013; 136:481 - 7; http://dx.doi.org/10.1016/j.biortech.2013.03.015; PMID: 23567720
- Osborne JM, Dehority BA. Synergism in degradation and utilization of intact forage cellulose, hemicellulose, and pectin by three pure cultures of ruminal bacteria. Appl Environ Microbiol 1989; 55:2247 - 50; PMID: 16348005
- Feng Y, Yu Y, Wang X, Qu Y, Li D, He W, Kim BH. Degradation of raw corn stover powder (RCSP) by an enriched microbial consortium and its community structure. Bioresour Technol 2011; 102:742 - 7; http://dx.doi.org/10.1016/j.biortech.2010.08.074; PMID: 20863696
- Okeke BC, Lu J. Characterization of a defined cellulolytic and xylanolytic bacterial consortium for bioprocessing of cellulose and hemicelluloses. Appl Biochem Biotechnol 2011; 163:869 - 81; http://dx.doi.org/10.1007/s12010-010-9091-0; PMID: 20859703
- Maki ML, Idrees A, Leung KT, Qin W. Newly isolated and characterized bacteria with great application potential for decomposition of lignocellulosic biomass. J Mol Microbiol Biotechnol 2012; 22:156 - 66; http://dx.doi.org/10.1159/000341107; PMID: 22832891
- Chaabouni Y, Drean J-Y, Msahli S, Sakli F. Morphological Characterization of Individual Fiber of Agave americana L. Text Res J 2006; 75:367 - 74; http://dx.doi.org/10.1177/0040517506061965
- Brenner K, You L, Arnold FH. Engineering microbial consortia: a new frontier in synthetic biology. Trends Biotechnol 2008; 26:483 - 9; http://dx.doi.org/10.1016/j.tibtech.2008.05.004; PMID: 18675483
- Ng TK, Ben-Bassat A, Zeikus JG. Ethanol Production by Thermophilic Bacteria: Fermentation of Cellulosic Substrates by Cocultures of Clostridium thermocellum and Clostridium thermohydrosulfuricum. Appl Environ Microbiol 1981; 41:1337 - 43; PMID: 16345787
- Saddler JN, Chan MK-H. Conversion of pretreated lignocellulosic substrates to ethanol by Clostridium thermocellum in mono- and co-culture with Clostridium thermosaccharolyticum and Clostridium thermohydrosulphuricurn.. Can J Microbiol 1983; 30:212 - 20; http://dx.doi.org/10.1139/m84-032
- Zuroff TR, Xiques SB, Curtis WR. Consortia-mediated bioprocessing of cellulose to ethanol with a symbiotic Clostridium phytofermentans/yeast co-culture. Biotechnol Biofuels 2013; 6:59; http://dx.doi.org/10.1186/1754-6834-6-59; PMID: 23628342
- Kato S, Haruta S, Cui ZJ, Ishii M, Igarashi Y. Stable coexistence of five bacterial strains as a cellulose-degrading community. Appl Environ Microbiol 2005; 71:7099 - 106; http://dx.doi.org/10.1128/AEM.71.11.7099-7106.2005; PMID: 16269746
- Yang S-J, Kataeva I, Hamilton-Brehm SD, Engle NL, Tschaplinski TJ, Doeppke C, Davis M, Westpheling J, Adams MW. Efficient degradation of lignocellulosic plant biomass, without pretreatment, by the thermophilic anaerobe “Anaerocellum thermophilum” DSM 6725. Appl Environ Microbiol 2009; 75:4762 - 9; http://dx.doi.org/10.1128/AEM.00236-09; PMID: 19465524
- Nazarpour F, Abdullah DK, Abdullah N, Zamiri R. Evaluation of Biological Pretreatment of Rubberwood with White Rot Fungi for Enzymatic Hydrolysis. Materials 2013; 6:2059 - 73; http://dx.doi.org/10.3390/ma6052059
- Ingram LO, Gomez PF, Lai X, Moniruzzaman M, Wood BE, Yomano LP, York SW. Metabolic engineering of bacteria for ethanol production. Biotechnol Bioeng 1998; 58:204 - 14; http://dx.doi.org/10.1002/(SICI)1097-0290(19980420)58:2/3<204::AID-BIT13>3.0.CO;2-C; PMID: 10191391
- Dùrre P. New insights and novel developments in clostridial acetone/butanol/isopropanol fermentation. Appl Microbiol Biotechnol 1998; 49:639 - 48; http://dx.doi.org/10.1007/s002530051226
- Rangaswamy S, Agblevor FA. Screening of facultative anaerobic bacteria utilizing D-xylose for xylitol production. Appl Microbiol Biotechnol 2002; 60:88 - 93; http://dx.doi.org/10.1007/s00253-002-1067-8; PMID: 12382046
- Mylsamy K, Rajendran I. Investigation on Physio-chemical and mechanical properties of raw and alkali-treated Agave Americana fibre. J Reinf Plast Comp 2010; 29:2925 - 35; http://dx.doi.org/10.1177/0731684410362817
- Xiao Z, Storms R, Tsang A. Microplate-based filter paper assay to measure total cellulase activity. Biotechnol Bioeng 2004; 88:832 - 7; http://dx.doi.org/10.1002/bit.20286; PMID: 15459905