Abstract
Electrospun fibers based on aliphatic polyesters, such as poly(ε-caprolactone) (PCL), have been widely used in regenerative medicine and drug delivery applications due to their biocompatibility, low cost and ease of fabrication. However, these aliphatic polyester fibers are hydrophobic in nature, resulting in poor wettability, and they lack functional groups for decorating the scaffold with chemical and biological cues. Current strategies employed to overcome these challenges include coating and blending the fibers with bioactive components or chemically modifying the fibers with plasma treatment and reactants. In the present study, we report on designing multifunctional electrospun nanofibers based on the inclusion complex of PCL-α-cyclodextrin (PCL-α-CD), which provides both structural support and multiple functionalities for further conjugation of bioactive components. This strategy is independent of any chemical modification of the PCL main chain, and electrospinning of PCL-α-CD is as easy as electrospinning PCL. Here, we describe synthesis of the PCL-α-CD electrospun nanofibers, elucidate composition and structure, and demonstrate the utility of functional groups on the fibers by conjugating a fluorescent small molecule and a polymeric-nanobead to the nanofibers. Furthermore, we demonstrate the application of PCL-α-CD nanofibers for promoting osteogenic differentiation of human adipose-derived stem cells (hADSCs), which induced a higher level of expression of osteogenic markers and enhanced production of extracellular matrix (ECM) proteins or molecules compared with control PCL fibers.
Introduction
A scaffold plays important roles in dictating cell functions and manipulating tissue development by providing structural support and biophysical and biochemical signals, and transporting nutrients and wastes.Citation1-Citation4 An ideal scaffold should have well-defined morphology, tunable degradation rate, sufficient mechanical strength for its intended application and a porous structure that has properties similar to those of the native extracellular matrix (ECM).Citation1-Citation4 In this context, scaffolds based on electrospun nanofibers have been studied for tissue engineering applications.Citation5-Citation8 These nano- and micro-scale fibers have mechanical strength similar to that of natural tissues and resemble the scale and arrangement of fibrous ECM components, in particular, collagen.Citation9,Citation10
The most widely employed electrospun nanofibrous scaffolds in tissue engineering and drug delivery are based on aliphatic polyesters, such as PCL or polylactide.Citation11 These materials have a number of useful properties, such as easy processing, biocompatibility and low cost; however, their biological applications are limited because they are hydrophobic and lack active natural cell recognition sites or functional groups along their polyester backbones.Citation10-Citation17 Therefore, depending upon the desired outcome, various polymer modification strategies, such as plasma treatment,Citation18-Citation20 coating,Citation19,Citation21 co-electrospinning with other polymers or bioactive componentsCitation22-Citation27 or chemically modifying the fibers with a strong reactant, such as sodium hydroxide, are employed. These treatments introduce functionalities,Citation28 improve hydrophilicityCitation29 and enhance cell function and tissue formation.Citation30,Citation31 Another important strategy for polyester functionalization is through copolymerizing polyester with functional monomers prior to polymerization; however, incorporating monomers makes it difficult to obtain high molecular weight polymers for fabricating tissue engineering nanofibrous scaffolds.Citation11,Citation32
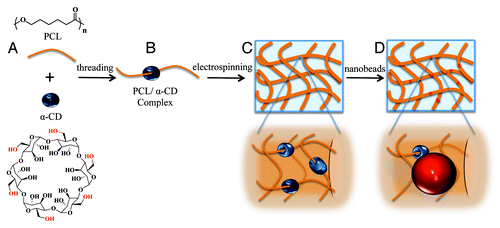
Here, we developed multifunctional electrospun nanofibers based on the inclusion complex (IC) of aliphatic polyester-α-cyclodextrin (e.g., PCL-α-CD)Citation33,Citation34 for tissue engineering applications (). α-CD is a six-member oligosaccharide doughnut ring structure with an inner cavity (diameter ~0.6 nm) and an outside diameter of ~1.4 nm.Citation35 α-CD rings physically thread onto the PCL chains via non-covalent interactions and resemble a molecular necklace structure ().Citation33-Citation35 α-CD bears hydroxyl groups that can be modified to create a variety of functionalities that also allow conjugation of multiple bioactive agents or ligands.Citation36-Citation40 Previously, we employed α-CDs to create a modular multifunctional hydrogel design for stem cell differentiation that can be modulated at the nanoscale level.Citation41 The versatility of this design enabled us to create precisely controlled 3D environments with independent mechanical, cell-adhesion and chemical properties. Here, we synthesized PCL-α-CD IC (), electrospun this material into nanofibers (), demonstrated the utility of functional groups on the nanofibers by conjugating a polymeric nanobead () and used the electrospun fiber as a scaffold for in vitro stem cell culture and differentiation for bone tissue formation. To our knowledge, this study demonstrates the first successful electrospinning of PCL-α-cyclodextrin nanofibers for creating tissue engineering scaffolds.
Results
Material characterization
PCL-α-CD IC was characterized for threading of α-CD on PCL chains by FTIR-ATR, WAXD and 1H NMR spectroscopy. FTIR-ATR screening of PCL-α-CD IC, PCL and α-CD showed three peaks at 1026 cm−1, 1079 cm−1 and 1158 cm−1 and confirmed the presence of α-CD. A distinct stretching band at 1735 cm−1 appeared as a result of the carbonyl bonds of PCL ().Citation42,Citation43 A broad band at 3382 cm−1 appeared because of the symmetric and antisymmetric OH stretching of α-CD in PCL-α-CD IC, which is absent in PCL.Citation43 Also, in contrast to the α-CD spectrum, a slight shift of the OH stretching band in the IC arose resulting from the formation of hydrogen bonds between α-CD and its guest polymer in the channel form.Citation38
Figure 2. WAXD spectra (A), FTIR-ATR spectra (B) and 1H-NMR spectra of α-CD, PCL and PCL-α-CD IC (C).
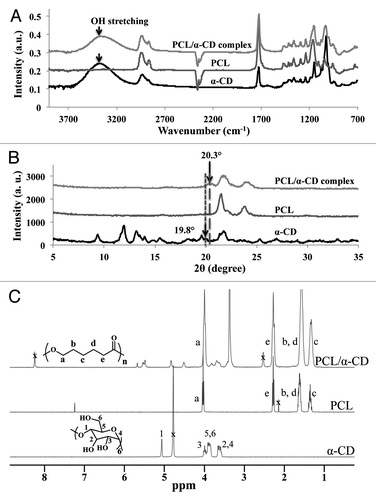
WAXD result showed that PCL exhibited two typical strong peak reflections at 2θ = 22° and 23.8°, while α-CD displayed a series of peaks at 9.9°, 12.2°, 14.5°, 19.8° and 21.9° as previously reported ().Citation44,Citation45 In PCL-α-CD IC, most crystalline diffraction peaks due to PCL disappeared, which indicated suppression of guest crystallization by formation of IC. New peaks at ~20° and ~22.5° appeared due to formation of IC.Citation46,Citation47 The molar ratio of the two components in PCL-α-CD IC was quantified by integration of resonances for the 1H NMR spectra of α-CD and PCL (shown in ).
Nanofiber synthesis, characterization and modification
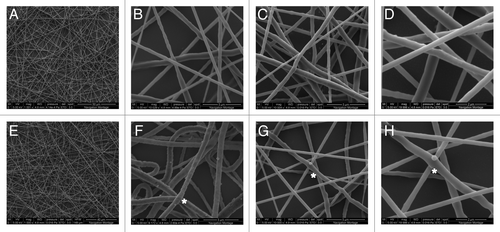
Unlike PCL alone, neither DMSO nor CH2Cl2 dissolved IC completely and, therefore, was unsuitable for electrospinning of the polymer solution. However, the polymer was successfully dissolved and electrospun in a mixture of DMSO/CH2Cl2 (3/2, v/v). These fibers were also tested for retention of threaded α-CD on PCL chains by utilizing the hydroxyl groups of α-CD on the surface for further chemical modifications and conjugations. First, both PCL and PCL-α-CD fibers were activated by CDI (); second, CDI-activated hydroxyl groups were modified to amine groups by reacting with a short length diamine (e.g., ethylenediamine) (). Subsequently, an amine-reactive fluorescent molecule, fluorescamine, was conjugated onto the fiber surfaces (). The fibers modified with fluorescamine turned blue under UV light exposure (). The hydroxyl groups on the nanofibers were also utilized to conjugate a structural component (amine-containing polystyrene nanobeads). SEM images at higher magnification showed no conjugated nanobeads on PCL fibers (); however, PCL-α-CD fibers were decorated with nanobeads via hydroxyl sites (). The CDI-untreated PCL or PCL-α-CD fibers did not conjugate to nanobeads.
Figure 3. (A) The hydroxyl groups of α-CD present in PCL-α-CD IC fibers can be conjugated with several biological or chemical moieties, including a fluorescent molecule. Step 1: Activation of α-CD with N,N’-carbonyldiimidazole (CDI) followed by its reaction with ethylenediamine. The hydroxyl groups are abundant and available for activation by N,N’- CDI in PCL-α-CD IC compared with only terminal hydroxyl groups of PCL. Step 2: Fluorescamine was conjugated to amine groups. (B) Optical microscope images of electrospun fibers of PCL before (i) and after fluorescamine labeling (ii); PCL/α-CD fibers before (iii) and after fluorescamine labeling (iv).
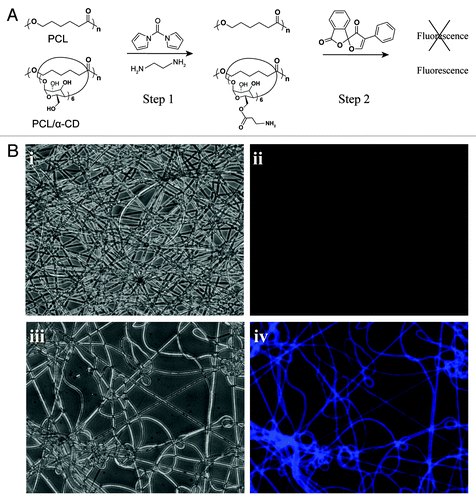
Figure 4. Electrospun fibers of PCL-10% (w/v) in CH2Cl2/DMSO (17/9, v/v) with magnification X1 (A), X10 (B) and (C), X20 (D); PCL-α-CD IC-10% (w/v) in CH2Cl2/DMSO (2/3 v/v); with magnification X1 (E), X10 (F) and (G), X20 (H). These fibers were chemically modified with N,N’-carbonyldiimidazole in acetonitrile followed by conjugation of amine functionalized polystyrene nanobeads (200 nm diameter size). *denotes beads
Cell response to PCL-α-CD nanofibers
hADSCs’ viability and spreading were studied over 3, 7, 14 and 21 d with LIVE/DEAD® (InvitrogenTM, Life Technologies) and F-actin staining. hADSCs attached to both PCL and PCL-α-CD fibers (), and exhibited an elongated fibroblast-like morphology after 3 d, which indicated a viable state. A continuous increase in cell number was visually observed for both fibers. After 21 d, LIVE/DEAD® staining determined the presence of 96.5 ± 2.5% and 97.1 ± 1.5% live cells for PCL and PCL-α-CD fibers, respectively.
Figure 5. hADSCs cultured on either PCL or PCL-α-CD fibers are stained for (A) LIVE/DEAD® Viability/Cytotoxicity (InvitrogenTM, Life Technologies, Grand Island, NY) using calcein AM, ethidium homodimer-1 and Hoechst dye; (B) F-actin using Texas Red-X® phalloidin, and Hoechst dye; (C) alizarin red for mineralization (calcium deposition) and ALP for alkaline phosphatase activity.
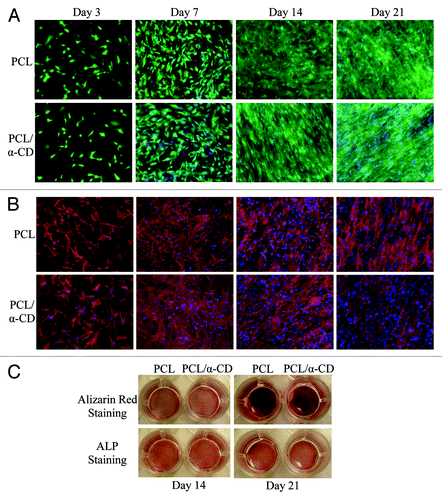
Positive staining with alizarin red and ALP () was observed on both fibers, which confirmed calcium deposition and mineralization. A substantial increase in the intensity of alizarin red staining was observed from day 14 to day 21 on both fibers, suggesting that by day 21, mineral deposition was greatly enhanced ().
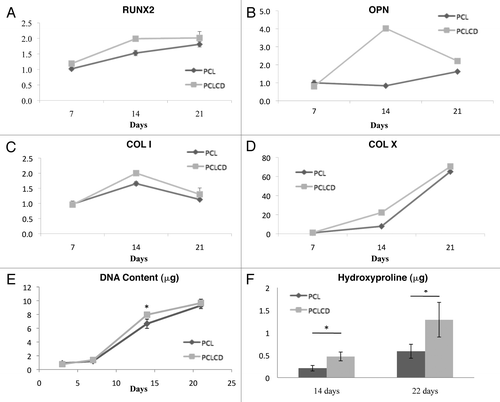
We then performed quantitative analysis of osteogenic gene-expression. Four osteogenic markers were selected for this study: osteogenesis transcription factor Runx2, and three bone collagen structural proteins: osteopontin, collagen type I and collagen type X. In general, PCL-α-CD fibers induced greater amounts of osteogenic gene expression compared with PCL fibers (). Similarly, relatively higher collagen deposition was obtained on PCL-α-CD fibers (). In summary, ADSCs proliferated at a similar rate on both types of fibers, while PCL-α-CD fibers enhanced osteogenesis.
Figure 6. The relative gene expression of some osteogenic markers during osteogenesis of hADSCs seeded on PCL and PCL-α-CD fibers, including RunX2 (A), osteopontin (B), collagen type I (C) and collagen type X (D); biochemical assays showing DNA content (E) and collagen deposition (F) on the fibers. Each gene expression level was compared with the expression level on day 7 for PCL samples. The DNA assay did not show any significant differences between the DNA values for PCL and PCL-α-CD samples during the time period of the experiment except on day 14; however, the collagen contents for these two samples were significantly different on both day 14 and day 21.
Discussion
Scaffolds based on aliphatic polyesters, such as PCL nanofibers, have been successfully used in biomedical applications, including stem cell culture and differentiation.Citation12,Citation15,Citation16,Citation48 However, depending on the desired cell function and tissue formation outcomes, aliphatic polyester nanofibers are often chemically modified with bioactive molecules and cell-recognizable ligands by mimicking natural ECM’s chemical and biological cues.
In this context, our reported PCL-α-CD-based electrospun nanofibrous scaffold has unique advantages: first, it is as easy to fabricate as PCL fibers; second, it has multiple functional sites for further conjugation and third, it is independent of the PCL-main chain modification as α-CD physically threads onto PCL chains. The ease of conjugation of various chemical and biological components to create user-specific unique cell environments without PCL modification, makes these nanofibers a powerful biomaterial tool for tissue engineering. For example, we showed the utility of the hydroxyl groups of the α-CD on the fiber surface by conjugating a fluorescent small molecule, fluorescamine, and a polystyrene nanobead ( and ). Similarly, we can conjugate various small molecules; cell-interactive peptides, such as the cell-binding peptide Arg-Gly-Asp (RGD) and other biological components to improve cell-binding capability of the nanofibers and provide necessary chemical and biological signals for cell functions. A previously reported strategy to improve cell adhesion on PCL nanofibers is co-electrospinning PCL with naturally derived materials, including gelatin or mineralized ECM.Citation19,Citation26 Another potential application of PCL-α-CD nanofibers can be for the controlled release of biological components from the fiber surface.
For desirable cellular functions, tissue engineering often needs to deliver bioactive agents via the scaffold. These components are incorporated into PCL fibers and can enhance desirable cell functions upon the agents’ leakage and release from the fibers. For example, Martins et al. incorporated dexamethasone, an osteogenesis inducer, into PCL fibers to enhance osteogenic differentiation of mesenchymal stem cells (MSCs).Citation48 However, they observed a burst of dexamethasone released as a result of uncontrolled surface erosion, as water penetrated the fiber surface first, leading to sudden drug release.Citation48 Bioactive components could be conjugated to our PCL-α-CD fiber via external-stimulus-sensitive bonds through functionalized CDs, such as hydrolyzable ester or photocleavable bonds.Citation49 This would allow the drug to have a greater sustained-release time profile, which is highly desirable in a scaffold design for controlled drug release.
We also investigated application of PCL-α-CD nanofibers as a 2D substrate for cell growth and osteogenic differentiation potential of hADSCs. Recently, there has been much attention focused on hADSCs because of their biological similarity to hBM-MSCs (human bone marrow-MSCs), ease of isolation through abundant and readily accessible adipose tissue, replication capability and multi-lineage differentiation potential. This makes hADSCs a promising source of adult stem cells for bone tissue engineering applications.Citation50-Citation54
We hypothesized that PCL-α-CD nanofibers can be employed as a scaffold for osteogenic differentiation of hADSCs, as can PCL nanofibers. Therefore, we cultured hADSCs onto 2D substrates of PCL and PCL-α-CD nanofibers in osteogenic media. Morphologically, cells were fully extended and elongated at early time points, indicating cell viability and adhesion, as shown in . By three weeks, hADSCs appeared to be completely integrated into the structure of the fibers.
We monitored the extent of osteogenic differentiation of hADSCs on nanofibers by gross-images of positive staining for calcium mineralization and alkaline phosphatase (ALP) activity (). ALP is an enzyme responsible for dephosphorylation of phosphates and initiating mineralization of ECM, which induces matrix mineralization by restricting matrix nucleation inhibitors.Citation55 However, ECM mineralization occurs in the later stage of osteogenic differentiation and requires long-term culture before any measurable matrix production occurs.Citation56 As such, ALP is regarded as an early-stage marker in osteogenesis, and its turning to plateau from upregulation is considered a signal for the initiation of mineralization.Citation57 We observed a substantial increase in the intensity of alizarin red staining from day 14 to day 21 on both fibers, suggesting that by day 21, mineral deposition was greatly enhanced (). However, ALP staining did not show much visual difference between PCL and PCL-α-CD samples. This might be due to a possible plateauing of ALP generation at the mid-to-later stage of osteogenesis.
PCR studies showed that ADSCs seeded onto PCL-α-CD fibers exhibited equal or marginally higher relative expressions of osteogenesis markers than on PCL fibers, as shown in . The selected markers are critical transcription factors or proteins involved in osteogenesis. Runx2 is an important transcription factor during osteogenesis, while osteopontin, collagen type I and collagen type X are main structural proteins of collagens found in bone. As we observed in this study, their higher expressions with time indicated a greater tendency to differentiate into bone-related cell types. We also found that while the DNA content of the two samples remained the same at each time point (), collagen deposition on PCL-α-CD fibers was significantly higher than on PCL fibers (). An increase in DNA content over time indicated that cells proliferated equally well on both PCL-α-CD and PCL fibers (). Collagen is a major organic component of mineralized ECM, comprising ~90% of all the organic material in bone, and it serves as a template for mineral deposition.Citation58 Our result suggests that the PCL-α-CD fibers could enhance collagen production, making a relatively better substrate to induce bone formation; possibly due to a change in the substrate hydrophilicity via the hydroxyl groups of α-CD. Furthermore, the chemical composition of the nanofiber with α-CD played an important role in cell growth and differentiation, while fiber morphology or topography was unchanged. Earlier, we showed that functionalized α-CDs in PEG hydrogels could enhance tissue formation.Citation41 These findings support the concept that α-CDs promote stem cell differentiation into musculoskeletal tissues, regardless of the type of polymers used for creating an artificial environment in the form of hydrogels or nanofibrous scaffolds.
Materials and Methods
Synthesis of PCL-α-CD IC
PCL (1.0 g, Mw 70 k ~ 90 k Da; Sigma-Aldrich) was dissolved in acetone (60 mL) and heated at 50°C in a silicon oil bath. α-CD (0.5 g, Sigma-Aldrich) was dissolved in 10 mL of dimethylformamide (DMF) and added dropwise to the heated PCL-acetone solution. After stirring for 2 h, the mixture was air-cooled to room temperature. This solution was poured into a glass flat-bottom PYREX® container (Corning Inc. Life Sciences) and stirred slowly overnight at room temperature to evaporate the acetone. A thin layer of PCL-α-CD film was formed, which was soaked in and washed multiple times with water to remove any unthreaded α-CD. PCL-α-CD IC was further characterized by 1H-NMR (300 or 400 MHz; Bruker), wide-angle X-ray diffraction (WAXD) from 2θ = 5° to 35° (PANalytical MPD Pro Diffractometer, Cu-Kα radiation; PANalytical B.V.) and Fourier transform infrared-attenuated total reflectance (FTIR-ATR) (Bruker, Vector 22 with a Pike Miracle ATR attachment) spectroscopy within a range of wavenumber 700–3,800 cm−1. WAXD and FTIR-ATR were also performed on PCL only and α-CD only samples, as controls.
Electrospinning of PCL and PCL-α-CD IC nanofibers
PCL was dissolved in a mixture of dichloromethane (DCM) and dimethyl sulfoxide (DMSO) (17/9, v/v) at a concentration of 10% (w/v).Citation59 The solution was drawn into a 1 mL syringe (Norm-Ject, Henke-Sass Wolf GmbH) with a 30 G needle (Becton, Dickinson and Co.) and electrospun at 8 kV and 5 mL/h. PCL-α-CD was dissolved in a mixed solvent of DCM and DMSO (2/3, v/v) at a concentration of 10% (w/v) and filled into the same kind of syringe and needle. The PCL-α-CD fibers (605 ± 85 nm, n = 100 nm) were electrospun at 5.5 kV and 6 mL/h to obtain fibers with similar diameter to those of PCL (617 ± 170 nm, n = 100). Fibers were collected onto aluminum foil covered with 15 mm diameter microscope coverslips (Thermo Fisher Scientific), which were kept at a distance of 16 cm from the tip of the syringe needle. Electrospun fibers-covered coverslips were cut off from the aluminum foil and kept for further use. Before seeding with cells, fiber samples were put into 24-well plates and sterilized by overnight UV exposure.
Fluorescamine conjugation to PCL-α-CD IC fibers
PCL and PCL-α-CD fibers on microscope coverslips (dia ~15 mm) were soaked in DMSO containing N,N′-carbonyldiimidazole (N,N′-CDI; Sigma-Aldrich) at room temperature. Ethylenediamine (Sigma-Aldrich) was added to these fibers, and after 30 min of shaking, both fibers were taken out, washed with fresh DMSO and soaked in fluorescamine-DMSO solution. After subsequent washing with fresh DMSO and water, fluorescence images of two different samples were taken on a Nikon DXM1200 microscope under both bright field and UV light.
Polystyrene nanobeads conjugation to PCL-α-CD IC fibers
PCL and PCL-α-CD fibers were soaked in N,N′-CDI/DMSO solution while undergoing shaking. After 1 h, both fibers were taken out, washed with fresh DMSO and soaked in DMSO containing polystyrene nanobeads with amine functional groups (0.2 μm dia; Invitrogen™, Life Technologies). After shaking for ~4 h, fibers were washed with ethanol to remove any unconjugated nanobeads that had settled on the fiber surface. The fibers on coverslips were placed vertically in both DMSO and ethanol to avoid any gravitational settling or physical adsorption of beads on the fibers. These fibers were vacuum dried, sputter coated (Anatech Hummer 6.2) with platinum and characterized by SEM (FEI Quanta 200).
Cell culture on nanofibers
Human adipose-derived stem cells (hADSCs) were isolated as previously described,Citation60 received via a material transfer agreement, and expanded up to passage 4 before usage. For expansion, cells were cultured in a medium consisting of low glucose (1.0 g/L) DMEM supplemented with 876 mg/L of L-glutamine, 10% fetal bovine serum (FBS), 100,000 U/L penicillin, 10 mg/L streptomycin and 1 µg/L basic fibroblast growth factor (InvitrogenTM, Life Technologies). For osteogenic induction, cells were seeded onto nanofibers at a cell density of 5,000/cm2 in an osteogenic medium composed of high glucose (4.5 g/L) DMEM supplemented with 100,000 U/L penicillin, 10 mg/L streptomycin, 10% FBS, 50 µM ascorbic acid, 0.1 µM dexamethasone and 10 mM glycerol-2-phosphate disodium salt. Cells were harvested and analyzed on days 7, 14 and 21.
Gene expression
Cellular mRNAs were extracted as previously described by Strehin et al.Citation61 Briefly, the mRNA was extracted with 1 mL trizol per well and then precipitated, washed with isopropanol and 75% ethanol, and redissolved in diethylpyrocarbonate-treated water (DEPC-treated water). This solution of mRNA was incubated at 60°C for 10 min and quickly put on ice. The concentration of mRNA was quantified using a NanodropTM 2000 spectrophotometer (Thermo Scientific). The cDNA was synthesized according to the manufacturer’s protocol for the Superscript 1st Strand System Kit (InvitrogenTM, Life Technologies). The cDNA was used for real-time polymerase chain reaction (PCR) with SYBR® Green PCR Master Mix (Applied Biosystems, Life Technologies) using the primers shown in with β-actin as a reference gene. The level of expression was calculated using the Pfaffl method.Citation62
Table 1. Primer sequences for real-time PCR
Biochemical assays
Biochemical assays were performed using a revised version of the method described by Strehin et al.Citation61 Briefly, after aspirating off media, samples were rinsed thrice with PBS, removed from the 24-well plate and lyophilized. After measuring the dry weight of the samples, they were incubated overnight at 60°C in 500 µL papainase buffer, which contained 1 M Na2HPO4, 10 mM disodium EDTA.2H2O, 10 M l-cysteine and 9.3 units/mL papain type III (Worthington Biochemical Corp.). Supernatants were collected after centrifugation and used for DNA and collagen assays.
For DNA assays, 30 µL of sample digest was mixed with 3 mL of pH 7.4 DNA buffer solution, which contained 100 µg/mL Hoechst 33258, 10 mM Tris base, 200 mM NaCl and 1 mM disodium EDTA.2H2O. The mixture was then analyzed with a DyNA Quant 200 Fluorometer (Hoefer, Inc.), with an excitation/emission of 365/460 nm. The measurements were analyzed with a calibration curve using DNA solutions made with calf thymus DNA (InvitrogenTM, Life Technologies).
For collagen assays, 100 µL of papain digest was added to 100 µL of 37% (v/v) conc. HCl and the mixture was hydrolyzed at 115°C for 18 h. Samples were neutralized with aq. NaOH and the volume was brought up to 3.5 mL with deionized water. Added to this solution (1 mL) was 0.5 mL of chloramine-T solution [69 mM chloramine-T in 89% (v/v) pH 6 buffer and 11% (v/v) isopropanol]; it was maintained at room temperature for 20 min. The pH 6.0 buffer solution contained 0.57 M NaOH, 0.16 M citric acid monohydrate, 0.59 M sodium acetate trihydrate, 0.8% (v/v) glacial acetic acid, 20% (v/v) isopropanol, 79.2% (v/v) dd H2O and 5 drops of toluene. Added to this solution was 0.5 mL of 4-(dimethylamino)benzaldehyde (pDAB) [1.17 M pDAB in 70% (v/v) isopropanol, 30% (v/v) of 60% perchloric acid in water]; and it was incubated at 60°C for 30 min. After cooling to room temperature, the samples were analyzed for their absorbance at 557 nm using a DU500 UV-Vis spectrophotometer (Beckman Coulter, Inc.) and compared with a standard solution of hydroxyproline.
Live/dead staining
Cells seeded on both fibers were stained with the LIVE/DEAD® Viability/Cytotoxicity Kit (InvitrogenTM, Life Technologies) as per the manufacturer’s protocol. Briefly, DMEM supplemented with 4 µM calcein-AM, 4 µM ethidium homodimer-1 and 4 µM Hoechst 33258 was added to cells and incubated at 37°C and 5% CO2 for 30 min. After rinsing the samples thrice with PBS, fluorescent images were taken with a Zeiss Axio optical microscope (HXP 120 fluorescent illuminator) (Carl Zeiss Microscopy). ImageJ (US National Institutes of Health) was used to merge images for further analysis.
F-actin staining
Cells were fixed with 4% paraformaldehyde for 10 min and incubated with 0.1% Triton™ X-100 at room temperature for 5 min. Subsequently, cells were rinsed with PBS, before adding 2.5% (v/v) Texas Red-X® phalloidin (200 U/mL; InvitrogenTM, Life Technologies) solution containing 4 μM Hoechst 33258. After being maintained in the dark for 30 min, samples were rinsed thrice with PBS. Images of these stained cells were taken with a Zeiss Axio optical microscope, merged and analyzed using ImageJ.
Alizarin red staining
The samples were rinsed twice with PBS after carefully aspirating off media from each well. Cells were fixed with 4% paraformaldehyde at room temperature for 15 min and rinsed thrice with deionized water. Subsequently, 1 mL of 40 mM alizarin red S solution (pH 4.1) was added to each well. Dye was aspirated off after 20 min, and fibers were rinsed thrice with distilled water. Images of the wells were taken with an Olympus C-765 camera (Olympus America).
Alkaline phosphatase (ALP) staining
Cells were rinsed with Tyrode’s balanced salt solution (TBSS, Sigma-Aldrich) twice, and fixed with a citrate-buffer acetone solution for 30 sec. The citrate-buffer acetone solution was composed of a 60% (v/v) citrate working solution and 40% (v/v) acetone. The citrate working solution was made by adding 2 mL of citrate concentrated solution (Sigma-Aldrich) to 98 mL of water. Cells were rinsed twice with PBS after removing the salt solution. One mL of fast violet-naphthol solution was added to each well, and incubated in the dark for 45 min at room temperature. Fast violet-naphthol solution was made by adding 0.5 mL of naphthol AS-MX alkaline solution (Sigma-Aldrich) to 12 mL of fast violet solution, which was made by dissolving one capsule of fast violet (Sigma-Aldrich) in 48 mL of water. Images of the stained cells were taken with an Olympus C-765 camera.
Statistical analysis
Data are expressed as mean ± standard deviation. Statistical analysis was performed using SPSS v.19 (IBM Corp.). One-way ANOVA was performed among groups to determine any statistically significant differences in values of means. Samples with equal variances and sizes were analyzed using Tukey’s post-hoc test, while the Games-Howell post-hoc test was used for samples with unequal variances and unequal sample sizes. p ≤ 0.05 was considered statistically significant.
Conclusions
In this work, we successfully electrospun PCL-α-CD nanofibers and showed the utility of the α-CD hydroxyl groups functional sites through labeling fluorescent small molecule and polymeric nanobead attachments. We also demonstrated that PCL-α-CD nanofibers supported hADSCs viability, induced a higher level of expression of osteogenic markers and enhanced production of ECM proteins or molecules, compared with control PCL fibers. We believe that by using our design approach, scaffolds based on the electrospun nanofibers with various functional groups, such as amine or carboxylic acid on the polymer chains of functionalized α-CDs, can easily be created to modulate material properties. Furthermore, cell-sensitive biological moieties can also be conjugated to and immobilized on the fibers through these functional sites.
Acknowledgments
This research was funded by the Maryland Stem Cells Research Foundation Postdoctoral fellowship to A.S., the National Science Foundation (CMMI 0948053), and NIH’s National Institute of Dental and Craniofacial Research (Grant No. R01DE016887).
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Author Contributions
A.S. and J.Z. conceived the project and experiments; J.Z., A.S., Z.Z. and L.H. designed and performed the experiments; and A.S., J.Z. and J.H.E. wrote the manuscript.
References
- Pan Z, Ding J. Poly(lactide-co-glycolide) porous scaffolds for tissue engineering and regenerative medicine. Interface Focus 2012; 2:366 - 77; http://dx.doi.org/10.1098/rsfs.2011.0123
- Drury JL, Mooney DJ. Hydrogels for tissue engineering: scaffold design variables and applications. Biomaterials 2003; 24:4337 - 51; http://dx.doi.org/10.1016/S0142-9612(03)00340-5; PMID: 12922147
- Hollister SJ. Porous scaffold design for tissue engineering. Nat Mater 2005; 4:518 - 24; http://dx.doi.org/10.1038/nmat1421; PMID: 16003400
- Chen GP, Ushida T, Tateishi T. Scaffold design for tissue engineering. Macromol Biosci 2002; 2:67 - 77; http://dx.doi.org/10.1002/1616-5195(20020201)2:2<67::AID-MABI67>3.0.CO;2-F
- Griffith LG, Naughton G. Tissue engineering--current challenges and expanding opportunities. Science 2002; 295:1009 - 14; http://dx.doi.org/10.1126/science.1069210; PMID: 11834815
- Li WJ, Laurencin CT, Caterson EJ, Tuan RS, Ko FK. Electrospun nanofibrous structure: a novel scaffold for tissue engineering. J Biomed Mater Res 2002; 60:613 - 21; http://dx.doi.org/10.1002/jbm.10167; PMID: 11948520
- Yoshimoto H, Shin YM, Terai H, Vacanti JP. A biodegradable nanofiber scaffold by electrospinning and its potential for bone tissue engineering. Biomaterials 2003; 24:2077 - 82; http://dx.doi.org/10.1016/S0142-9612(02)00635-X; PMID: 12628828
- Ma ZW, Kotaki M, Inai R, Ramakrishna S. Potential of nanofiber matrix as tissue-engineering scaffolds. Tissue Eng 2005; 11:101 - 9; http://dx.doi.org/10.1089/ten.2005.11.101; PMID: 15738665
- Barnes CP, Sell SA, Boland ED, Simpson DG, Bowlin GL. Nanofiber technology: designing the next generation of tissue engineering scaffolds. Adv Drug Deliv Rev 2007; 59:1413 - 33; http://dx.doi.org/10.1016/j.addr.2007.04.022; PMID: 17916396
- Sill TJ, von Recum HA. Electrospinning: applications in drug delivery and tissue engineering. Biomaterials 2008; 29:1989 - 2006; http://dx.doi.org/10.1016/j.biomaterials.2008.01.011; PMID: 18281090
- Jiao Y-P, Cui F-Z. Surface modification of polyester biomaterials for tissue engineering. Biomed Mater 2007; 2:R24 - 37; http://dx.doi.org/10.1088/1748-6041/2/4/R02; PMID: 18458475
- Lim SH, Mao HQ. Electrospun scaffolds for stem cell engineering. Adv Drug Deliv Rev 2009; 61:1084 - 96; http://dx.doi.org/10.1016/j.addr.2009.07.011; PMID: 19647024
- Liang D, Hsiao BS, Chu B. Functional electrospun nanofibrous scaffolds for biomedical applications. Adv Drug Deliv Rev 2007; 59:1392 - 412; http://dx.doi.org/10.1016/j.addr.2007.04.021; PMID: 17884240
- Pham QP, Sharma U, Mikos AG. Electrospinning of polymeric nanofibers for tissue engineering applications: a review. Tissue Eng 2006; 12:1197 - 211; http://dx.doi.org/10.1089/ten.2006.12.1197; PMID: 16771634
- Li WJ, Tuli R, Huang X, Laquerriere P, Tuan RS. Multilineage differentiation of human mesenchymal stem cells in a three-dimensional nanofibrous scaffold. Biomaterials 2005; 26:5158 - 66; http://dx.doi.org/10.1016/j.biomaterials.2005.01.002; PMID: 15792543
- Li WJ, Tuli R, Okafor C, Derfoul A, Danielson KG, Hall DJ, et al. A three-dimensional nanofibrous scaffold for cartilage tissue engineering using human mesenchymal stem cells. Biomaterials 2005; 26:599 - 609; http://dx.doi.org/10.1016/j.biomaterials.2004.03.005; PMID: 15282138
- Woodruff MA, Hutmacher DW. The return of a forgotten polymer-Polycaprolactone in the 21st century. Prog Polym Sci 2010; 35:1217 - 56; http://dx.doi.org/10.1016/j.progpolymsci.2010.04.002
- Prabhakaran MP, Venugopal J, Chan CK, Ramakrishna S. Surface modified electrospun nanofibrous scaffolds for nerve tissue engineering. Nanotechnology 2008; 19:455102; http://dx.doi.org/10.1088/0957-4484/19/45/455102; PMID: 21832761
- Ma ZW, He W, Yong T, Ramakrishna S. Grafting of gelatin on electrospun poly(caprolactone) nanofibers to improve endothelial cell spreading and proliferation and to control cell Orientation. Tissue Eng 2005; 11:1149 - 58; http://dx.doi.org/10.1089/ten.2005.11.1149; PMID: 16144451
- Martins AE, Pinho ED, Faria S, Pashkuleva I, Marques AP, Reis RL, et al. Surface modification of electrospun polycaprolactone nanofiber meshes by plasma treatment to enhance biological performance. Small 2009; 5:1195 - 206; PMID: 19242938
- Araujo JV, Martins A, Leonor IB. Pinho Ed, Reis RL, Neves NM. Surface controlled biomimetic coating of polycaprolactone nanofiber meshes to be used as bone extracellular matrix analogues. J Biomat Sci-Polym Ed 2008; 19:1261-78.
- Prosecká E, Buzgo M, Rampichová M, Kocourek T, Kochová P, Vysloužilová L, et al. Thin-layer hydroxyapatite deposition on a nanofiber surface stimulates mesenchymal stem cell proliferation and their differentiation into osteoblasts. J Biomed Biotechnol 2012; 2012:428503; http://dx.doi.org/10.1155/2012/428503; PMID: 22319242
- Zhang YZ, Venugopal J, Huang ZM, Lim CT, Ramakrishna S. Characterization of the surface biocompatibility of the electrospun PCL-collagen nanofibers using fibroblasts. Biomacromolecules 2005; 6:2583 - 9; http://dx.doi.org/10.1021/bm050314k; PMID: 16153095
- Heydarkhan-Hagvall S, Schenke-Layland K, Dhanasopon AP, Rofail F, Smith H, Wu BM, et al. Three-dimensional electrospun ECM-based hybrid scaffolds for cardiovascular tissue engineering. Biomaterials 2008; 29:2907 - 14; http://dx.doi.org/10.1016/j.biomaterials.2008.03.034; PMID: 18403012
- Ghasemi-Mobarakeh L, Prabhakaran MP, Morshed M, Nasr-Esfahani MH, Ramakrishna S. Electrospun poly(epsilon-caprolactone)/gelatin nanofibrous scaffolds for nerve tissue engineering. Biomaterials 2008; 29:4532 - 9; http://dx.doi.org/10.1016/j.biomaterials.2008.08.007; PMID: 18757094
- Jo JH, Lee EJ, Shin DS, Kim HE, Kim HW, Koh YH, et al. In vitro/in vivo biocompatibility and mechanical properties of bioactive glass nanofiber and poly(epsilon-caprolactone) composite materials. J Biomed Mater Res B Appl Biomater 2009; 91:213 - 20; http://dx.doi.org/10.1002/jbm.b.31392; PMID: 19422050
- Chong EJ, Phan TT, Lim IJ, Zhang YZ, Bay BH, Ramakrishna S, et al. Evaluation of electrospun PCL/gelatin nanofibrous scaffold for wound healing and layered dermal reconstitution. Acta Biomater 2007; 3:321 - 30; http://dx.doi.org/10.1016/j.actbio.2007.01.002; PMID: 17321811
- Lam CX, Hutmacher DW, Schantz JT, Woodruff MA, Teoh SH. Evaluation of polycaprolactone scaffold degradation for 6 months in vitro and in vivo. J Biomed Mater Res A 2009; 90:906 - 19; http://dx.doi.org/10.1002/jbm.a.32052; PMID: 18646204
- Ma Z, Mao Z, Gao C. Surface modification and property analysis of biomedical polymers used for tissue engineering. Colloids Surf B Biointerfaces 2007; 60:137 - 57; http://dx.doi.org/10.1016/j.colsurfb.2007.06.019; PMID: 17683921
- Place ES, George JH, Williams CK, Stevens MM. Synthetic polymer scaffolds for tissue engineering. Chem Soc Rev 2009; 38:1139 - 51; http://dx.doi.org/10.1039/b811392k; PMID: 19421585
- Cipitria A, Skelton A, Dargaville TR, Dalton PD, Hutmacher DW. Design, fabrication and characterization of PCL electrospun scaffolds-a review. J Mater Chem 2011; 21:9419 - 53; http://dx.doi.org/10.1039/c0jm04502k
- Seyednejad H, Ghassemi AH, van Nostrum CF, Vermonden T, Hennink WE. Functional aliphatic polyesters for biomedical and pharmaceutical applications. 2011; 152:168 - 76
- Kawaguchi Y, Nishiyama T, Okada M, Kamachi M, Harada A. Complex formation of poly(epsilon-caprolactone) with cyclodextrins. Macromolecules 2002; 33:4472 - 7; http://dx.doi.org/10.1021/ma992103b
- Shin KM, Dong T, He Y, Taguchi Y, Oishi A, Nishida H, et al. Inclusion complex formation between alpha-cyclodextrin and biodegradable aliphatic polyesters. Macromol Biosci 2004; 4:1075 - 83; http://dx.doi.org/10.1002/mabi.200400118; PMID: 15586392
- van de Manakker F, Vermonden T, van Nostrum CF, Hennink WE. Cyclodextrin-based polymeric materials: synthesis, properties, and pharmaceutical/biomedical applications. Biomacromolecules 2009; 10:3157 - 75; http://dx.doi.org/10.1021/bm901065f; PMID: 19921854
- Khan AR, Forgo P, Stine KJ, D’Souza VT. Methods for selective modifications of cyclodextrins. Chem Rev 1998; 98:1977 - 96; http://dx.doi.org/10.1021/cr970012b; PMID: 11848955
- Li J, Loh XJ. Cyclodextrin-based supramolecular architectures: syntheses, structures, and applications for drug and gene delivery. Adv Drug Deliv Rev 2008; 60:1000 - 17; http://dx.doi.org/10.1016/j.addr.2008.02.011; PMID: 18413280
- Harada A, Takashima Y, Yamaguchi H. Cyclodextrin-based supramolecular polymers. Chem Soc Rev 2009; 38:875 - 82; http://dx.doi.org/10.1039/b705458k; PMID: 19421567
- Haiqing D, Yongyong L, Lan L, Donglu S. Cyclodextrins/polymer based (pseudo) polyrotaxanes for biomedical applications. Prog Chem 2011; 23:914 - 22
- Yui N, Katoono R, Yamshita A. Functional cyclodextrin polyrotaxanes for drug delivery. Inclusion Polymers 2009; 222:55 - 77; http://dx.doi.org/10.1007/12_2008_8
- Singh A, Zhan J, Zhaoyang Y, Elisseeff JH. Modular multifunctional poly(ethylene glycol) hydrogels for stem cell differentiation. Adv Func Mat 2012; 10.1002/adfm.201201902.
- Nelles G, Weisser M, Back R, Wohlfart P, Wenz G, Neher SM. Controlled orientation of cyclodextrin derivatives immobilized on gold surfaces. J Am Chem Soc 1996; 118:5039 - 46; http://dx.doi.org/10.1021/ja9539812
- Wenz G. Cyclodextrins as building-blocks for supramolecular structures and functional units. Angew Chem Int Ed Engl 1994; 33:803 - 22; http://dx.doi.org/10.1002/anie.199408031
- Mori T, Dong T, Yazawa K, Inoue Y. Preparation of highly transparent and thermally stable films of alpha-cyclodextrin/polymer inclusion complexes. Macromol Rapid Commun 2007; 28:2095 - 9; http://dx.doi.org/10.1002/marc.200700502
- Shuai XT, Wei M, Porbeni FE, Bullions TA, Tonelli AE. Formation of and coalescence from the inclusion complex of a biodegradable block copolymer and alpha-cyclodextrin. 2: A novel way to regulate the biodegradation behavior of biodegradable block copolymers. Biomacromolecules 2002; 3:201 - 7; http://dx.doi.org/10.1021/bm015609m; PMID: 11866574
- Huang L, Allen E, Tonelli AE. Study of the inclusion compounds formed between alpha-cyclodextrin and high molecular weight poly(ethylene oxide) and poly(epsilon-caprolactone). Polymer (Guildf) 1998; 39:4857 - 65; http://dx.doi.org/10.1016/S0032-3861(97)00568-5
- Harada AS, Suzuki S, Okada M, Kamachi M. Preparation and characterization of inclusion complexes of polyisobutylene with cyclodextrins. Macromolecules 1996; 29:5611 - 4; http://dx.doi.org/10.1021/ma960428b
- Martins A, Duarte AR, Faria S, Marques AP, Reis RL, Neves NM. Osteogenic induction of hBMSCs by electrospun scaffolds with dexamethasone release functionality. Biomaterials 2010; 31:5875 - 85; http://dx.doi.org/10.1016/j.biomaterials.2010.04.010; PMID: 20452016
- Deans TL, Singh A, Gibson M, Elisseeff JH. Regulating synthetic gene networks in 3D materials. Proc Natl Acad Sci U S A 2012; 109:15217 - 22; http://dx.doi.org/10.1073/pnas.1204705109; PMID: 22927376
- Lin Y, Wang T, Wu L, Jing W, Chen X, Li Z, et al. Ectopic and in situ bone formation of adipose tissue-derived stromal cells in biphasic calcium phosphate nanocomposite. J Biomed Mater Res A 2007; 81:900 - 10; http://dx.doi.org/10.1002/jbm.a.31149; PMID: 17236222
- Liu Q, Cen L, Yin S, Chen L, Liu G, Chang J, et al. A comparative study of proliferation and osteogenic differentiation of adipose-derived stem cells on akermanite and beta-TCP ceramics. Biomaterials 2008; 29:4792 - 9; http://dx.doi.org/10.1016/j.biomaterials.2008.08.039; PMID: 18823660
- Halvorsen YD, Franklin D, Bond AL, Hitt DC, Auchter C, Boskey AL, et al. Extracellular matrix mineralization and osteoblast gene expression by human adipose tissue-derived stromal cells. Tissue Eng 2001; 7:729 - 41; http://dx.doi.org/10.1089/107632701753337681; PMID: 11749730
- Gimble JM, Katz AJ, Bunnell BA. Adipose-derived stem cells for regenerative medicine. Circ Res 2007; 100:1249 - 60; http://dx.doi.org/10.1161/01.RES.0000265074.83288.09; PMID: 17495232
- Schäffler A, Büchler C. Concise review: adipose tissue-derived stromal cells--basic and clinical implications for novel cell-based therapies. Stem Cells 2007; 25:818 - 27; http://dx.doi.org/10.1634/stemcells.2006-0589; PMID: 17420225
- Liao J, Guo X, Nelson D, Kasper FK, Mikos AG. Modulation of osteogenic properties of biodegradable polymer/extracellular matrix scaffolds generated with a flow perfusion bioreactor. Acta Biomater 2010; 6:2386 - 93; http://dx.doi.org/10.1016/j.actbio.2010.01.011; PMID: 20080214
- McCullen SD, Zhu Y, Bernacki SH, Narayan RJ, Pourdeyhimi B, Gorga RE, et al. Electrospun composite poly(L-lactic acid)/tricalcium phosphate scaffolds induce proliferation and osteogenic differentiation of human adipose-derived stem cells. Biomed Mater 2009; 4:035002; http://dx.doi.org/10.1088/1748-6041/4/3/035002; PMID: 19390143
- Takagishi Y, Kawakami T, Hara Y, Shinkai M, Takezawa T, Nagamune T. Bone-like tissue formation by three-dimensional culture of MG63 osteosarcoma cells in gelatin hydrogels using calcium-enriched medium. Tissue Eng 2006; 12:927 - 37; http://dx.doi.org/10.1089/ten.2006.12.927; PMID: 16674304
- Jang JH, Castano O, Kim HW. Electrospun materials as potential platforms for bone tissue engineering. Adv Drug Deliv Rev 2009; 61:1065 - 83; http://dx.doi.org/10.1016/j.addr.2009.07.008; PMID: 19646493
- Luong-Van E, Grøndahl L, Chua KN, Leong KW, Nurcombe V, Cool SM. Controlled release of heparin from poly(epsilon-caprolactone) electrospun fibers. Biomaterials 2006; 27:2042 - 50; http://dx.doi.org/10.1016/j.biomaterials.2005.10.028; PMID: 16305806
- Mitchell JB, McIntosh K, Zvonic S, Garrett S, Floyd ZE, Kloster A, et al. Immunophenotype of human adipose-derived cells: temporal changes in stromal-associated and stem cell-associated markers. Stem Cells 2006; 24:376 - 85; http://dx.doi.org/10.1634/stemcells.2005-0234; PMID: 16322640
- Strehin IA, Elisseeff JH. Characterizing ECM production by cells encapsulated in hydrogels. Methods Mol Biol 2009; 522:349 - 62; http://dx.doi.org/10.1007/978-1-59745-413-1_23; PMID: 19247606
- Pfaffl MW. A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Res 2001; 29:e45; http://dx.doi.org/10.1093/nar/29.9.e45; PMID: 11328886