Keywords: :
Inhibitor of DNA-binding (Id) proteins comprise a family of proteins that heterodimerizes with basic helix-loop-helix (bHLH) transcription factors to inhibit their binding to DNA.Citation1 Id proteins also contain the HLH-dimerization domain but lack the basic DNA-binding domain. They are thereby considered dominant negative regulators of bHLH transcription factors. Their main protein partners are the bHLH transcriptional activators known as E proteins. These proteins form transcriptionally active complexes with tissue specific bHLH activators.Citation2 Four vertebrate Id gene products have been identified (Id-1 to Id-4), which are closely related in their HLH regions and show similar affinities for the various E proteins.Citation3 However, they differ in their expression patterns.Citation4 Id-1 represents the most studied Id protein in neural stem cells (NSC) biology. Indeed, it is intimately involved both in brain development and cancer, preventing cell differentiation while inducing cell proliferation.Citation3,Citation4
Using in vitro experiments of specific knockdown, we sought to investigate the role of Id-1, the most studied Id protein in brain cancer and development,Citation4 in the regulation of transcriptional programs involved in cell adhesion processes of neural stem cells (NSC).
We found a previously undisclosed link between Id-1 and Rap1 (Ras-related protein 1), a protein that belongs to the Ras family of small G-proteins, widely recognized as a central regulator of cell adhesion.Citation5 Indeed, we found a new bHLH target: Rap1GAP (GTPase-activating protein 1), a protein that is able to specifically block Rap1-mediated signaling by converting active Rap1-GTP to inactive Rap1-GDP.Citation6
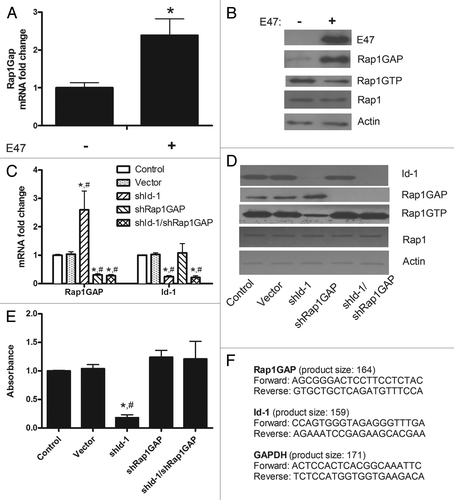
We first demonstrated that in presence of a common bHLH protein, E47, a known interactor of Id-1,Citation1,Citation2,Citation4 there is a significant increase in Rap1GAP mRNA () and protein () levels in primary murine neural stem cells (mNSC). These mNSC were isolated from E13.5 mouse telencephalon and cultured in the presence of EGF and FGF-2 (20 ng/ml each), as previously described.Citation2,Citation7,Citation8
Figure 1. (A) In primary cultured mNSC, Rap1GAP mRNA levels increase in presence of purified E47 protein. The expression levels were analyzed by real time RT-PCR (Applied Biosystems). The average of three independent analyses for each gene and sample was calculated and was normalized to the endogenous reference control gene GAPDH. Data are expressed as mean ± SE; *, p < 0.05, ANOVA. (B) E47 induces also an increase in Rap1GAP protein levels, assessed by a specific monoclonal antibody (Abcam), accompanied by a decrease in Rap1 activity, which was assayed using a pull-down assay (Thermo Fisher Scientific) in which isolation of Rap1-GTP was followed by western blotting and probing with an anti-Rap1 antibody (Thermo Fisher Scientific), according to the manufacturer’s instructions. Actin was used as loading control. Standard methods were used for immunoblotting. The blots shown are representative of three experiments. (C) Rap1GAP and Id-1 mRNA levels were assessed in mNSC as described in panel (A) after specific knockdown of Id-1, Rap1GAP or both (data are expressed as mean ± SE; *p < 0.01 vs. control; #, p < 0.01 vs. vector, ANOVA). (D) The expression levels of Id-1 and Rap1GAP and Rap1 activity were evaluated in mNSC after silencing of Id-1, Rap1GAP or both. Actin was used as loading control. Representative blots (performed in triplicate) are shown. (E) The cell adhesion assay was performed in quadruplicate using a commercially available kit (ScienCellTM) according to the manufacturer’s instructions (data are expressed as mean ± SE; *, p < 0.01 vs. control; #, p < 0.01 vs. vector, ANOVA). (C–E) Lentiviral vector-expressed shRNA sequences were CATGAACGGCTGTTACTCA for Id-1 and TTGGTGTGTGAAGACGTCA for Rap1GAP. (F) Sequences (5′-3′) and product length of the primers used to perform real time RT-PCR of Rap1GAP, Id-1 and GAPDH.
Subsequently, we applied a specific gene knockdown technique to silence Id-1 and/or Rap1GAP, using viral-mediated RNA interference, as described.Citation3 Briefly,shRNAs targeting murine Id-1 and Rap1GAP were designed, screened and cloned directly into a U6 promoter-containing vector. The entire U6 promoter-shRNA cassette was transferred into lentiviral vectors by gateway clonase recombination reaction (Life Technologies Ltd.).
Interestingly, the efficient Id-1 silencing induced a significant increase of mRNA () and protein () levels of Rap1GAP compared with control and to vector, paralleled by a decrease in Rap1 activity (). Furthermore, Rap1GAP knockdown caused an increase in Rap1 activity (), which was no more present after the contemporary silencing of Id-1 and Rap1GAP ().
Our findings show that Id-1 is able to repress the bHLH-mediated activation of Rap1GAP, thus serving to maintain the GTPase activity of Rap1. To further investigate the consequences mediated by this new molecular link on cell adhesion, we performed a specific adhesion assay on mNSC. We found that Id-1 silencing induced a significant decrease in cell adhesion ()
In the present study, we discovered that Rap1GAP represents a new downstream target engaged by Id-1 in mNSC. Indeed, Id-1 is able to repress the bHLH-mediated activation of Rap1GAP, serving thereby to maintain the GTPase activity of Rap1, a key mediator of cell adhesion. Preventing the elevation of Rap1GAP efficiently countered the consequences of Id-1 knockdown. Given the recognized importance of cell adhesion in brain development and cancer, our results open new fields of investigation in both these areas, especially concerning the study of NSC function and the potential future strategies to treat brain tumors.Citation9
References
- Bhattacharya A, et al. Cell 2011; 147:881 - 92; http://dx.doi.org/10.1016/j.cell.2011.08.055; PMID: 22078884
- Jung S, et al. Stem Cells Dev 2010; 19:831 - 41; http://dx.doi.org/10.1089/scd.2009.0093; PMID: 19757990
- Hirai S, et al. EMBO J 2012; 31:1190 - 202; http://dx.doi.org/10.1038/emboj.2011.486; PMID: 22234186
- Manthey C, et al. BMC Cell Biol 2010; 11:2; http://dx.doi.org/10.1186/1471-2121-11-2; PMID: 20070914
- Gloerich M, et al. Trends Cell Biol 2011; 21:615 - 23; http://dx.doi.org/10.1016/j.tcb.2011.07.001; PMID: 21820312
- Vuchak LA, et al. Cell Adhes Migr 2011; 5:323 - 31; http://dx.doi.org/10.4161/cam.5.4.17041; PMID: 21785277
- Park JH, et al. Neurosci Lett 2012; 506:50 - 4; http://dx.doi.org/10.1016/j.neulet.2011.10.046; PMID: 22044874
- Sirko S, et al. Stem Cells 2010; 28:775 - 87; http://dx.doi.org/10.1002/stem.309; PMID: 20087964
- Anido J, et al. Cancer Cell 2010; 18:655 - 68; http://dx.doi.org/10.1016/j.ccr.2010.10.023; PMID: 21156287