Abstract
Purpose: Phage display was used to select novel peptides that specifically bind the TAG-72 antigen and with properties suitable for imaging TAG-72 positive cancers. Results: After three rounds of selection against TAG-72 and using two different elution conditions including a long elution, the consensus sequences FRERCDKHPQKCTKFL and DPRHCQKRVLPCPAWL were expressed on phages G3-15 and T3-15 respectively. ELISA, fluorescence-activated cell sorting analysis and fluorescence microscopy provided evidence that both phages specifically bound TAG-72 in vitro. Both peptides are stable in 37oC serum. By a cell binding competition assay, the IC50 for T3-15 was measured as 0.29 nM and therefore 36-fold higher affinity than G3-15 at 10.32 nM. The biodistribution in mice carrying LS-174T tumors in one thigh were similar for both 99mTc-peptides at 30 min, but at 90 min the 99mTc-T3-15 peptide accumulated almost three times higher in the tumor. The SPECT/CT images were consistent with the biodistribution results. Procedures: The f88-4/Cys6 phage library and two different elution conditions were used to identify two new higher affinity binding peptides for the TAG-72 antigen. One, was a single brief elution with pH 2.2 glycine buffer, and the second began with the glycine elution but was followed with a longer elution with Tris buffered saline (TBS) at pH 7.4. The phages that bound TAG-72 were evaluated by fluorescence-activated cell sorting analysis using TAG-72 positive LS-174T cells and confirmed by immunofluorescence imaging. The consensus peptides displayed on the selected phages were synthesized and conjugated with NHS-MAG3 for radiolabeling with 99mTc. The IC50 for TAG-72 binding was evaluated by cell binding competition in vitro while binding affinity was evaluated in vivo by necropsy and SPECT/CT imaging in a tumor mouse model. Conclusion: We have identified a peptide with a sub nanomolar inhibition constant for the TAG-72 antigen that may have application in cancer imaging.
Introduction
The tumor-associated mucin-like glycoprotein TAG-72 is overexpressed in most epithelial-derived cancers, such as colonic adenocarcinomas, invasive ductal carcinomas of the breast, non-small cell lung carcinomas, common epithelial ovarian carcinomas and pancreatic, gastric and esophageal cancers, but is seldom observed in normal tissues. Thus TAG-72 has often been a target for the diagnosis and therapy of a variety human cancersCitation1,Citation2 using anti-TAG-72 monoclonal antibodies such as B72.3 and CC49.Citation3–Citation5 However, the drawbacks to using intact antibodies that include slow pharmacokinetics and the generation of human anti-mouse antibody (HAMA) immune response in patients are well known.Citation6–Citation8 To overcome some of these problems antibody fragments with lower molecular weight have been successfully engineered by selection from combinatorial libraries to enhance their therapeutic potential.Citation9–Citation11 Compared with monoclonal antibodies and engineered antibody fragments, antitumor peptides are even smaller, which potentially permits them to access tumors more easily and to clear more rapidly from non-target tissues.Citation12,Citation13
Combinatorial phage display peptide libraries have been used widely and successfully to identify various high affinity binding peptides against tumor cells.Citation14–Citation16 Tumor antigen, intact tumor cells, or even tissues in vivo can be used as targets for the selection process.Citation17,Citation18 Several peptides selected from phage libraries are already in clinical trials.Citation19 In our previous study, using the f88-4/Cys6 phage display peptide library expressing 16 mer cysteine constrained peptides, two phages were identified with high binding affinity for TAG-72 with sequences NPG TCK DKW ECL LIN G (A3-10)Citation20 and GGV SCM QTS PVC ENN L (A2-6).Citation21,Citation22 Both phage-bound peptides were investigated against TAG-72 positive tumor cells in vitro, while only the A2-6 was investigated as the free peptide and demonstrated specific binding in vitro and tumor accumulation in vivo.
We have again identified phages expressing peptides with binding affinity for TAG-72 by screening the same display peptide library used by us previously.Citation21 However, in our previous report, phages bound to the TAG-72 wells were eluted with glycine buffer pH 2.2. The objective of this present study was to select for phages that bind more strongly to TAG-72. Based on off-rate kinetics, longer incubations are likely to elute more strongly bound and therefore higher binding affinity phages from the target protein.Citation23 Therefore two elution conditions were used, the first was the common brief elution with pH 2.2 glycine buffer and the second was a longer elution with Tris buffered saline (TBS) at pH 7.4. The neutral pH of the second elution was necessary since the phages cannot withstand exposure to low pH for an extended time.
We have again used the f88-4/Cys6 phage display peptide library, with different buffer elution conditions that are expected to identify new and higher affinity binding peptides for the TAG-72 antigen. As is described below, after three rounds of selection, two consensus phages, named G3-15 (glycine buffer) and T3-15 (TBS buffer) were identified and both were shown by phage ELISA, flow cytometry and fluorescence microscopy to bind specifically to TAG-72. Thereafter, the free peptides were studied in cell culture and, after radiolabeling with technetium-99m (99mTc), their biodistribution in tumored mice were evaluated by necropsy and SPECT/CT imaging.
Results
Selection of TAG-72 specific phages.
In this investigation two different elution conditions were used, the brief glycine elution method as before and a longer elution method using TBS that was expected to select for higher binding affinity phages. Of the ELISA positive phages eluted with the glycine buffer, 33% showed the sequence FRE RCD KHP QKC TKF L and was named G3-15, whereas the longer elution in TBS produced 30% of the phages with the sequence DPR HCQ KRV LPC PAW L and was named T3-15. The phage M3-15, from an unrelated selection and used as a control, had the sequence TVM LCN PME QGC RWM C. The results of the ELISA assay presented in demonstrate that phages G3-15 and T3-15 show nearly identical levels of binding to TAG-72 and both show 100-fold higher binding than the control phage M3-15 that is near background.
Phage tumor cell binding.
The binding of phages G3-15 and T3-15 to LS-174T colon cancer cells, known to express high levels of TAG-72, were evaluated by flow cytometry. Phages were added to the cells followed by the anti-fd-bacteriophage antibody and PE-conjugated goat anti-rabbit F(ab')2 antibody. As shown by flow cytometry in , both phages G3-15 and T3-15 displayed statistically higher (p < 0.01) binding to LS-174T cells than to control HT-29 cells and higher binding to LS-174T cells relative to the negative control phage M3-15 (p < 0.01). Fluorescence microscopy was used to further verify binding and to determine the binding location to LS-174T cells. The results presented in show that both phages G3-15 and T3-15 specifically bind to the LS-174T cells, with no signal observed for the negative control phage M3-15 and cells with buffer only as a further control. Furthermore, the signal with the two study phages was observed largely restricted to the cell membrane and not internalized. The blue stain (DAPI) indicates the nucleus.
Peptide stability.
The peptides expressed by phages G3-15 and T3-15 were synthesized, radiolabeled with 99mTc and their binding to TAG-72 characterized. After labeling and P2 column purification, both peptides showed by C18 reversed phase chromatography a single peak and a radiochemical purity of more than 90%. The stability to enzymatic degradation was evaluated by size exclusion HPLC following incubation of the 99mTc-labeled peptides in serum. Both peptides showed the same single peak when incubated in 37°C buffer for 5 min ( and D) and in 37°C serum ( and E). Even after 1 h in 37°C serum, no lower molecular weight peaks were seen as evidence of stability, but for both peptides, a small percentage of the label eluted as two peaks at earlier retention times therefore higher molecular weight ( and F). The higher molecular weight peaks are probably due to protein binding. Radioactivity recovery was always greater than 90%.
Peptide tumor cell binding.
The LS-174T cells were incubated with each 99mTc-radiolabeled peptide in the presence of increasing concentrations of the respective unlabeled peptide. The results presented in show a decrease in radiolabel bound with increasing unlabeled peptide for both peptides expected for specific binding. Analysis provided an IC50 of 10.32 nM for 99mTc-G3-15 and 0.29 nM for 99mTc-T3-15.
Biodistribution of the 99mTc-peptides.
Nude mice bearing LS-174T tumors in one thigh received by tail vein 2 µg (about 10 µCi) of either 99mTc-labeled G3-15 or T3-15 peptides. The results are reported in as the percent of the injected dose per gram of tissue (%ID/g) at 5, 30 and 90 min. Both labeled peptides show rapid clearance, however, the 99mTc-G3-15 peptide accumulated at lower levels in most tissues. Also, the higher accumulations in stomach and small and large intestine for 99mTc-T3-15 may indicate that these two peptides have different routes of clearance. Even so, both peptides show improved ratios for tumor thigh/normal thigh and tumor thigh/blood from 5–90 min. SPECT/CT imaging was performed in another set of mice receiving 20 µg peptide radiolabeled with 200 µCi with acquisitions started at 20 and 60 min (). The 20 min scan shows that both peptides have cleared from circulation with the kidneys being the primary route of excretion, although some activity is observed in the intestinal tract, gall bladder and urinary bladder. At 20 min the tumor in the left thigh is clearly visible for both peptides. However, by 60 min, the tumor with 99mTc-G3-15 is nearly at background, but remains obvious for the 99mTc-T3-15.
Discussion
In previous studies from this laboratory using the same phage library, peptides A3-10,Citation20 and A2-6,Citation21,Citation22 were identified that demonstrated high binding properties for TAG-72. The A3-10 peptide was evaluated only in its phage form and showed a 4.3-fold higher binding to the LS-174T cells over an irrelevant control phage, and when incubated with LS-174T tumor cubes, the A3-10 99mTc-labeled phage demonstrated saturation, an indication of specific binding. In addition, the phage competed with the B72.3 antibody for binding to the tumor cubes thus suggesting that the B72.3 antibody and the A3-10 phage may bind to the same site. In another study,Citation21 the A2-6 phage also showed high binding to LS-174T cells also with evidence of specific binding as the free A2-6 peptide bound to TAG-72 positive LS-174T cells and showed positive staining of LS-174T tumor xenografts by immunohistochemistry. Fluorescence microscopy demonstrated that the A2-6 phages bound to the cell membrane of LS-174T cells. Furthermore, in subsequent studies the 99mTc labeled A2-6 peptide IC50 was determined to show binding affinity to TAG-72 in the nanomolar range.Citation22 The peptide proved to be stable in serum with fast clearance in mice, with tumor accumulation at 3 h of 0.11% injected dose per gram.
To identify peptides with higher affinities, we have now introduced an overnight elution after the subtraction selection steps used previously. After three rounds of selection, phage G3-15 was isolated based on the previous method with a brief incubation in low pH glycine buffer, while phage T3-15 was isolated based on an additional longer elution time in neutral pH buffer. Compared to a phage from an irrelevant study as a negative control, phages G3-15 and T3-15 both showed high binding to TAG-72 in an ELISA phage assay (). Further, by flow cytometry both study phages bound specifically to TAG-72 positive cells compared to the control phage and compared to the TAG-72 negative control cells (). The background binding observed for the study phages to the negative control cell line is likely due to the properties of the phage itself, and not the peptide displayed on the surface. Fluorescence microscopy demonstrated that the two study phages bound to the surface of the TAG-72 positive LS-174T cells (). The peptides displayed on the phage protein coat for G3-15 and T3-15 were synthesized and their binding to TAG-72 was characterized in vitro and in vivo after radiolabeling with 99mTc. The 99mTc-MAG3-peptides after purification were over 90% radiochemically pure. No evidence for instability of either radiolabeled peptide in serum was observed and both showed no more than 10% shift in the radioactivity profile to higher molecular weight probably due to serum protein binding ( and F). No degradation products were apparent (). The 99mTc-labeled peptides in a cell binding competition study showed that both 99mTc-labeled G3-15 and T3-15 bound to TAG-72 positive LS-174T cells and that the binding could be inhibited with increasing concentrations of the unlabeled peptides. The IC50 for 99mTc-T3-15 was 36 times lower at 0.29 nM than for 99mTc-G3-15 at 10.32 nM (), and both were lower than for the peptides that we selected previously, A2-6 and A3-10, at about 50 nM and 400 nM, respectively (data not published). Generally, the IC50 for peptides selected from phage display are in the nM to µM range.Citation27,Citation28 Compared with these peptides, the low IC50 for peptide T3-15 is likely due to the selection method and longer elution conditions established here, and may also indicate that it has a higher binding affinity for the TAG-72 over the peptides selected by us previously since the IC50 is directly proportional to the affinity constant.
Although the peptide 99mTc-T3-15 has a lower IC50 than 99mTc-G3-15 for TAG-72, these two peptides showed similar biodistributions by necropsy in mice bearing TAG-72 positive tumors in one thigh (). The percent injected dose per gram for kidneys () shows no apparent decrease from 5–90 min for both peptides, suggesting that a portion of activity may be retained in this organ. A more complete evaluation of activity in kidneys is outside the focus of this current report. Both peptides showed an increase in tumor to normal thigh and tumor to blood ratio over time. But since tumor accumulation did not increase from 5–90 min, the improved ratios are likely due to the faster clearance of the peptide from blood and normal thigh. At 30 min both of these peptides showed similar tumor accumulation, but by 90 min the 99mTc-T3-15 was almost three times higher in tumor than the 99mTc-G3-15. Similarly, by imaging, both peptides showed fast clearance from circulation to the kidneys and small intestines and excretion to bladder was obvious at 20 min. The scans clearly showed the tumor at 20 min following injection and, as in agreement with the biodistribution study, the signal in tumor decreased at the later time (60 min) for both peptides.
In conclusion, by using a longer elution method, we identified a higher affinity peptide that may have applications in imaging and possibly radiotherapy of cancers.
Materials and Methods
Reagents and cell lines.
Both the f88-4/Cys6 phage library and Escherichia coli strain K91 BlueKan were gifts from Dr. George Smith (University of Missouri at Columbia, Columbia, MO). The TAG-72 glycoprotein was purchased from Sigma-Aldrich Co., (St. Louis, MO) as partially purified from human fluids and the B72.3 antibody was obtained from the NCI Biological Resource Branch Preclinical Repository (Rockville, MD). The primer used for sequencing was 5′-AGT AGC AGA AGC CTG AAG A-3′ (Quigen, Alameda, CA). All other chemicals were from standard suppliers. The LS-174T and HT-29 cells were obtained from the American Type Culture Collection (ATCC, Manassas, VA) and were grown in Dulbecco's modified Eagle's medium (MEM), supplemented with 10% fetal bovine serum (FBS) containing 1% penicillin and streptomycin and 1% non-essential amino acids. The 99mTc pertechnetate was eluted from a 99Mo-99mTc generator (Perkin-Elmer, Billerica, MA). The NHS-MAG3 was synthesized in house.Citation24 The peptides were purchased as the uniform L isomers with an amino hexyl linker on the amino end and 95% purity by New England Peptide Company (Gardner, MA). Swiss male nude mice at 6 weeks of age were purchased from Taconic Farms (Germantown, NY).
Purification of TAG-72.
The TAG-72 was purified using methods described by us previously.Citation20,Citation21 Briefly, the B72.3 monoclonal antibody was added to a Protein-L (Pierce Biotechnology, Inc., Rockford, IL) affinity column and was used to purify the partially purified TAG-72. After excessively washing, the retained TAG-72 was eluted with 0.05 M diethylamine containing 3 M sodium iodide, pH 11.5. The TAG-72 was then further purified by overnight dialysis at 4°C against phosphate buffered saline (PBS).
Selection of TAG-72 binding phages.
Selection of peptides was accomplished by a subtraction selection strategy with different elution buffers and elution times for the final step. The first strategy included a single brief elution with pH 2.2 glycine buffer and the second strategy began with the glycine elution but was followed with a longer elution with Tris buffered saline (TBS) at pH 7.4. As described previously,Citation20,Citation21 6-well plates (35 mm in diameter, BD Falcon) were coated with the wash from the Protein-L-B72.3 affinity column that was free of TAG-72, with 0.5% BSA or with 1.2 µg purified TAG-72 in 0.1 M bicarbonate buffer, pH 8.5. Three wells within each plate were coated with one of the three samples. The plates were left at 4°C for 24 h then the coating solution was removed and blocking buffer containing 0.5% BSA was added. After 2 h at room temperature, the blocking buffer was removed and the wells were washed extensively with TBS containing 0.05% Tween-20 (TBS/Tween). To begin the selection, about 5.6 × 1010 phages in 400 µl TBS/Tween contain 1 mg/ml BSA were added to the well coated with the TAG-72 free wash from the Protein-L-B72.3 affinity column and incubated at room temperature for 30 min. The solution containing unbound phages was then transferred to the second well, coated with 0.5% BSA and incubated at room temperature for 30 min. Finally, the solution with unbound phages was transferred to the third well coated with the purified TAG-72. After 2 h incubation, the unbound phages were removed and the well was washed 15 times with TBS/Tween, before the bound phages were eluted with a brief incubation in 0.2 M glycine buffer, pH 2.2 (400 µl) for 10 min for the first elution condition. Finally this phage eluant was neutralized with 50 µl of 1 M Tris/HCl, pH 9.2.
For the second elution condition the TAG-72 coated well was first eluted briefly with glycine buffer as above, and then washed with pH 7.4 TBS three times before 400 µl of TBS was added and left undisturbed for 24 h. Finally, this overnight TBS eluant was recovered and, along with the brief glycine buffer eluant, the phages therein were amplified separately in Escherichia coli strain K91 BlueKan and titered as described previouslyCitation20,Citation21 in preparation for the next round of selection. In brief, after amplification the bacteria were removed by centrifugation, and the phages were recovered by precipitation after an overnight incubation at 4°C with 17.6% polytheylene glycol 8000 (PEG) and 3.3 M sodium chloride (NaCl) (at a volume ratio of 1:6). The precipitated phages were recovered by centrifugation at 13,000 rpm for 20 min and the pellet was resuspended in 1 ml TBS and then re-spun at 10,000 rpm to remove any remaining bacteria. The phages were precipitated a second time with PEG/NaCl (volume ratio of 1:6) on ice for 1 h, then spun again at 13,000 rpm for 10 min to recover the phage pellet which was finally resuspend in 100 µl TBS containing 0.2% sodium azide and used for the next round of selection. The entire selection and purification procedure was repeated two additional times.
Phage analysis by ELISA
After three rounds of selection, 20 phage clones, randomly selected from the plates used for titer, were amplified in an overnight incubation at 37°C in NZY medium with 20 µg/ml tetracycline. The bacteria were removed by centrifugation and the phages were precipitated with PEG/NaCl as above. A phage ELISA was then used to identify those phages that demonstrated the highest binding to TAG-72. Wells of a 96-well plate were coated with TAG-72 as before, the wells were blocked with 4% non-fat milk for 1 h and 1012 phages of each of the 20 amplified phages was added to wells in triplicate. The plates were left for 1 h at room temperature, and then washed three times with PBS, containing 0.1% Tween-20 (PBS/Tween), and then three times with PBS alone. To evaluate the number of bound phages, 100 µl of horseradish peroxidase-conjugated anti-M13 phage antibody (Amersham, NJ) diluted 1:1,000 was added to each well and left for 1 h at room temperature. Finally, the wells were washed three times with PBS/Tween and three times with PBS before the addition of 3,3′,5,5′-tetramethylbenzidine, the substrate for horseradish peroxidase, and the absorbance was measured at 405 nm in a microplate plate reader (SpectraMax M5/M5, Molecular Devices Corporation, Sunnyvale, CA).
DNA sequencing.
Single-stranded phage DNA was prepared from the ELISA-positive clones using the method standard in this laboratory. Briefly, 0.1 ml containing 4 M sodium iodide in 10 mM Tris-HCl, pH 8.0 and 1 mM ethylenediaminetetraacetic acid (EDTA) was added to the phage pellet followed by 0.25 ml of cold 100% ethanol to precipitate the DNA. The DNA pellet was washed with 70% ethanol and dried using a vacuum concentrator (SpeedVac Savant Instruments Inc., Holbrook, NY). The DNA was sequenced using the primer 5′-AGT AGC AGA AGC CTG AAG A-3′ on an automated fluorescent DNA sequencer (Applied Biosystems 3100, Foster City, CA).
Phage cell binding.
Flow cytometry analysis. From the 20 clones evaluated, the phage expressing a consensus sequence obtained with the glycine buffer was named G3-15 while the phage expressing a consensus sequence obtained with the TBS buffer was named T3-15. These two plus a control phage, M3-15, from an unrelated selection study, were analyzed by flow cytometry for binding to TAG-72 positive colon cancer cells LS-174T and TAG-72 negative cells HT-29 cells.Citation21,Citation25 The phages (1012 plaque tetracycline units = PTU) were added to 105 cells suspended in MEM medium containing 1% FBS in triplicate. After 1 h at room temperature, the samples were washed with PBS containing 3% BSA (PBSB) before 100 µl of a 1:1,000 dilution of rabbit anti-fd-bacteriophage antibody (Sigma) was added and the samples left for 1 h at room temperature. Thereafter the cells were washed three times with PBSB and 100 µl of a 1:250 dilution of PE-conjugated goat anti-rabbit F(ab')2 antibody (Sigma) was added. After the samples were incubated at room temperature for 30 min the cells were washed three times with PBSB and fixed with 0.5 ml cold 0.5% paraformaldehyde solution for flow cytometry analysis on a fluorescence activated cell sorter Caliber Cytometer (Becton Dickinson, Franklin Lakes, NJ) using CellQuest software (Becton Dickinson).
Fluorescence microscopy. Fluorescence microscopy was performed to evaluate the positive clones for cellular binding and localization. The phages G3-15, T3-15 or the control phage (1012 PTU) were added to separate chambers of an eight-well chambered slide (Nunc, NY), with the LS-174 T cells grown to 80–90% confluence and incubated for 1 h. After washing with PBS three times, the cells were fixed with 3.7% paraformaldehyde for 10 min, washed again with PBS three times and then blocked with 3% BSA for 30 min. The bound phages were indicated with the sequential addition of rabbit anti-fd-bacteriophage antibody (200 µl of a 1:1,000 dilution) and after washing as above, PE-conjugated goat anti-rabbit F(ab')2 antibody (200 µl of a 1:250 dilution). After 30 min the slide was washed three times with PBS and the nuclei were stained with 100 nM 4′,6-diamidino-2-phenylindole (DAPI, Molecular Probes, Eugene, OR) in PBS for 5 min. All incubations were at room temperature. The cells were observed on a Nikon Eclipse TE 2000-S microscope (Nikon Instruments Inc., Melville, NY) equipped with a Cy5.5 filter and CCD camera and the images were processed using IPLab software (BD Biosciences).
Peptide radiolabeling with 99mTc.
The peptides from the two phages G3-15 and T3-15 that showed high TAG-72 binding, were synthesized commercially as the uniform L isomers with an amino hexyl linker on the amino end and 95% purity and with preserved cysteine constraints by New England Peptide Company (Gardner, MA). The peptides were prepared for radiolabeling by conjugation with NHS-MAG3 following the methods standard in this laboratory.Citation26 Briefly, 20 µl of water with 100 µg of peptide was adjusted to pH 8 with 2 µl 0.1 M sodium bicarbonate solution pH 9. With agitation, NHS-MAG3 was added at a 20-fold molar excess to the peptide and incubated 1 h. The conjugated peptide was separated from free MAG3 on a 0.7 × 20 cm P-2 column (Bio-Rad) using D-PBS (Dulbecco's PBS) as eluant. To radiolabel with 99mTc, 200 µl containing about 50 µg MAG3 conjugated peptide was added to an aliquot of sodium tartrate (50 mg/ml) in 0.5 M sodium bicarbonate, 0.25 M ammonium acetate and 0.175 M ammonium hydroxide buffer (pH 9.2) such that the final concentration of tartrate was 7 mg/ml. After addition of about 0.6 mCi (for biodistribution) or 2 mCi (for imaging) of 99mTc pertechnetate generator eluant, 20 µl of a fresh solution of SnCl2·2H2O (4 µg/µl in 10 mM HCl) was added. The labeled peptide was purified on a P-2 column. The final sample was analyzed by C18 reversed phase chromatography (Vydac, The Nest Group, Southborough, MA) using a linear gradient of acetonitrile in 0.08% trifluoroacetic acid from 15–90% over 45 min at a flow rate of 1 ml/min (Waters 515 solvent delivery system, Waters, Milford, MA).
Peptide stability.
Stability of both 99mTc-labeled peptides towards peptidases was evaluated after incubation at 37°C in serum. A 100 µl aliquot of each 99mTc-labeled peptide solution containing about 2 µg peptide was added to 100 µl of buffer or serum and incubated at 37°C. Samples were removed at 5 min and 1 h for analysis by size exclusion HPLC on a Superose-12 column (Amersham Pharmacia Biotech, Piscataway, NJ). The running solution was 0.1 M Tris-HCl (pH 8.0) containing 20% acetonitrile using a flow rate of 0.6 ml/min. Peaks appearing after the peptide (i.e., lower molecular weight) would indicate peptide degradation.
Peptide cell binding.
A competitive cell binding assay was performed with the 99mTc-labeled G3-15 and T3-15 peptides against LS-174T cells. Approximately 1.5 × 104 cells in MEM medium with 10% FBS were divided into 96 well filtration system plates (Multiscreen HTS, Millipore, MA) and the cells grown at 37°C for 24 h. The cells were washed twice with ice-cold TBS and 0.02 µg radiolabeled peptide in TBS was added to each well in the presence of increasing concentrations of the unlabeled peptide from 10−13 to 10−5 M. After incubation at room temperature for 1.5 h, the cells were washed three times with TBS using a Multiscreen vacuum manifold (Millipore). The bottom filter of each well was punched directly into plastic tubes for measure of radioactivity in a gamma well counter Na (TI) (Cobra II Auto-Gamma, Packard Instrument Co., Downers Grove, IL). The IC50 values for the peptides were calculated using Prism version 5 (GraphPad) software.
Biodistribution of the 99mTc-peptides.
All animal studies were performed with the approval of the university's Institutional Animal Care and Use Committee. Male 6-weeks-old nude mice were injected subcutaneously into the left thigh with 106 LS-174T cells. Approximately two weeks later, when the tumors were about 0.5–1.0 cm in one dimension, the mice received about 2 µg (about 10 µCi) of either 99mTc-labeled G3-15 or T3-15 peptide through a tail vein. In groups of four, the animals were sacrificed at 5, 30 or 90 min after injection. At sacrifice, blood and the entire liver, heart, kidneys, spleen, lungs, stomach, small intestine, large intestine and samples of muscle, thigh with tumor and contralateral thigh were removed, weighed and the radioactivity in each organ was counted in the gamma well NaI(TI) counter against a standard of the injectate. The percentage of the injected dose administered per gram of tissue (%ID/g) was then calculated.
In a separate study, SPECT/CT images were acquired on a NanoSPECT/CT™ small animal camera (Bioscan, Inc., Washington, DC) with administration of about 20 µg (carrying 200 µCi) of either 99mTc-labeled peptide. The larger amount of peptide was needed to achieve the higher activity required for high resolution imaging at later times. This was especially necessary since the peptide was anticipated to have fast clearance. Scans were acquired at 20 min and 60 min after administration with SPECT parameters of 24 projections with 60 sec per projection and a whole body CT scan for rcoegistration was obtained before each SPECT image, thus requiring a total acquisition time of 30 min. Animals were anesthetized for imaging with 1–2% isoflurane in oxygen. The image reconstruction was performed using Bioscan's InVivoScope 1.37 software.
Figures and Tables
Figure 1 The results of phage ELISA measurements of relative binding of G3-15, T3-15 and the control phage M3-15 to TAG-72. Both study phages showed similar binding to TAG-72 compared to the control. The data is presented as the mean of three replicates with one SD represented by the error bars.
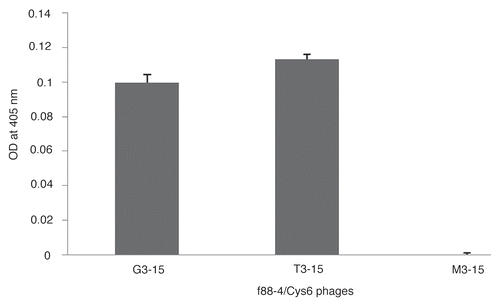
Figure 2 (A) Relative binding of G3-15, T3-15 and M3-15 control phages to LS-174T and HT29 cells by flow cytometry. The data is presented as the percentage of phage binding to the cell total. The data is presented as the mean of three replicates with one SD represented by the error bars. (B) Fluorescence microscopy of LS-174T cells incubated with the two study phages G3-15 and T3-15, the control phage M3-15 and buffer only. The red fluorescence indicates bound phages and blue fluorescence (DAPI) indicates the nucleus. Phages are shown bound to the cell membrane (red stain) for the two study phages with no signal observed for the control phage and buffer alone (Magnification x400).
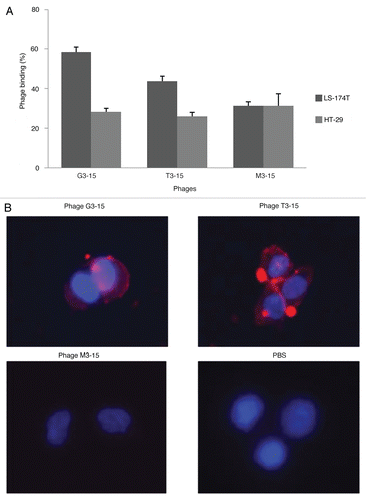
Figure 3 Size exclusion HPLC radiochromatograms of the 99mTc-labeled G3-15 and T3-15 peptides incubated in PBS (A and D) and in 37°C serum for 5 min (B and E) and 1 h (C and F).
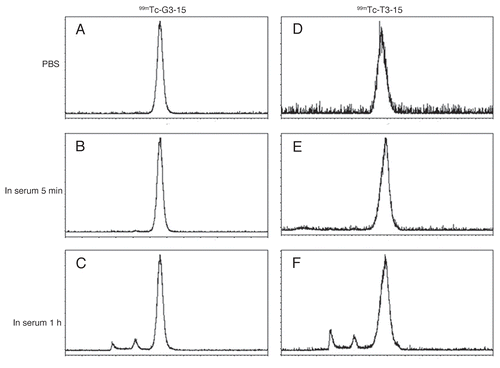
Figure 4 The results of competitive binding assay in LS-174T cells incubated with 99mTc-G3-15 (A) or 99mTc-T3-15 (B) peptides showing in both cases decreased binding with increasing concentrations of unlabeled peptide from 10−13 to 10−5 M. Data presented as the mean of four measurments with one SD indicated by the error bars.
Figure 5 Anterior views of the SPECT/CT fused projections for 99mTc-G3-15 and 99mTc-T3-15 in mice bearing LS-174T tumors in the left thigh. Acqusitions were obtained at 20–40 min and 60–80 min post injection. Arrows show the tumors.
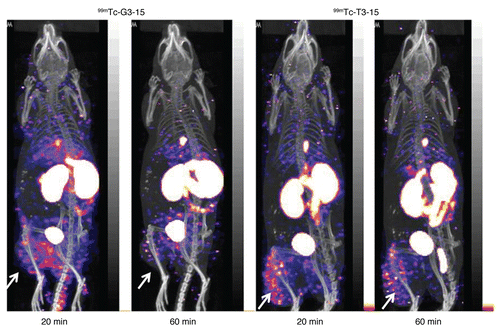
Table 1 Presenting the biodistributions by necropsy at 5, 30 and 90 min post administratin of 99mTc-G3-15 and 99mTc-T3-15 in mice with LS-174T tumor in one thigh
Acknowledgements
We thank Dr. George Smith and Susan Deutcher for providing the phage display peptide library and Escherichia coli strain K91 BlueKan, Dr. Alexei Bogdanov Jr., for use of the fluorescence microplate reader and the NCI Biological Resource Branch Preclinical Repository (Rockville, MD) for providing the B72.3 monoclonal antibody. Funding was provided in part by the NIH under CA111606.
References
- Thor A, Ohuchi N, Szpak CA, Johnston WW, Schlom J. Distribution of oncofetal antigen tumor-associated glycoprotein-72 defined by monoclonal antibody B72.3. Cancer Res 1986; 46:3118 - 3124
- Stramignoni D, Bowen R, Atkinson BF, Schlom J. Differential reactivity of monoclonal antibodies with human colon adenocarcinomas and adenomas. Int J Cancer 1982; 31:543 - 552
- Nuti M, Teramoto YA, Mariani-Costantini R, Hand PH, Colcher D, Schlom J. A monoclonal antibody (B72.3) defines patterns of distribution of a novel tumor-associated antigen in human mammary carcinoma cell populations. Int J Cancer 1982; 29:539 - 545
- Divigi CR, Scott AM, Mcdermott K, Fallone PS, Hilton S, Siler K, et al. Clinical comparison of radiolocalization of two monoclonal antibodies (mAbs) against the TAG-72 antigen. Nucl Med Biol 1994; 21:9 - 15
- Divigi CR, Scott AM, Gulec S, Broussard EK, Levy N, Young C, et al. Pilot radioimmunotherapy trail with 131I-labeled murine monoclonal antibody CC49 and deoxyspergualin in metastatic colon carcinoma. Clin Cancer Res 1995; 1:1503 - 1510
- Fischman AJ, Khaw BA, Strauss HW. Quo vadis radioimmune imaging. J Nucl Med 1989; 30:1911 - 1915
- Michele JL, Robert LD, Marc M, Robert MS, David MG. Human immune response to anti-carcinoembryonic antigen murine monoclonal antibodies. Cancer Res 1990; 50:1055 - 1058
- Reilly RM, Sandhu J, Alvarez-Diez TM, Gallinger S, Kirsh J, Stern H. Problems of delivery of monoclonal antibodies. Pharmaceutical and pharmacokinetic solutions. Clin Pharmacokinet 1995; 28:126 - 142
- Kashmiri SV, Shu L, Padlan EA, Milenic DE, Schlom J, Hand PH. Generation, characterization and in vivo studies of humanized anticarcinoma antibody CC49. Hybridoma 1995; 14:461 - 473
- Kim SJ, Hong HJ. Guided selection of human antibody light chains against TAG-72 using a phage display chain shuffling approach. J Microbiol 2007; 45:572 - 577
- Ridgway JB, Eric N, Kern JA, Lee J, Brush J, Goddard A, et al. Identification of a human anti-CD55 single-chain Fv by subtractive panning of a phage library using tumor and nontumor cell lines. Cancer Res 1999; 59:2718 - 2723
- Liu S, Edward S, Barrett J. 99mTc labeling of highly potent small peptides. Bioconjugate Chem 1997; 8:621 - 636
- Ania OH, Sroka TC, Chen ML, Lam KS. Therapeutic cancer targeting peptides. Biopolymers 2002; 66:184 - 199
- Smith GP. Filamentous fusion phage: novel expression vectors that display cloned antigens on the virion surface. Science 1985; 228:1315 - 1317
- Lee TY, Lin CT, Kuo SY, Chang DK, Wu HC. Peptide-mediated targeting to tumor blood vessels of lung cancer for drug delivery. Cancer Res 2007; 67:10958 - 10965
- Oyama T, Sykes KF, Samli KN, Minna JD, Johnston SA, Brown KC. Isolation of lung tumor specific peptides from a random peptide library: generation of diagnostic and cell-targeting reagents. Cancer Lett 2003; 202:219 - 230
- Pasqualini R, Ruoslahti E. Organ targeting in vivo using phage display peptide libraries. Nature 1996; 380:364 - 366
- Zitzmann S, Kramer S, Mier W, Hebling U, Altmann A, Rother A, et al. Identification and evaluation of a new tumor cell-binding peptide, FROP-1. J Nucl Med 2007; 48:965 - 972
- Lindon LA, Zou J, Deutscher SL. Is phage display technology on target for developing peptide-based cancer drugs. Curr Drug Discov Technol 2004; 1:113 - 132
- Rusckowski M, Gupta S, Liu GZ, Dou SP, Hnatowich DJ. Evidence of specificity of radiolabeled phage display peptides for the TAG-72 antigen. Cancer Biother Radiopharm 2007; 22:564 - 572
- Chen L, Wang Y, Liu XR, Dou SP, Liu GZ, Hnatowich DJ, et al. A new TAG-72 cancer marker peptide identified by phage display. Cancer Lett 2008; 272:122 - 132
- Chen L, Wang Y, Liu XR, Liu GZ, Xiao N, Dou SP, et al. Evaluation of a new TAG-72 binding peptide. J Nucl Med 2008; 49 supplement 1 332p
- Yuan B, Schulz P, Liu R, Sierks RM. Improved affinity selection using phage display technology and off-rate based selection. J Biotechnol 2006; 9:171 - 175
- Wang Y, Liu XR, Donald J, Hnatowich. An improved synthesis of NHS-MAG3 for conjugation and radiolabeling of biomolecules with 99mTc at room temperature. Nat Protoc 2007; 2:972 - 978
- Fang LY, Battisti RF, Cheng H, Reigan P, Xin Y, Shen J, et al. Enzyme specific activation of benzoquinone ansamycin prodrug using antibody-β-galactosidase conjugates. J Med Chem 2006; 49:6290 - 6297
- Wang Y, Liu XR, Zhang YM, Liu GZ, Rusckowski M, Hnatowich DJ. Nonspecific cellular accumulation of 99mTc-labeled oligonucleotides in culture is influenced by their guanine content. Nucl Med Biol 2007; 34:47 - 54
- Paradis-Bleau C, Lioyd A, Sanschagrin F, Clarke T, Blewett A, Bugg T, et al. Phage display-derived inhibitor of the essential cell wall biosynthesis enzyme MurF. BMC Biochem 2008; 9:33
- Desjobert C, Soultrait VR, Faure A, Parissi V, Litvak S, Litvak L, et al. Identification by phage display selection of a short peptide able to inhibit only the strand transfer reaction catalyzed by human immunodeficiency virus type 1 integrase. Biochemistry 2004; 43:13097 - 13105