Abstract
The role of tumor suppressors and cell cycle factors in gastric carcinogenesis are well understood; however, the post-transcriptional changes that affect gene expression in gastric cancer are poorly characterized. MiR-135a has been shown to play a role in Hodgkin lymphoma. The aim of this study was to investigate the expression and role of miR-135a in gastric cancer. Quantitative real-time PCR demonstrated that miR-135a expression is downregulated in the majority of human primary gastric cancer tissues (8/11; 73%), compared with pair-matched adjacent non-tumor tissues. Furthermore, compared with the nonmalignant gastric cell line, GES-1, miR-135a expression was substantially downregulated in gastric cancer cell lines of various degrees of differentiation. Target analysis indicated miR-135a directly regulates Janus kinase 2 (JAK2), a cytoplasmic tyrosine kinase involved in cytokine receptor signaling pathways. Overexpression of miR-135a significantly downregulated the expression of JAK2 protein and also reduced gastric cancer cell proliferation and colony formation in vitro. MiR-135a-mediated JAK2 downregulation also reduced p-STAT3 activation and cyclin D1 and Bcl-xL protein expression. This study suggests that miR-135a may function as a tumor suppressor via targeting JAK to repress p-STAT3 activation, reduce cyclin D1 and Bcl-xL expression and inhibit gastric cancer cell proliferation. These results imply that novel treatment approaches targeting miR-135a may potentially benefit patients with gastric cancer.
Keywords: :
Introduction
MicroRNAs (miRNAs) are small noncoding RNAs which post-transcriptionally regulate gene expression by binding to the 3′ untranslated region (3′ UTR) of target mRNAs, to trigger mRNA degradation or translational repression.Citation1,Citation2 Like small interfering RNAs (siRNAs), miRNAs are loaded into Argonaute-containing RNA induced silencing complexes, also termed miRNA ribonucleoprotein complexes (miRNPs).Citation3 Over the last several years, it has become clear that miRNAs contribute to most, if not all, basic biological processes including embryonic stem cell differentiation, metabolism, cell proliferation, apoptosis and signal transduction.Citation4 Precise chronological and topological regulation of post-transcriptional gene silencing by miRNAs is essential during animal development and tissue differentiation.Citation5 Abnormal expression of miRNAs is associated with various human disorders including cancer;Citation6 Compared with transcription factors, the functions of individual microRNAs in cancer initiation and progression are poorly understood. In recent years, microRNA expression profiles have been used to classify human cancers and measure the expression levels of many microRNAs,Citation7 and it has become clear that microRNAs can play a role in the development of cancer as oncogenes and tumor suppressors.
Gastric cancer is one of the most common cancers worldwide.Citation8 Recent research has suggested that miRNAs are involved in gastric cancer,9 as a substantial number of miRNAs are differentially expressed in gastric cancer tissues. The role of miRNAs in gastric cancer has not yet been fully characterized and the relationship between signal transduction pathways and miRNAs is currently the focus of intense research. MiR-152,Citation10 miR-218Citation11 and miR-125a-5pCitation12 are reported to exert tumor suppressor activity by suppressing expression of mRNA targets which inhibit apoptosis or promote cell-cycle progression. MiR-218 has been shown to induce apoptosis in gastric cancer cells, through direct targeting of the epidermal growth factor receptor-co-amplified and overexpressed protein which is a positive regulator of NF-kB transcriptional activity.Citation13
The JAK/STAT signaling pathway is a common signaling pathway for many cytokinesCitation14, and plays an important role in proliferation, differentiation and apoptosis. JAK proteins are activated by cytokine and growth factor receptor signaling to subsequently activate downstream proteins, including the signal transducers and activators of transcription. The JAKs, consisting of JAK1, JAK2, JAK3 and TYK2, share structural and functional homologies. When cytokines bind to a cell-surface receptor, the receptor dimerizes and leads to phosphorylation and activation of JAK tyrosine kinases, which in turn phosphorylate the receptor to allow binding and phosphorylation of cytoplasmic STAT proteins. There are seven STATs, all of which contain a DNA-binding domain. STAT3 and STAT5 are the preferred downstream targets of phosphorylated JAK2 and regulate expression of genes related to cell survival, proliferation, angiogenesis and immune evasion.Citation15 Recent studies have shown that the JAK/STAT signaling pathway is regulated at several levels, allowing fine control of the signal strength, duration and specificity.Citation16 The JAK2/STAT3 pathway has been intensely investigated in gastricCitation17 and other types of cancer.Citation18,Citation19 STAT3 is constitutively activated in a variety of human tumors and possesses oncogenic potential and anti-apoptotic activities.Citation20
In this study, we examined the expression of miR-135a in gastric cancer, and observed that miR-135a is significantly downregulated in gastric cancer tissues compared with pair-matched adjacent non-tumor tissues. MiRanda, TargetScan and PicTar indicated that the oncogene JAK2 may be a target of miR-135a; therefore, we investigated whether miR-135a can function as a tumor suppressor by targeting JAK2 to influence the proliferation of gastric cancer cells.
Results
MiR-135a is frequently downregulated in gastric cancer
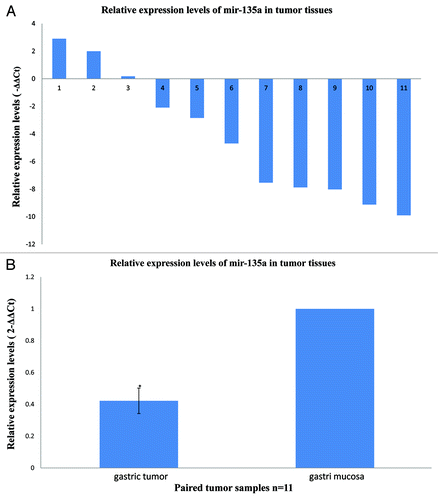
We examined miR-135a expression in primary gastric cancer tissues and pair-matched adjacent non-tumor tissues using quantitative real-time PCR, to determine whether miR-135a expression is associated with gastric cancer. Compared with the non-tumor tissues, miR-135a expression was decreased in 8/11 (73%) of the cancer tissues (). The mean ± SEM expression of miR-135a in gastric cancer tissues (0.4228 ± 0.08) was significantly lower than normal tissues (1.0 ± 0, p < 0.01; ).
Figure 1. MiR-135a is downregulated in human gastric cancer. Quantitative real-time PCR for miR-135a was performed in 11 human gastric cancer surgical specimens and the matched adjacent non-tumor mucosal tissue. (A) The relative expression levels of miR-135a in individual gastric cancer samples expressed relative to the matched adjacent non-tumor tissue, miR-135a was downregulated in 8/11 (73%) of the tumor samples. (B) The mean ± SEM relative expression level of miR-135a in gastric cancer and non-tumor samples (*p < 0.01).
MiR-135a is downregulated in gastric cancer cell lines
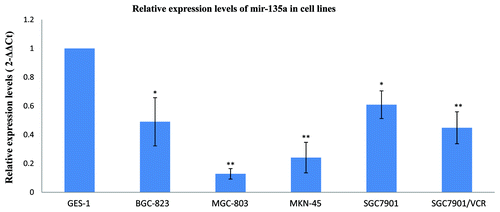
To confirm the association between miR-135a expression and gastric cancer, we examined miR-135a expression in several gastric cancer cell lines using stem-loop qRT-PCR. Five cell lines derived from gastric cancers at various degrees of differentiation were selected, including moderately differentiated SGC-7901, poorly differentiated MGC-803 and MKN-45, undifferentiated BGC-823 and the MDR (Multidrug resistance) cell line SGC-7901/VCR. Compared with the nonmalignant gastric cell line GES-1, miR-135a expression was substantially reduced in all of the gastric cancer cell lines (p < 0.05; ).
Figure 2. MiR-135a is downregulated in malignant gastric cancer cell lines. Real-time quantification of miR-135a using stem-loop RT-PCR in a variety of malignant gastric cancer cell lines and the nonmalignant gastric mucosa cell line GES-1. Expression of miR-135a was normalized to U6 snRNA and expressed relative to GES-1 cells. Values are mean ± SEM of triplicate experiments; **p < 0.01, *p < 0.05.
Overexpression of miR-135a represses JAK2 protein expression
Putative human protein-coding gene targets of miR-135a were identified using miRanda, TargetScan and PicTarCitation22 to characterize the function of miR-135a in gastric carcinogenesis. JAK2 was predicted by both TargetScan and PicTar and was selected for further analysis.
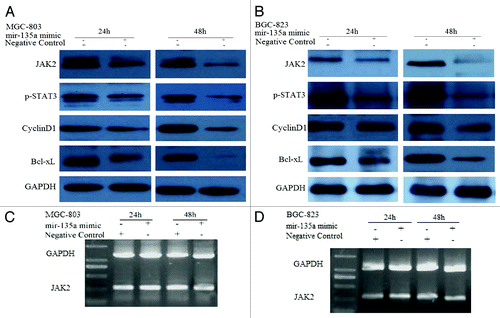
To further investigate the role of miR-135a in gastric carcinogenesis, we transfected the gastric cell lines MGC-803 and BGC-823 with miRNA-135a mimic to overexpress miR-135a. The qRT-PCR analysis confirmed that expression of mature miR-135a was significantly increased in cells transfected with miR-135a mimic, compared with the negative control cells (data not shown). Overexpression of miR-135a in MGC-803 and BGC-823 cells lead to significantly reduced expression of JAK2 protein (); however, JAK2 mRNA expression was not significantly affected (). These results suggest that miR-135a may suppress JAK2 protein expression via translational repression of JAK2 mRNA.
Figure 3. MiR-135a targets JAK2. (A, B) western blot of MGC-803 and BGC-823 cells transfected with the 40 nM miR-135a mimic or the negative control mimic at two time points. GAPDH was used as a loading control. (C, D) RT-PCR of JAK2 in MGC-803 and BGC-823 cells transfected with 40 nM miR-135a mimic or negative control mimic at two time points. GAPDH was used as a control.
MiR-135a regulates p-STAT3, cyclin D1 and Bcl-xL expression via JAK2
STAT3 is a downstream target of JAK2, which is activated by JAK2 to p-STAT3. Overexpression of miR-135a significantly reduced the expression of p-STAT3 in MGC-803 and BGC-823 cells. As STAT3 activation upregulates expression of cyclin D1, which promotes cell cycle progression and expression of the anti-apoptotic protein Bcl-xL, we investigated effect of miR-135a on cyclin D1 and Bcl-xL protein expression using western blotting. Overexpression of miR-135a downregulated expression of both cyclin D1 and Bcl-xL in MGC-803 and BGC-823 cells ().
MiR-135a overexpression inhibits gastric cancer cell proliferation
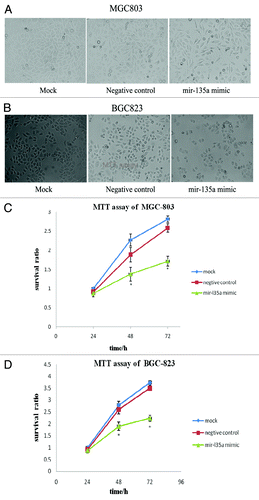
Phase-contrast microscopy () and MTT assays () demonstrated that overexpression of miR-135a significantly inhibited the proliferation of MGC-803 and BGC-823 cells, compared with miR-135a mock transfected and control cells (p < 0.05). Collectively, these results indicate that miR-135a may play an important role in the regulation of gastric cancer cell proliferation.
Figure 4. Overexpression of miR-135a inhibits the proliferation of gastric cancer cells in vitro. (A, B) Phase contrast microscopy analysis of MGC-803 cells (A) and BGC-823 cells (B) 48 h post-transfection with 40 nM miR-135a mimic, 40 nM negative control mimic and untransfected control cells; (C, D) MTT assay of MGC-803 cells (C) and BGC-823 cells (D) 24, 48 and 72 h post-transfection with 40 nM miR-135a mimic, 40 nM negative control mimic and untransfected control cells. Data are mean ± SEM of three independent experiments; *p < 0.05 compared with the negative control mimic.
MiR-135a overexpression inhibits gastric cancer cell colony formation
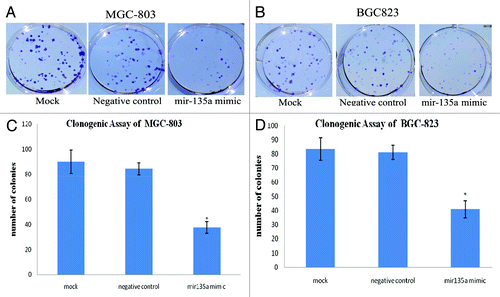
Clonogenic assays indicated that the ability of gastric cancer cells to form colonies was significantly reduced in miR-135a overexpressing cells (). Compared with negative control cells and mock transfected cells, miR-135a overexpressing MGC-803 and BGC-823 cells had a significantly lower colony formation ability, with a 55–58% reduction in the number of colonies observed in cells transfected with 40 nM miR-135a mimic (p < 0.01; ).
Figure 5. Overexpression of miR-135a reduces the colony formation ability of gastric cancer cells in vitro. Anchorage-independent growth was assessed by the in vitro colony formation assay. (A, B) Representative colony formation of miR-135a mimic, negative control mimic and untransfected control MGC-803 (A) and BGC-823 (B) cells at 2 weeks. (C, D) Number of colonies containing ≥ 50 cells in miR-135a mimic, negative control mimic and untransfected control MGC-803 (C) and BGC-823 (D) cells at 2 weeks. Data are mean ± SE of three independent experiments; *p < 0.05 compared with the negative control cells.
Discussion
Gastric cancer is one of the most common cancers worldwide, and is often detected at the later stages, leading to a 5 y survival rate of less than 20%.Citation23 Therefore, it is important to understand the etiology and progression of gastric cancer. The importance of bacterial,Citation24 environmental and host genetic risk factorsCitation25 have been extensively studied; however, little is known about the molecular progression of gastric carcinogenesis. The relationship between gastric cancer and miRNAs is currently the focus of much research.Citation9
Although miRNA signatures have been established for gastric cancer,Citation26 few studies have described the role of deregulated miRNA expression in gastric carcinogenesis. In this study, we observed that miR-135a is frequently downregulated in gastric cancer tissues and cell lines, suggesting that reduced expression of miR-135a is common in human gastric cancer. Navarro et al.Citation27 identified that miR-135a is frequently downregulated in classical Hodgkin lymphoma, and that miR-135a can modulate apoptosis and survival in L-428 lymphoma cells via targeting JAK2 mRNA. Navarro et al. also reported that miR-135a could potentially be used as a novel biologic prognostic marker of relapse in classic Hodgkin lymphoma. Liu et al. reported that miR-135a is highly overexpressed in portal vein tumor thrombosis and the hepatocellular carcinoma cell line, CSQT-2, and that blockade of miR-135a can significantly reduce the incidence of portal vein tumor thrombosis.Citation28
Our study provides strong evidence that miRNAs, specifically miR-135a, can regulate the JAK/STAT signaling pathway in gastric cancer cells, suggesting miR-135a functions as a potential tumor suppressor. The JAK/STAT signaling pathway is regulated at several steps via a number of distinct mechanisms, and deregulation of the JAK/STAT signaling pathway is associated with several aspects of tumorigenesis, including increased proliferation and reduced apoptosis.Citation29 Constitutive activation of the JAK/STAT signaling pathway is observed in several immune diseases and a variety of cancers,Citation16 including human gastric cancer.Citation17 The results of this study indicate that reduced expression of miR-135a in gastric cancer may lead to constitutive activation of the JAK/STAT signaling pathway.
To investigate the effect of miR-135a on gastric carcinogenesis, we overexpressed miR-135a in gastric cancer cell lines. Overexpression of miR-135a significantly inhibited gastric cancer cell proliferation and colony formation in vitro. Overexpression of miR-135a downregulated JAK, reduced p-STAT3 activation and lead to a subsequent downregulation of cyclin D1 and Bcl-xL protein expression. These observations indicate that miR-135a demonstrates tumor suppressor activity. Downregulation of miR-135a in gastric cancer cells may play a role in gastric carcinogenesis by constitutively activating the JAK/STAT3 signaling pathway. RT-PCR and western blotting indicated that expression of miR-135a leads to post-translational silencing of JAK mRNA translation, rather than JAK mRNA degradation.
Overexpression of several STAT3 gene products, including cyclin D1 and Bcl-xL, are recognized as critical steps in the development of cancer;Citation18 therefore, targeting of the JAK/STAT3 pathway has been considered as a potential anticancer strategy. The discovery of activating mutations in the JAK/STAT3 pathway lead to a great deal of optimism for the identification and development of JAK2 inhibitors for the treatment of cancer,Citation30 and a number of JAK2 inhibitors are currently being developed for the treatment of myeloproliferative neoplasms.Citation31 Given that JAK2 may play an important role in cancer carcinogenesis, our discovery that miR-135a inhibits JAK activity and is commonly downregulated in gastric cancer may provide a new therapeutic opportunity, as miRNAs could potentially be used to modulate JAK2, either alone or in combination with JAK2 inhibitors.
In conclusion, we have identified that miR-135a modulates gastric cancer cell proliferation and colony formation by targeting JAK2, which in turn regulates pSTAT3 activation, and cyclin D1 and Bcl-xL expression. Overexpression of synthetic miR-135a oligonucleotides may provide a potential therapeutic approach in gastric cancer. The application of miRNAs in the treatment of gastric cancer should be investigated in future studies.
Materials and Methods
Clinical samples
Eleven adult patients with gastric cancer who were diagnosed at the JiangSu Province Academy of Traditional Chinese Medicine, NanJing, People’s Republic of China between September 2010 and June 2011 were included in the analysis. Subsequently, tumor samples were resected from each patients during surgery (n = 11), and at the same time non-tumor samples from the macroscopic tumor margin were isolated and used as the matched adjacent non-tumor tissue. All the samples were divided into two parts; one was immediately snap frozen and stored in liquid nitrogen for RNA analysis and the other was stored in formalin for pathology. Standard histopathological investigations including H&E staining were independently performed by two pathologists and clinical data were obtained from the medical records of the patients.
Cell lines and culture
The GES-1, BGC-823, MGC-803, MKN-45 and SGC-7901 human gastric carcinoma cell lines were purchased from the National Institute of Cells, Shanghai, China. The SGC7901/VCR human gastric carcinoma cell line was obtained from the State Key Laboratory of Cancer Biology and Institute of Digestive Diseases, Xijing Hospital, Fourth Military Medical University, Xian, China. Cultured cells were grown as monolayers in RPMI 1640 media supplemented with 10% fetal bovine serum (Gibco, 16000-044), 100 U/ml penicillin and 100 mg/ml streptomycin in a humidified incubator containing 5% CO2 at 37°C.
Cell proliferation assay
Cells were plated at 80% confluence and 100 nM miR-135a mimic or 100 nM negative control mimic were transfected using Lipofectamine 2000 (Invitrogen, 11668-019) for 6 h, then the cells were trypsinized and seeded into 96-well culture plates at a density of 5000 cells/well in growth medium supplemented with 10% serum. The MTT [3-(4, 5-dimethylthiazol-2-yl)-2, 5-diphenyl-tetrazoliumbromide] assay was performed 24, 48 and 72 h post-transfection and absorbance was measured using a spectrophotometer at 490 nm. Each assay was performed in triplicate with three independent replicates.
RNA extraction and miRNA qRT-PCR
RNA was extracted from the cell lines using TRIzol total RNA isolation reagent (Invitrogen, 15596-026) following the manufacturer’s protocol. Total RNA (10 ng) was reverse transcribed and mature miRNA expression was quantified using stem-loop RTCitation21 and normalized to U6 snRNA, following a standard microRNA real-time PCR assay (Roche, 04913850001) protocol on the Rotor-Gene 3000 real-time PCR Detection System using SYBR green. The ΔCt method was used for relative quantization using the formula ΔCt = Ct miR135a - Ct U6 snRNA. The expression of miR-135a (ΔΔCt) in the tumor tissues and cancer cell lines were expressed relative to the expression of miR-135a in the paired non-tumor tissues and GES-1 cells, respectively.
JAK2 RT-PCR analysis
Total RNA was used to synthesize cDNA using the cDNA Reverse Transcription Kit (Takara, 2680A) according to the manufacturer’s protocol. RT-PCR was performed using JAK2 specific primers (forward: 5′- ATGGGAATGGCCTGCCTTACGATGAC -3′ and reverse: 5′- 5′-ACTCATCTATATGGAAGACATGG -3′) and GAPDH specific primers (forward: 5′- ATGGGGAAGGTGAAGGTCG -3′ and reverse: 5′- TTACTCCTTGGAGGCCATGTG -3′).
Western blotting
Whole cell protein extracts were denatured, 50 μg was separated on 10% SDS PAGE gels, transferred to PVDF membranes, blocked in 1% FBS in TBS-T and probed overnight at 4°C with primary antibodies diluted in TBS-T. The primary antibodies were purchased from Cell Signaling Technology: JAK2, 3230; p-STAT3, 9145; cyclin D1, 2978 and Bcl-xL, 2764 and used at 1:1,000 dilutions. After incubation with the appropriate HRP-conjugated secondary antibodies, the bands were detected using Western Lightning Chemiluminescence Reagent (Millipore, WBLUR010).
Clonogenic assay
Cells were transfected with miR-135a mimic or control mimic as previously described and plated into 6 well plates at a density of 200 cells per well, incubated at 37°C for 2 weeks, fixed and stained with crystal violet. The mean ± SEM number of colonies containing > 50 cells were counted under a microscope from three independent replicates.
Statistical analysis
Data are presented as mean ± SEM of at least three independent experiments. Statistical analysis was performed using SPSS version 13. Groups were compared using one-way ANOVA and the Student t-test was used to analyze the tumor and non-tumor surgical samples; p < 0.05 was considered significant.
| Abbreviations: | ||
| 3′ UTR | = | 3′ untranslated region |
| JAK2 | = | janus kinase 2 |
| MiRNAs | = | microRNAs |
| MiRNPs | = | ribonucleoprotein complexes |
| MDR | = | multidrug resistance |
| qRT-PCR | = | quantitative polymerase chain reaction |
| SEM | = | standard error of the mean |
| TBS-T | = | Tris buffered solution containing tween 20 |
Acknowledgments
We thank Dr Xueting Cai and Dr Wuguang Lu (Laboratory of Cellular and Molecular Biology, Jiangsu Province Institute of Traditional Chinese Medicine) for their kind help and suggestions in this study. The authors are grateful to the fund support by the National Natural Science Foundation of China (30840095).
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
References
- Kato M, Slack FJ. MicroRNAs: small molecules with big roles - C. elegans to human cancer. Biol Cell 2008; 100:71 - 81; http://dx.doi.org/10.1042/BC20070078; PMID: 18199046
- Mattick JS, Makunin IV. Non-coding RNA. Hum Mol Genet 2006; 15:R17 - 29; http://dx.doi.org/10.1093/hmg/ddl046; PMID: 16651366
- Yoda M, Kawamata T, Paroo Z, Ye X, Iwasaki S, Liu Q, et al. ATP-dependent human RISC assembly pathways. Nat Struct Mol Biol 2010; 17:17 - 23; http://dx.doi.org/10.1038/nsmb.1733; PMID: 19966796
- Ahmed FE. Role of miRNA in carcinogenesis and biomarker selection: a methodological view. Expert Rev Mol Diagn 2007; 7:569 - 603; http://dx.doi.org/10.1586/14737159.7.5.569; PMID: 17892365
- Alvarez-Garcia I, Miska EA. MicroRNA functions in animal development and human disease. Development 2005; 132:4653 - 62; http://dx.doi.org/10.1242/dev.02073; PMID: 16224045
- Sayed D, Abdellatif M. MicroRNAs in development and disease. Physiol Rev 2011; 91:827 - 87; http://dx.doi.org/10.1152/physrev.00006.2010; PMID: 21742789
- Farazi TA, Spitzer JI, Morozov P, Tuschl T. miRNAs in human cancer. J Pathol 2011; 223:102 - 15; http://dx.doi.org/10.1002/path.2806; PMID: 21125669
- Patel SH, Kooby DA. Gastric adenocarcinoma surgery and adjuvant therapy. Surg Clin North Am 2011; 91:1039 - 77; http://dx.doi.org/10.1016/j.suc.2011.06.009; PMID: 21889029
- Wu WK, Lee CW, Cho CH, Fan D, Wu K, Yu J, et al. MicroRNA dysregulation in gastric cancer: a new player enters the game. Oncogene 2010; 29:5761 - 71; http://dx.doi.org/10.1038/onc.2010.352; PMID: 20802530
- Chen Y, Song Y, Wang Z, Yue Z, Xu H, Xing C, et al. Altered expression of MiR-148a and MiR-152 in gastrointestinal cancers and its clinical significance. J Gastrointest Surg 2010; 14:1170 - 9; http://dx.doi.org/10.1007/s11605-010-1202-2; PMID: 20422307
- Gao CP, Zhang ZY, Cai GH, Liu WZ, Xiao SD, Lu H. Reduced expression of miR-218 and its significance in gastric cancer. Zhonghua Zhong Liu Za Zhi 2010; 32:249 - 52; PMID: 20510072
- Nishida N, Mimori K, Fabbri M, Yokobori T, Sudo T, Tanaka F, et al. MicroRNA-125a-5p is an independent prognostic factor in gastric cancer and inhibits the proliferation of human gastric cancer cells in combination with trastuzumab. Clin Cancer Res 2011; 17:2725 - 33; http://dx.doi.org/10.1158/1078-0432.CCR-10-2132; PMID: 21220473
- Gao C, Zhang Z, Liu W, Xiao S, Gu W, Lu H. Reduced microRNA-218 expression is associated with high nuclear factor kappa B activation in gastric cancer. Cancer 2010; 116:41 - 9; PMID: 19890957
- Li WX. Canonical and non-canonical JAK-STAT signaling. Trends Cell Biol 2008; 18:545 - 51; http://dx.doi.org/10.1016/j.tcb.2008.08.008; PMID: 18848449
- Ingley E, Klinken SP. Cross-regulation of JAK and Src kinases. Growth Factors 2006; 24:89 - 95; http://dx.doi.org/10.1080/08977190500368031; PMID: 16393697
- Murray PJ. The JAK-STAT signaling pathway: input and output integration. J Immunol 2007; 178:2623 - 9; PMID: 17312100
- Yu LF, Zhu YB, Qiao MM, Zhong J, Tu SP, Wu YL. Constitutive activation and clinical significance of Stat3 in human gastric cancer tissues and cell lines. Zhonghua Yi Xue Za Zhi 2004; 84:2064 - 9; PMID: 15730617
- Vera J, Rateitschak K, Lange F, Kossow C, Wolkenhauer O, Jaster R. Systems biology of JAK-STAT signalling in human malignancies. Prog Biophys Mol Biol 2011; 106:426 - 434; http://dx.doi.org/10.1016/j.pbiomolbio.2011.06.013; PMID: 21762720
- Constantinescu SN, Girardot M, Pecquet C. Mining for JAK-STAT mutations in cancer. Trends Biochem Sci 2008; 33:122 - 31; http://dx.doi.org/10.1016/j.tibs.2007.12.002; PMID: 18291658
- Valentino L, Pierre J. JAK/STAT signal transduction: regulators and implication in hematological malignancies. Biochem Pharmacol 2006; 71:713 - 21; http://dx.doi.org/10.1016/j.bcp.2005.12.017; PMID: 16426581
- Benes V, Castoldi M. Expression profiling of microRNA using real-time quantitative PCR, how to use it and what is available. Methods 2010; 50:244 - 9; http://dx.doi.org/10.1016/j.ymeth.2010.01.026; PMID: 20109550
- Witkos TM, Koscianska E, Krzyzosiak WJ. Practical aspects of microRNA target prediction. Curr Mol Med 2011; 11:93 - 109; http://dx.doi.org/10.2174/156652411794859250; PMID: 21342132
- Hartgrink HH, Jansen EP, van Grieken NC, van de Velde CJ. Gastric cancer. Lancet 2009; 374:477 - 90; http://dx.doi.org/10.1016/S0140-6736(09)60617-6; PMID: 19625077
- Rathbone M, Rathbone B. Helicobacter pylori and Gastric Cancer. Recent Results Cancer Res 2011; 185:83 - 97; http://dx.doi.org/10.1007/978-3-642-03503-6_5; PMID: 21822821
- Bornschein J, Malfertheiner P. Gastric carcinogenesis. Langenbecks Arch Surg 2011; 396:729 - 42; http://dx.doi.org/10.1007/s00423-011-0810-y; PMID: 21611816
- Ueda T, Volinia S, Okumura H, Shimizu M, Taccioli C, Rossi S, et al. Relation between microRNA expression and progression and prognosis of gastric cancer: a microRNA expression analysis. Lancet Oncol 2010; 11:136 - 46; http://dx.doi.org/10.1016/S1470-2045(09)70343-2; PMID: 20022810
- Navarro A, Diaz T, Martinez A, Gaya A, Pons A, Gel B, et al. Regulation of JAK2 by miR-135a: prognostic impact in classic Hodgkin lymphoma. Blood 2009; 114:2945 - 51; http://dx.doi.org/10.1182/blood-2009-02-204842; PMID: 19666866
- Liu S, Guo W, Shi J, Li N, Yu X, Xue J, et al.. MicroRNA-135a contributes to the development of portal vein tumor thrombus by promoting metastasis in hepatocellular carcinoma. J Hepatol 2011; In press http://dx.doi.org/10.1016/j.jhep.2011.08.008; PMID: 21888875
- Smirnova OV, Ostroukhova TY, Bogorad RL. JAK-STAT pathway in carcinogenesis: is it relevant to cholangiocarcinoma progression?. World J Gastroenterol 2007; 13:6478 - 91; http://dx.doi.org/10.3748/wjg.13.6478; PMID: 18161917
- Santos FP, Verstovsek S. JAK2 inhibitors: what's the true therapeutic potential?. Blood Rev 2011; 25:53 - 63; http://dx.doi.org/10.1016/j.blre.2010.10.004; PMID: 21095048
- Quintás-Cardama A, Verstovsek S. New JAK2 inhibitors for myeloproliferative neoplasms. Expert Opin Investig Drugs 2011; 20:961 - 72; http://dx.doi.org/10.1517/13543784.2011.579560; PMID: 21521147