Abstract
Apoptosis is one of the core signaling pathways disrupted in pancreatic ductal adenocarcinoma (PDA). Death receptor 5 (DR5) is a member of the tumor necrosis factor (TNF)-receptor superfamily that is expressed in cancer cells. Binding of TNF-related apoptosis-inducing ligand (TRAIL) to DR5 is a potent trigger of the extrinsic apoptotic pathway, and numerous clinical trials are based on DR5-targeted therapies for cancer, including PDA. Human antigen R (HuR), an RNA-binding protein, regulates a select number of transcripts under stress conditions. Here we report that HuR translocates from the nucleus to the cytoplasm of PDA cells upon treatment with a DR5 agonist. High doses of DR5 agonist induce cleavage of both HuR and caspase 8. HuR binds to DR5 mRNA at the 5′-untranslated region (UTR) in PDA cells in response to different cancer-associated stressors and subsequently represses DR5 protein expression; silencing HuR augments DR5 protein production by enabling its translation and thus enhances apoptosis. In PDA specimens (n = 53), negative HuR cytoplasmic expression correlated with elevated DR5 expression (odds ratio 16.1, p < 0.0001). Together, these data demonstrate a feedback mechanism elicited by HuR-mediated repression of the key apoptotic membrane protein DR5.
Introduction
Pancreatic cancer, the fourth leading cause of cancer-related death in this country, has a dismal 5-y survival rate. One core phenotype shared by all pancreatic ductal adenocarcinomas (PDA) is their ability to avoid programmed cell death (apoptosis),Citation1 which aids in the increased ability of PDA cells to survive, grow and develop drug resistance mechanisms. Apoptotic modulatory genes are regulated via different mechanisms (e.g., transcriptional, post-transcriptional and post-translational events).Citation2,Citation3 Although it is well established that apoptosis is dysregulated in pancreatic tumorigenesis,Citation1 the molecular mechanism behind the aberrant apoptotic response of PDA cells is relatively unknown.
Death Receptor 5 (DR5, also known as TRAIL-receptor 2, TRAIL-R2 and TNFRSF10B) is the main, initiating component of the death factor/death receptor (extrinsic) apoptosis pathway. Upon ligand exposure, DR5 triggers apoptosis in a caspase-dependent manner and is currently being explored as a ‘druggable’ target in multiple cancers, including pancreatic cancer.Citation4 TNF-related apoptosis ligand (TRAIL) is the natural ligand for DR5 and binding induces death in malignant cell lines while normal cells are spared, making TRAIL an attractive cytotoxicity agent.Citation5,Citation6 Several TRAIL ligand variants and antibodies specific to DR5 (agonists) are currently being pursued in pre-clinical experiments and clinical trials (three trials specifically in the pancreas).Citation7 While several DR5 agonists have made their way to clinical trials, an unidentified, ubiquitous resistance mechanism is believed to be the reason why targeting these molecules have been relatively ineffective in the clinic. Multiple cancer cell lines both in culture and in vivo have proven to be resistant to TRAIL therapy.Citation8 Several hypotheses behind the cause of this resistance include: 1) mutations in the TRAIL receptorCitation9; 2) competitive binding at the receptor by decoy receptorsCitation10; and 3) inhibition of caspases and apoptosis via different gene products.Citation11 Yet, despite these hypotheses, the field has been hampered by an unexplained cancer cell resistance to DR5 targeting and the inability to define a stratifying predictive marker for DR5-directed therapy.Citation12 Thus, before we can dissect the regulatory mechanisms responsible for resistance, there is pressing need to more fully understand the post-transcriptional regulation of DR5—acutely as resistance sets in—which is the main focus of this current study.
HuR (ELAV1) is an RNA-binding protein that regulates post-transcriptional gene regulation and was previously shown to be implicated as part of pro-survival and anti-apoptotic networks.Citation2,Citation3,Citation13 HuR is primarily localized in the nucleus. In response to specific stressors, HuR can transiently traffic to the cytoplasm, where it facilitates the stabilization and translation of mRNAs encoding key survival proteins.Citation14 Mechanistically, HuR regulates mRNA cargos that typically contain U- or AU-rich sequences in the 3′-untranslated region (UTR).Citation14 HuR affects several of these target mRNAs specifically related to the tumorigenic process including VEGF, p21, cyclins A and B1, COX-2, HIF-1α and p53 mRNAs.Citation3,Citation13 Notably, there are also examples of HuR-repressed mRNAs, such as those that encode IGF-1RCitation15 and p27.Citation16 HuR associates with the 5′UTR instead of the 3′UTR of these two mRNAs and represses the translation of the encoded proteins. In general, the influence of HuR on target mRNAs provides a survival advantage for the cell.Citation17
In relation to anti-cancer drug efficacy, HuR was recently linked to the historically important nucleoside analog, gemcitabine (Gemzar), used for the treatment of pancreatic cancer. Gemcitabine continues to be backbone of standard of care and experimental regimens for the treatment of PDA, regardless of pathologic staging.Citation18 The enzyme deoxycytidine kinase (dCK) phosphorylates and metabolizes the prodrug, gemcitabine, to its active metabolites, which then target cancer cells for cell death. HuR, has been linked to gemcitabine metabolism, by binding to dCK mRNA and affecting dCK protein levels thereby changing gemcitabine efficacy in PDA cells.Citation19 These data have clinical implications as high cytoplasmic HuR status in patient samples correlated strongly with increased overall survival in patients receiving gemcitabine-based therapy.Citation19,Citation20
In cancer, HuR provides an advantage to a cell under stress by supporting an anti-apoptotic network.Citation13,Citation21 For example, HuR can bind to the mRNAs encoding the pro-survival proteins SIRT1, Bcl-2, Mcl-1 and ProTα and enhance their expression.Citation13 More recently, HuR was shown to mediate the expression of the intrinsic cellular caspase inhibitor (XIAP), and this connection was linked to HuR’s ability to protect against cell death.Citation22 Another study showed that HuR regulates an anti-apoptotic program via its direct effect on MEK-1.Citation21 These studies underscore the importance of HuR activation as a master post-transcriptional regulator of pro-survival/anti-apoptotic pathways.Citation13 Previous research has shown that caspase-3 cleaves HuR and potentially results in increased apoptotic signaling.Citation23 However, HuR’s regulatory role in the extrinsic apoptotic pathway has not been clearly elucidated. Thus in order to understand completely the connection between HuR and the extrinsic apoptotic pathway, we investigated HuR’s interaction primarily with DR5, the main trigger of this pathway.
Results
HuR associates with the 5′ UTR of DR5 mRNA after stress
Baseline levels of DR5 were established in PL5, PL45 and MiaPaCa2 PDA cell lines by RT-qPCR after RNA extraction. PL45 cells had the highest level of endogenous DR5 expression but all tested cell lines had some baseline DR5 expression (). The association of HuR with endogenous DR5 mRNA was assessed by ribonucleoprotein-immunoprecipitation (RIP) assays as previously described.Citation19,Citation26 The results were normalized to GAPDH or 18S, both accepted internal controls for total RNA levels.Citation19,Citation27 As a positive control, cells were treated with gemcitabine and the levels of dCK, a known mRNA binder of HuR, were shown to be upregulated (12-fold) in the HuR-IP group compared with the IgG (control) group.Citation19 Gemcitabine treatment also resulted in an increase in DR5 mRNA levels in the HuR-IP group (, top). Similarly, after treatment with DR5/TRAILR2 monoclonal antibody, DR5 mRNA levels were strongly enriched (12- to 48-fold) in the HuR-IP group as compared with the IgG IP group (, bottom). By isolating RNA bound to HuR and comparing it to RNA bound to IgG, this data demonstrates that DR5 mRNA is bound to HuR when stressed by either DR5/TRAILR2 antibody or by gemcitabine.
Figure 1. HuR binds DR5 mRNA and affects DR5 levels. A. RT-qPCR of cell line RNA analyzing baseline DR5 levels. B. After treatment with gemcitabine (top) or DR5/TRAILR2 monoclonal antibody (R&D Systems, Minneapolis, MN, USA) (bottom), there are increased levels of DR5 in the HuR IP group (white bars) compared with the IgG IP group (black bars). Relative binding of HuR to mRNA was calculated based on RT-qPCR after RNP-IP and normalized to GAPDH, an established internal control. C. Treatment with siRNA causes an increase in levels of DR5 mRNA after 24 h. Data were calculated using RT-qPCR and normalized to 18S, a ubiquitously expressed rRNA used as a control. D. PARP-1 cleavage in cells exposed to DR5 antibody for 4 h; β-actin used as a loading control.
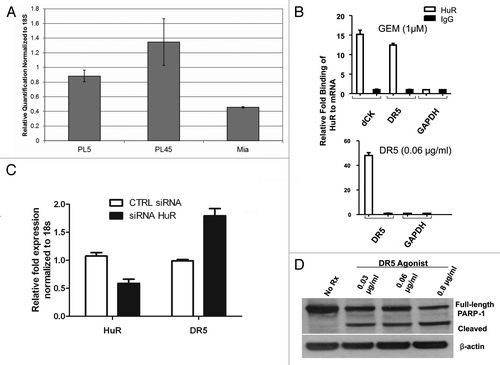
Treatment of MiaPaCa2 cells with HuR siRNA resulted in a 50% reduction in HuR mRNA levels (). This modest reduction in HuR levels led to an upregulation of DR5 mRNA 24 h after transfection (). At 36 h after treatment with HuR siRNA, the level of DR5 mRNA is similar to the levels seen after treatment with control siRNA (data not shown) signifying that this increase in DR5 mRNA may be a transient effect. PARP-1 cleavage was utilized to validate that treatment with DR5/TRAILR2 monoclonal antibody (0.03–0.08 μg/ml) triggered apoptosis ().
Silencing HuR upregulates DR5 protein expression in PDA cells
Having discovered an association between HuR and DR5 mRNA, we next examined HuR regulation of DR5 protein expression. MiaPaCa2, BxPC3 and PL5 cell lines were treated with HuR siRNA. Two PDA cell lines, MiaPaCa2 and BxPC3, exhibited a dramatic 4-fold increase in DR5 protein levels at 48 h post silencing when HuR levels are decreased by HuR siRNA treatment (). However, HuR silencing in PL5 cells did not affect DR5 protein levels compared with control transfections (). To confirm these results, flow cytometry was used to analyze the cell surface expression of DR5. In both the MiaPaCa2 and PL45 cell lines, there was an approximately 10% higher mean fluorescence intensity (MFI) of DR5 in the HuR siRNA group compared with the control siRNA group (). This trend was lost in the PL5 cell line with no difference in the MFI between the two groups (), consistent with the Immunoblot data (). These results show that in three of the PDA lines studied (i.e., except for PL5 cells where HuR did not regulate DR5 protein levels) HuR has an inverse relationship with DR5 protein expression. Similarly these findings appear to correlate with DR5-agonist sensitivity and resistance.
Figure 2. HuR negatively regulates DR5 protein levels. A. Western immunoblot of DR5 protein levels in control and HuR siRNA transfected cell lines (PL45, MiaPaCa2 and PL5). Shown is the 48 h post transfection time point. B. PDA cell lines at 48 h post tranfection with control or HuR siRNA were immunostained with PE-anti-DR5 mAb or PE-isotype control (CTLR) and analyzed by flow cytometry. Representative results are displayed as mean fluorescence intensity (MFI). C. Same as (B), showing results using PL5 cells in two separate experiments.
HuR binds to the 5′ UTR of DR5 mRNA
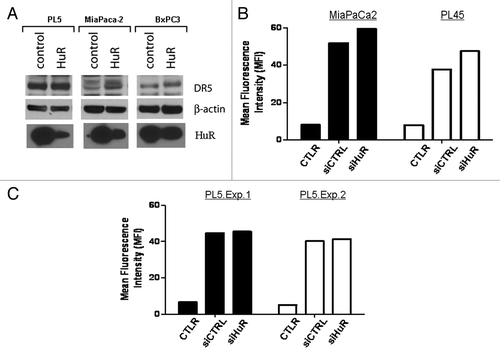
In order to determine if HuR regulates DR5 mRNA through direct interaction and to map the region of interaction, the DR5 5′-and 3′-UTRs () were subcloned into luciferase expression constructs and transfected into MiaPaCa2 cells. A positive control construct was designed using a CAG promoter to ubiquitously drive luciferase. The 5′UTR-DR5 construct (pCHEK.5UTR) showed decreased luciferase expression () compared with both an empty vector control and two previously defined DR5–3′UTR fragments (pCHEK.3UTRa and pCHEK.3UTRb) indicating that HuR regulated DR5 expression through the DR5 5′UTR.Citation28 MiaPaCa2 cells were then treated with DR5/TRAIL antibody causing further reduction of luciferase activity at the 5′UTR (, left). These results were confirmed using RT-qPCR with a luciferase probe; the lowest luciferase expression was in the group correlating with binding to the 5′UTR (, right). Next the cells were treated with HuR siRNA that showed a modest decrease in the luciferase activity in an untreated group and a reversal of the trend in the group treated with DR5 antibody (). RIP-SEQ data corroborated an interaction between HuR and DR5 mRNA at the 5′UTR (see methods for details and ) following treatment of the cells with the established HuR stressor, gemcitabine.Citation19
Figure 3. DR5 expression is controlled through the 5′UTR. A. Design and schematic of DR5 mRNA and the luciferase DR5 reporter constructs. The control is the top construct designed with a CaG repeat sequence. B. Results of luciferase assay in which cells were transfected with the luciferase constructs shown in 3A demonstrating that HuR binds to the 5′UTR region. C. Left: Results of luciferase assays in cells treated with 0.06 μg/ml of DR5 agonist for 4 h. Right: RT- qPCR analysis of luciferase mRNA in cells treated under the same conditions as described in the left panel. D. MiaPaCa2 cells were treated with HuR siRNA or control siRNA and co-transfected with luciferase constructs demonstrating that luciferase activity was lower in the HuR siRNA group. In the group treated with DR5/TRAILR2 antibody, the trend was reversed. See for all DR5 mRNA:HuR protein interactions.
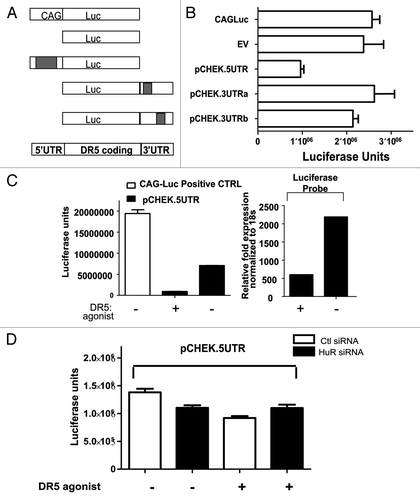
Table 1. HuR binds to the DR5 5′-UTR following gemcitabine treatment
Treatment of pancreatic cancer cells with DR5 agonist increases cytoplasmic HuR abundance and HuR cleavage
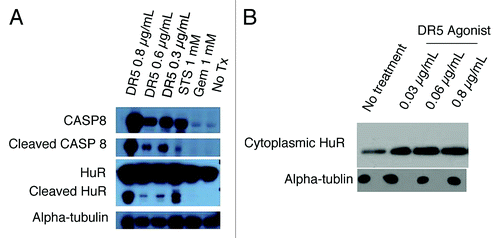
PDA cells were treated with staurosporine (positive control) or varying doses of DR5 agonist; 24 h later, analysis of whole-cell lysates by western blotting revealed cleavage of HuR (CP1 24 kDA) and Caspase 8 (CASP8) (p43/p41) at several doses of DR5 agonists (); by contrast, even at extremely high doses (above any accepted clinical doses) of gemcitabine (1 mM), there was no cleavage of HuR. Twenty-four h after cells were treated with staurosporine [1 mM, included as an established positive controlCitation23] or DR5 agonist at varying doses, cytoplasmic extracts were obtained using digitonin solution. Western blot analysis revealed that HuR translocated to the cytoplasm in a dose-dependent manner (). In the extrinsic apoptotic pathway, CASP8 leads to activation of caspase-3 (CASP3), which has been shown to cleave HuR resulting in CP1 and CP2.Citation23 Our data show that at high doses of DR5 agonist, HuR is cleaved, likely in a caspase-dependent manner (),Citation23 and at treatment with lower doses of DR5/TRAILR2 antibody, HuR translocates to the cytoplasm only without cleavage ().
Figure 4. Effect of treating PDA cells with DR5 agonist upon HuR. A. Western blot analysis of whole-cell protein lysates prepared from PDA cells after treatment with Staurosporine (positive control) or varying doses of DR5 agonist. B. Western blot analysis of HuR levels in PDA cytoplasmic lysates after treatment with DR5 agonist (0.03 µg/ml and 0.06µg/ml). Alpha-tubulin is used as a cytoplasmic loading control.
Functional relevance of HuR-regulation upon DR5 agonist treatment of pancreatic cancer cells
Since silencing HuR expression () increases DR5 protein levels, we hypothesized that silencing HuR expression could alter the effectiveness of the extrinsic apoptotic pathway after agonism with DR5/TRAIL. Using MiaPaCa2 cells, HuR was silenced by 88% (). Silencing HuR led to an ~11% increase in apoptosis as measured by ANNEXIN V staining in MiaPaCa2 cells exposed to DR5 agonist (0.06 μg/ml, 8 h; ). To determine the results of treating multiple cell lines with DR5 agonist, a long-term drug sensitivity assay was performed. After treating MiaPaCa2, PL45, BxPC3 and PL5 cells with a range of DR5 agonist doses, PL5 cells had the greatest resistance to the drug (). Even in the presence of high doses, PL5 cells never approached a 50% inhibitory concentration (IC50); however, MiaPaCa2 cells had an IC50 of ~0.03 μg/mL while PL45 cells had an IC50 of ~0.09 μg/mL (). The resistance of PL5 cells to the DR5 agonist may be related to their ability to avoid HuR regulation of DR5 expression (), consistent with our finding above ().
Figure 5. Influence of HuR on DR5 toxicity. A. HuR level as measured by RT-qPCR after treatment with HuR siRNA or control siRNA. B. Levels of cell death after silencing HuR in MiaPaCa2 cells treated with DR5 agonist (0.06 μg/ml) as measured by Annexin V assay. C. Percentages of cell survival in four PDA cell lines after treatment with DR5 agonists, as assessed using Picogreen analysis. D. Percentages of cell survival after overexpression of HuR and treatment with DR5 agonists, as assessed using Picogreen analysis.
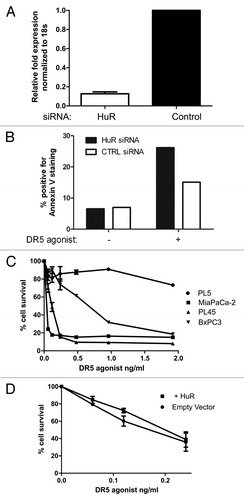
In order to verify that HuR affects DR5 sensitivity, HuR was overexpressed in MiaPaca2 cells using GFP tagged HuR plasmid and then a long-term drug sensitivity assay was performed. Cells with increased levels of HuR had a slight decrease in sensitivity to DR5/TRAILR2 antibody compared with the control group ().
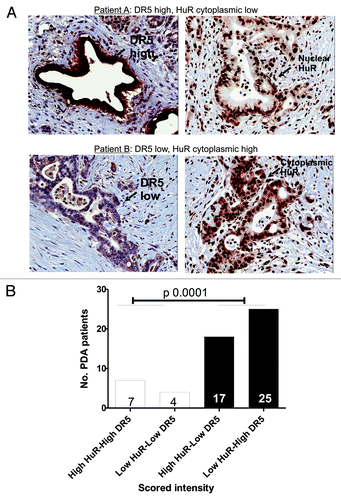
To begin to test the clinical relevance of our findings, we used a set of PDA clinical specimens and identified a strong correlation between HuR cytoplasmic expression and DR5 levels in nearly 80% (42 of 53) of the specimens (see Materials and Methods for description of scoring, as described previously for HuR).Citation19 PDA samples were obtained from resected PDA patients at TJU under IRB consent. We have previously published a similar percentage of cytoplasmic HuR positive patients in naively treated resected patients.Citation19,Citation20 Cytoplasmic levels of HuR in these PDA patients are most likely due to hypoxic events induced by the tumor microenvironment.Citation29,Citation30 The top panel () is a representative example of a PDA specimen with low or absent cytoplasmic HuR (right) that has high DR5 intensity (left). The bottom panel is a representative example of a PDA specimen with elevated cytoplasmic HuR (right) that has low DR5 intensity (left) (). This observation is strikingly consistent with our in vitro data () and provides evidence for a primary role of HuR in the extrinsic apoptotic pathway (). Statistical analysis revealed an odds ratio of 16.1 (4.1,63.2; p < 0.0001) ().
Figure 6. Correlation between HuR and DR5 in patient samples. A. The levels of cytoplasmic HuR and DR5 were assessed by immunohistochemistry (20× magnifications). The top panel (patient A) is a representative PDA specimen with low or absent cytoplasmic HuR (right) and a high DR5 intensity in the same tumor cells (left). The bottom panel (patient B) demonstrates a PDA specimen with elevated cytoplasmic HuR (right) in the tumor cells that also have a low DR5 intensity in the same cells (left) (). B. Scores from 53 PDA specimens were calculated. Significant correlation was observed between DR5 intensity and cytoplasmic HuR levels (p 0.0001).
Discussion
We have presented evidence that HuR plays a role in a core apoptotic pathway disrupted in PDA cellsCitation1 at least in part by binding to the DR5 mRNA and regulating DR5 protein expression. Our data demonstrate that HuR binds to DR5 mRNA at the 5′UTR as previously described for other HuR-repressed targets such as IGF-1R and p27 mRNAsCitation16 thus inhibiting DR5 translation. This is supported by the data that low levels of HuR obtained by treating cells with HuR siRNA correlates with higher levels of DR5 protein.
Furthermore, our data support the previous notion that HuR enhances an anti-apoptotic network.Citation13 Cells depleted of HuR have increased apoptosis (). In addition, overexpression of HuR correlates with decreased sensitivity to DR5/TRAILR2 antibody (). Therefore, HuR may act to acutely inhibit the extrinsic apoptotic pathway through its inhibition of DR5 protein production. This, in turn, may lead to decreased initiation of the caspase cascade. Our results indicate that when DR5 is severely stressed (high doses DR5/TRAILR2 antibody), HuR is cleaved based on previous work in a caspase-dependent manner.Citation23 Therefore, by inhibiting DR5 protein expression, HuR may in fact be protecting itself from caspase-dependent cleavage. This notion would support HuR as functioning in a feedback mechanism.
In a scenario of physiologic relevant stress (such as hypoxia) DR5 is engaged by its ligand and following this engagement, elevated levels of cytoplasmic HuR may act in a negative feedback loop and downregulate DR5 expression levels. These data may be most relevant under harsh tumor microenvironment conditions, where PDA specimens with high cytoplasmic HuR levels correlate with a poor pathologic feature (T staging).Citation20 In this circumstance, downregulation by HuR may be advantageous for the cancer cell to escape apoptosis so these cells may be selectively increased in the tumor.Citation3,Citation13 The precise role and/or relevance of HuR cleavage (CP1) in this pathway is currently under investigation, although others have linked this event to pp32’s induction of apoptosis.Citation23,Citation31,Citation32
Clinically, we have previously shown that cytoplasmic HuR levels in PDA specimens can be a useful predictive marker for gemcitabine therapy.Citation19,Citation20 Based on our current results, the cytoplasmic HuR levels could also be a predictive marker for DR5-targeted therapy, given the inverse relationship between cytoplasmic HuR abundance and DR5 intensity in patient tumors (p < 0.0001, ). For example, low cytoplasmic HuR levels in a patient’s tumor could predict a positive response to DR5-based therapy, while elevated cytoplasmic HuR levels would be better stratified to gemcitabine therapy.Citation19,Citation20 In addition, the correlation between low DR5 and high HuR seen in patient samples supports the data in vitro that HuR represses DR5 protein expression. Of note, one cell line (PL5) demonstrated resistance to DR5-agonist and HuR regulation (Figs. Two and 5C). These cells may have either a defect in the extrinsic pathway or may represent cells that have an HuR-independent drug resistance mechanism.
Ongoing experiments include identifying if the association of HuR with DR5 mRNA leading to the regulation of DR5 is dependent on cell lineage, the type of stressor and/or the contribution of other RNA-binding proteins or microRNAs.Citation28 These studies will uncover additional mechanistic aspects of post-transcriptional gene regulation of DR5 and its impact upon DR5 protein levels. Other ongoing work includes attempting to further understand the function of CP1 in cells and uncover the exact signal by which CP1 may provide feedback to HuR and DR5 mRNA.Citation32
Our data along with others show that HuR can assert its influence and be influenced at many points in this powerful apoptotic pathway.Citation13 Confirmatory studies in other tumor systems will determine if all cancer cells select for HuR’s ability to modulate or put the brakes on DR5-directed signaling in an effort to avoid apoptosis. Based on the results uncovered here and the mounting evidence that HuR modulates apoptosis,Citation23,Citation31 further investigation of HuR as a predictive marker and target in PDA cells is warranted.
Materials and Methods
Cell culture
PDA cell lines MiaPaCa2, BxPC3, PL45 and PL5 were purchased from the ATCC and cultured at 37°C in Dulbecco’s modified Eagle’s medium (Gibco/Invitrogen) supplemented with 10% fetal bovine serum (Gibco/Invitrogen) 1% L-glutamine (Gibco/Invitrogen), and 1% penicillin-streptomycin (Invitrogen).
Transfections
PCR products corresponding to the 5′ and 3′UTR of DR5 were purified and cloned into a Promega psiCheck2 luciferase vector (Promega). MiaPaCa2 cells were then plated to 60% confluency in 6-well plates and transfected using Lipofectamine 2000 (Gibco/Invitrogen) and Optimem (Gibco/Invitrogen); double transfections were performed where indicated. After 48 h, cells were lysed and luciferase activity was assayed using a luciferase assay reporter kit (Promega). For double transfections, the cells were transfected first with 1 µM of HuR siRNA or control siRNA followed 24 h by the luciferase construct transfection described above.
For siRNA analysis, 8 million MiaPaCa2 cells were plated to ~50% confluency in T75 flasks. The cells were washed with Opti-mem (Gibco/Invitrogen) and transfected with 1 μM of HuR siRNA or control siRNA (Applied Biosystems/Ambion) using Oligofectamine (Invitrogen).
For the Green Fluorescent Protein (GFP)-tagged HuR vector (donated to us from Dr. Imed Gallouzi, McGill University), we performed stable transfections as previously described.Citation19
Ribonucleoprotein immunoprecipitation (RIP) analysis
Twenty million MiaPaCa2 cells were plated to ~65% confluency in T-150 flasks. Cells were treated with 1 μM Gemcitabine (Eli Lilly), 75 μM ABT-888 (Abbot Laboratories), and/or 0.06 μg/mL DR5/TRAILR2 monoclonal antibody (R&D Systems) for 4 h. Cells were then trypsinized and washed with DPBS (Gibco/Invitrogen). After lysis, immunoprecipitation was performed using anti-HuR and IgG antibodies (MBL International) as described.Citation19 The RNA was quantified by RT-qPCR using an ABI 7500 machine; GAPDH mRNA was included as loading control.Citation19
Whole cell and cytoplasmic protein extracts for western blotting
MiaPaCa2 cells were treated for 3 h with 1 mM gemcitabine (Eli Lilly), 1 mM staurosporine (STS) (Sigma Aldrich), and 0.03 μg/mL, 0.06 μg/mL or 0.8 μg/mL DR5/TRAILR2 monoclonal antibody (R&D Systems). Whole-cell lysates were fractionated by using 10% Bis-Tris Gels (Gibco/Invitrogen). After transfer, membranes were incubated with a primary polyclonal antibody recognizing Caspase-8 (Cell Signaling) or a primary monoclonal antibody recognizing HuR (Santa Cruz Inc.) at a dilution of 1:1000 in 5% BSA (Cell Signaling). The corresponding secondary antibodies were used at 1:10,000 dilutions (Santa Cruz Inc.).
To study the influence of TRAIL2/DR5 on HuR subcellular localization, MiaPaCa2 cells were plated to ~70% confluency in T-75 flasks and treated with 0, 0.03, 0.06, or 0.8 μg/mL of TRAILR2/DR5 monoclonal antibody (DR5 agonist, R&D Systems) for 4 h. Cytoplasmic protein lysates were collected using the NE-PER Nuclear and Cytoplasmic Extraction Kit (Thermo Scientific). Lysates were loaded onto 10% Bis-Tris gels. After electrophoresis and transfer, membranes were incubated with primary anti-HuR mouse antibody (at a dilution of 1:1000) for 12 h in 5% milk. Secondary antibody incubations (1:10,000 goat anti-mouse antibodies) were performed for 1 h in 5% milk. Alpha tubulin served as loading control.
Expression of DR5 in PDA cells after HuR silencing
Pancreatic cancer cell lines cultured for 48 h following transfection with HuR or control siRNA were harvested with TrypLE Express (Invitrogen), immunostained with a phycoerythrin (PE)-conjugated anti-human DR5 (clone DJR2–4) mAb (BioLegend) or PE-conjugated isotype control antibody, and analyzed for DR5 expression by flow cytometry. Cells were analyzed using FlowJo software (Tree Star, Inc.), calculating the relative mean fluorescence intensity (MFI) of each histogram.
RNA isolation and qPCR analysis after HuR siRNA
RNA was isolated using the RNA minikit (Gibco/Invitrogen) and was used for RT-qPCR analysis as explained above, with 10–75 ng of cDNA per reaction using primer pairs for 18S (internal control), HuR and DR5 mRNAs.
Membrane bound protein isolation and DR5 western blotting
For protein isolation after HuR siRNA transfection, cells were pelleted and resuspended in 50 μl RIPA buffer supplemented with protease inhibitors and 10 mM Iodoacetamide, and placed on ice for 2 h with vortexing every 30 min. Cells were then pelleted at 12,000 rpm for 10 min and the supernatant was transferred into a fresh tube for protein quantitation (BCA assay). Lysates were size-fractionated on a 4–12% Bis-Tris gel (Gibco/Invitrogen) and transferred for 16 h, whereupon membranes were blocked and incubated with DR5 Ab (1:1000, Pro-Sci Cat# 2019).
Cell death assessment
Eight million MiaPaCa2 cells were plated to ~50% confluency in T75 flasks. Cells were washed with Opti-mem and transfected with 1 μM of HuR or control siRNA. Five hours later, cells were further cultured with 10% fetal bovine serum with no additional treatment or with 0.8 μg/mL of TRAILR2/DR5 monoclonal antibody for 36 h.
For ANNEXIN V staining, MiaPaCa2 cells at 60% confluency were transfected with 1 µM of HuR siRNA or control siRNA; 48 h later, cells were treated with 0.06 µg/mL of TRAILR2/DR5 monoclonal antibody (R&D Systems) for 8 h compared with control cells. Apoptotic activity was then measured using the Vybrant Apoptosis Assay Kit #10 (Invitrogen) and calculated using flow cytometry.
Pico-Green analysis
Cells were plated at a confluence of 1 × 104 cells per well in a 96-well plate. They were then subjected to multiple treatments of the DR5 agonist ranging from 0.06 μg/mL to 1.92 μg/mL. For Pico-Green analysis, cells were washed twice in PBS, lysed using deionized water, and stained using Quant-iTTM PicoGreen® dsDNA reagent (Invitrogen) at 1:200. The plates were analyzed using a plate reader.
Patient samples and immunohistochemical staining (HuR and DR5)
The Thomas Jefferson Office of Human Research which includes the Thomas Jefferson University-Institutional Review Board (IRB) approval and informed consent were obtained for all samples from patients included in this study. This approval was given in written form for all patients used in this study. All samples were blinded and were in complete compliance with IRB. We performed these studies under an IRB approved tissue banking protocol. Formalin-fixed, paraffin-embedded blocks were processed as previously described.Citation19,Citation20 Immunostaining was performed on 53 resected PDA specimens from the Thomas Jefferson University pathology archives, with IRB approval. Patients received gemcitabine, alone or in combination with Xeloda, radiation therapy or no treatment. DR5 and HuRCitation19 antibodies were applied to slides and incubated as described.Citation19 Staining intensity (strong vs. weak) and cellular localization (nuclear vs. cytoplasmic) were scored. Based on the percentage of stained cells (> 70% vs. < 70%) the expression was scored as described. Specifically for DR5, staining intensity and the proportion of immunopositive cells were examined. Intensity of staining was graded on a scale range from 0 to 3, according to the following semi-quantitative assessment: 0 = no detectable staining, 1 = weak staining, 2 = moderate staining, 3 = strong staining. The percentage of immunopositive cells was scored as a percentage of positive cells present in 200 tumor cells and divided in four categories: 0 ≤ 5%; 1 = 6–30%; 2 = 31–50%; 3 ≥ 50%. The final staining score was derived from the sum of the intensity score added to the percentage of staining cells score. Cases with score equal to or higher than 7 were considered as showing high DR5 expression.Citation19,Citation20,Citation24
RIP-sequencing
Less than 100 ng of RNA was sequenced on the SOLiD 4 sequencer (Life Technologies) following the manufacturer’s protocols. Sequence reads were mapped to the human genome assembly hg19 using the BWA algorithm.Citation25 Each read’s own quality value was used to perform quality trimming. Additionally a maximum of two mismatches were allowed, and only those sequence reads uniquely mapping to the genome were further considered for expression analysis. The uniquely mapped sequence reads were aligned to the coding regions of the ENSEMBL genome build 37.The sequencing was performed on the SOLiD in the Cancer Genomics Genomics Lab, TJU.
To estimate the HuR occupancy at each transcript, the following scheme was used. For each transcript we determined the total number of uniquely mapped reads intersecting with its exon. The resulting read count for exon was normalized to the length of the corresponding coding region for which it mapped to resulting in the number of sequence per exon length. These values were further normalized to the corresponding value of the 5–28S ribosomal subunit to normalize the sequence value between the gemcitabine treated and untreated samples. The base-2 logarithm was calculated by the 5–2S normalized value of gemcitabine treated relative to the non-treated sample.
For genes to have significant differential HuR occupancy, they were required to meet a criteria that required minimum number of reads at a given site as well as a minimum of 2x differential expression between HuR-IP gemcitabine treated and HuR-IP untreated (control) (note: for these RIP-assays, we utilized an established HuR stressor, gemcitabine).Citation19
Additional material
Download Zip (60.9 KB)Acknowledgments
J.R.B. and A.K.W. are supported by a W.W. Smith Charitable Trust grant, a Research Scholar Grant (American Cancer Society, ACS-RSG and ACS-IRG 08-060-01 grants), a Pancreatic Cancer Action Network-AACR grant, and support from "Fund A Cure." M.G. was supported by the NIA-IRP, NIH.
Notes
† These authors contributed equally to this work.
References
- Jones S, Zhang X, Parsons DW, et al. Core signaling pathways in human pancreatic cancers revealed by global genomic analyses. Science (New York, NY 2008;321(5897):1801-6.
- López de Silanes I, Fan J, Yang X, Zonderman AB, Potapova O, Pizer ES, et al. Role of the RNA-binding protein HuR in colon carcinogenesis. Oncogene 2003; 22:7146 - 54; http://dx.doi.org/10.1038/sj.onc.1206862; PMID: 14562043
- López de Silanes I, Lal A, Gorospe M. HuR: post-transcriptional paths to malignancy. RNA Biol 2005; 2:11 - 3; http://dx.doi.org/10.4161/rna.2.1.1552; PMID: 17132932
- Yang A, Wilson NS, Ashkenazi A. Proapoptotic DR4 and DR5 signaling in cancer cells: toward clinical translation. Current opinion in cell biology.
- Ashkenazi A, Pai RC, Fong S, Leung S, Lawrence DA, Marsters SA, et al. Safety and antitumor activity of recombinant soluble Apo2 ligand. J Clin Invest 1999; 104:155 - 62; http://dx.doi.org/10.1172/JCI6926; PMID: 10411544
- Walczak H, Miller RE, Ariail K, Gliniak B, Griffith TS, Kubin M, et al. Tumoricidal activity of tumor necrosis factor-related apoptosis-inducing ligand in vivo. Nat Med 1999; 5:157 - 63; http://dx.doi.org/10.1038/5517; PMID: 9930862
- Rajeshkumar NV, Rasheed ZA, García-García E, López-Ríos F, Fujiwara K, Matsui WH, et al. A combination of DR5 agonistic monoclonal antibody with gemcitabine targets pancreatic cancer stem cells and results in long-term disease control in human pancreatic cancer model. Mol Cancer Ther 2010; 9:2582 - 92; http://dx.doi.org/10.1158/1535-7163.MCT-10-0370; PMID: 20660600
- Di Pietro R, Zauli G. Emerging non-apoptotic functions of tumor necrosis factor-related apoptosis-inducing ligand (TRAIL)/Apo2L. J Cell Physiol 2004; 201:331 - 40; http://dx.doi.org/10.1002/jcp.20099; PMID: 15389537
- Lee SH, Shin MS, Kim HS, Lee HK, Park WS, Kim SY, et al. Somatic mutations of TRAIL-receptor 1 and TRAIL-receptor 2 genes in non-Hodgkin’s lymphoma. Oncogene 2001; 20:399 - 403; http://dx.doi.org/10.1038/sj.onc.1204103; PMID: 11313970
- Pan G, Ni J, Wei YF, Yu G, Gentz R, Dixit VM. An antagonist decoy receptor and a death domain-containing receptor for TRAIL. Science (New York, NY 1997;277(5327):815-8.
- Deveraux QL, Roy N, Stennicke HR, Van Arsdale T, Zhou Q, Srinivasula SM, et al. IAPs block apoptotic events induced by caspase-8 and cytochrome c by direct inhibition of distinct caspases. EMBO J 1998; 17:2215 - 23; http://dx.doi.org/10.1093/emboj/17.8.2215; PMID: 9545235
- Mahalingam D, Oldenhuis CN, Szegezdi E, et al. Targeting Trail Towards the Clinic. Current drug targets.
- Abdelmohsen K, Lal A, Kim HH, Gorospe M. Posttranscriptional orchestration of an anti-apoptotic program by HuR. Cell cycle (Georgetown, Tex 2007;6(11):1288-92.
- Hinman MN, Lou H. Diverse molecular functions of Hu proteins. Cell Mol Life Sci 2008; 65:3168 - 81; http://dx.doi.org/10.1007/s00018-008-8252-6; PMID: 18581050
- Meng Z, King PH, Nabors LB, Jackson NL, Chen CY, Emanuel PD, et al. The ELAV RNA-stability factor HuR binds the 5′-untranslated region of the human IGF-IR transcript and differentially represses cap-dependent and IRES-mediated translation. Nucleic Acids Res 2005; 33:2962 - 79; http://dx.doi.org/10.1093/nar/gki603; PMID: 15914670
- Kullmann M, Göpfert U, Siewe B, Hengst L. ELAV/Hu proteins inhibit p27 translation via an IRES element in the p27 5’UTR. Genes Dev 2002; 16:3087 - 99; http://dx.doi.org/10.1101/gad.248902; PMID: 12464637
- Abdelmohsen K, Gorospe M. Posttranscriptional regulation of cancer traits by HuR. Wiley interdisciplinary reviews;1(2):214-29.
- Burris HA 3rd, Moore MJ, Andersen J, Green MR, Rothenberg ML, Modiano MR, et al. Improvements in survival and clinical benefit with gemcitabine as first-line therapy for patients with advanced pancreas cancer: a randomized trial. J Clin Oncol 1997; 15:2403 - 13; PMID: 9196156
- Costantino CL, Witkiewicz AK, Kuwano Y, Cozzitorto JA, Kennedy EP, Dasgupta A, et al. The role of HuR in gemcitabine efficacy in pancreatic cancer: HuR Up-regulates the expression of the gemcitabine metabolizing enzyme deoxycytidine kinase. Cancer Res 2009; 69:4567 - 72; http://dx.doi.org/10.1158/0008-5472.CAN-09-0371; PMID: 19487279
- Richards NG, Rittenhouse DW, Freydin B, et al. HuR status is a powerful marker for prognosis and response to gemcitabine-based chemotherapy for resected pancreatic ductal adenocarcinoma patients. Annals of surgery 2010;252(3):499-505; discussion -6.
- Wang PY, Rao JN, Zou T, Liu L, Xiao L, Yu TX, et al. Post-transcriptional regulation of MEK-1 by polyamines through the RNA-binding protein HuR modulating intestinal epithelial apoptosis. Biochem J 2010; 426:293 - 306; http://dx.doi.org/10.1042/BJ20091459; PMID: 20001965
- Durie D, Lewis SM, Liwak U, Kisilewicz M, Gorospe M, Holcik M. RNA-binding protein HuR mediates cytoprotection through stimulation of XIAP translation. Oncogene 2011; 30:1460 - 9; http://dx.doi.org/10.1038/onc.2010.527; PMID: 21102524
- Mazroui R, Di Marco S, Clair E, von Roretz C, Tenenbaum SA, Keene JD, et al. Caspase-mediated cleavage of HuR in the cytoplasm contributes to pp32/PHAP-I regulation of apoptosis. J Cell Biol 2008; 180:113 - 27; http://dx.doi.org/10.1083/jcb.200709030; PMID: 18180367
- Vigneswaran N, Baucum DC, Wu J, Lou Y, Bouquot J, Muller S, et al. Repression of tumor necrosis factor-related apoptosis-inducing ligand (TRAIL) but not its receptors during oral cancer progression. BMC Cancer 2007; 7:108; http://dx.doi.org/10.1186/1471-2407-7-108; PMID: 17592646
- Li H, Durbin R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics 2009; 25:1754 - 60; http://dx.doi.org/10.1093/bioinformatics/btp324; PMID: 19451168
- Brody JR, Gonye, G.E. HuR’s Role in Gemcitabine Efficacy: An exception or opportunity? WIREs RNA 2010.
- Wang W, Furneaux H, Cheng H, Caldwell MC, Hutter D, Liu Y, et al. HuR regulates p21 mRNA stabilization by UV light. Mol Cell Biol 2000; 20:760 - 9; http://dx.doi.org/10.1128/MCB.20.3.760-769.2000; PMID: 10629032
- Kandasamy K, Kraft AS. Proteasome inhibitor PS-341 (VELCADE) induces stabilization of the TRAIL receptor DR5 mRNA through the 3′-untranslated region. Mol Cancer Ther 2008; 7:1091 - 100; http://dx.doi.org/10.1158/1535-7163.MCT-07-2368; PMID: 18483298
- Mazan-Mamczarz K, Hagner PR, Corl S, Srikantan S, Wood WH, Becker KG, et al. Post-transcriptional gene regulation by HuR promotes a more tumorigenic phenotype. Oncogene 2008; 27:6151 - 63; http://dx.doi.org/10.1038/onc.2008.215; PMID: 18641687
- Levy NS, Chung S, Furneaux H, Levy AP. Hypoxic stabilization of vascular endothelial growth factor mRNA by the RNA-binding protein HuR. J Biol Chem 1998; 273:6417 - 23; http://dx.doi.org/10.1074/jbc.273.11.6417; PMID: 9497373
- von Roretz C, Gallouzi IE. Protein kinase RNA/FADD/caspase-8 pathway mediates the proapoptotic activity of the RNA-binding protein human antigen R (HuR). J Biol Chem 2010; 285:16806 - 13; http://dx.doi.org/10.1074/jbc.M109.087320; PMID: 20353946
- von Roretz C, Macri AM, Gallouzi IE. Transportin 2 regulates apoptosis through the RNA-binding protein HuR. J Biol Chem 2011; 286:25983 - 91; http://dx.doi.org/10.1074/jbc.M110.216184; PMID: 21646354