Abstract
The pathogenesis of sporadic colorectal cancer involves distinct pathways, with characteristic genomic alterations. The first pathway, chromosome instability (CIN), is driven by APC mutations and is typified by Kras mutations, p53 mutation/loss of heterozygosity, and deletions at chromosome 18q. The second pathway is referred to as microsatellite instability (MSI), a genetic hallmark of the accumulated mutations that occur as a consequence of derangements in the mismatch repair genes. Finally, proximal colon cancers may involve methylation of a number of genes, which is frequently referred to as the CpG island methylator phenotype (CIMP), and are associated with B-raf mutations. The ability to stratify colorectal cancers by risk would be facilitated by the identification of polymorphisms that might be utilized as biomarkers. LIN28B is an RNA binding protein that is overexpressed in colon cancers. We find that LIN28B rs314277 is associated with significant recurrence of colorectal cancer in Stage II disease, which may have translational therapeutic implications.
Introduction
The LIN28 gene encodes two homologous RNA-binding proteins LIN28A and LIN28B, originally discovered in Caenorhabditis elegans as lin-28, a heterochronic regulator of larval and vulval development.Citation1 LIN28A and LIN28B both contain conserved RNA-binding domains, termed the cold-shock domain (CSD) and a retroviral type CCHC zinc-knuckle domain (ZKD). Through these domains, LIN28A/B mediate post-transcriptional downregulation of the let-7 microRNA family, and ultimately, the suppression of differentiation. Specifically, these two RNA-binding domains are important for binding to pri- or pre-let-7, and inhibiting miRNA maturation. The Lin28 CSD binds to pre-let-7 and remodels its terminal loop (including the Dicer cleavage site), which facilitates subsequent binding of the Lin28 ZKD to the conserved GGAG motif.Citation2,Citation3 Recently, it has been shown that LIN28A recruits Zcchc11/TUT4 to let-7 precursors to block processing by Dicer.Citation4 Unlike LIN28A, LIN28B acts exclusively in the nucleus by sequestering primary let-7 transcripts and inhibiting their processing by the microprocessor, a function that is independent of the Zcchc11/TUT4 terminal uridylase.Citation4
LIN28A and LIN28B have been shown to have roles in stem cells, aging and cancer. LIN28A is associated with undifferentiated human embryonic stem cells and cooperates with other genes, namely OCT4, SOX2 and KLF4, for the induction of inducible pleuripotent stem (iPS) cells from human somatic cells. Transgenic mice that express Lin28a have increased body size, abnormal length, and delayed onset of puberty, consistent with the finding of LIN28B genetic variation, specifically rs314276, in association with the timing of puberty,Citation5 and noted as well in independent studies (rs314280 and rs7759938 near the LIN28B gene).Citation6-Citation8 LIN28B expression is more widespread in diverse tissues and may be overexpressed in a variety of tumors, especially of epithelial origin including colon cancer.Citation4,Citation9 LIN28A is overexpressed in some cancers as well.Citation4,Citation9-Citation11 In the context of cancers, LIN28B has been demonstrated to enhance cell migration, invasion and metastasis.Citation12-Citation14 LIN28B is associated with aggressive subtypes of cancers, for example, high-grade serous ovarian cancers,Citation15 esophageal cancer,Citation16 and colon cancer.Citation4,Citation17 Furthermore, LIN28B overexpression correlates with reduced patient survival and increased probability of tumor recurrence in colon cancers.Citation17
The relationship of LIN28B polymorphisms to cancer development, progression and metastasis, and hence prognosis, has not been investigated in any particular detail. However, six SNPs from the DROSHA, FMR1, LIN28A and LIN28B genes, including rs12194974 (G > A), a SNP in a putative transcription factor binding site in the LIN28B promoter region, have been associated with susceptibility to epithelial ovarian cancer.Citation18 As a result, we sought to explore the relationship of LIN28B SNPs and colorectal cancer in well-annotated training and validation sets. To that end, the LIN28B SNP rs314277 showed significant association with risk of recurrence in 118 patients with stage II colorectal cancer. The variant containing genotype had a 2.65-fold increased risk of recurrence (95% CI = 1.18–5.95) compared with the common homozygous genotype. Interestingly, the LIN28B variant containing genotype of rs61764370 had a significantly reduced risk of recurrence (HR = 0.28, 95% CI = 0.09–0.92) in the combined stages II and III of colorectal cancer patients.
Results
Patient characteristics
The association of demographic, clinical, and treatment characteristics with OS, RFS, and PFS are shown in . Among the 745 patients in the training set, there were 221 (30%) deaths and 254 (34%) events for PFS and RFS during the follow-up. The majority of the patients were Caucasians (82.6%) and male (62%). No significant association with OS in the training set was observed for age (p = 0.83), smoking pack year (p = 0.76), sex (p = 0.10), race (p = 0.83), and tumor location (p = 0.08). However, stage, histological grade, those who underwent curative surgery of primary tumor, and those who received fluoropyrimidine-based chemotherapy were significantly associated with OS (p < 0.05). No significant association with PFS and RFS in the training set was observed for smoking pack year (p = 0.80), sex (p = 0.56), race (p = 0.47), and tumor location (p = 0.23). However, age, stage, histological grade, those who underwent curative surgery of primary tumor, and those who received fluoropyrimidine-based chemotherapy were significantly associated with PFS and RFS (p < 0.05). The median survival time was 111.8 mo and the median follow-up time was 32.7 mo in the training set. Among 358 patients in the testing set, a higher rate of recurrence, progression, and death was observed since the patients with a long history of prior CRC came to MD Anderson Cancer Center for potential tumor recurrence or progression. There were 218 (61%) deaths and 317 (89%) events for PFS and RFS during the follow-up (). The analysis focused on colorectal cancer patients with stage II, III or IV disease since patients with stage I disease were mainly treated with surgery alone and carry a good prognosis. Among those with stage I disease, there were only 2 patients (3%) who died and 5 patients (8%) had events for RFS during the follow-up in the training set (). To minimize the effect of treatment heterogeneity, we further restricted the analysis to stage II and stage III patients receiving surgery and adjuvant fluoropyrimidine-based chemotherapy (training set: 118 stage II and 180 stage III; testing set: 53 stage II and 109 stage III) and stage IV disease treated with first-line fluoropyrimidine-based chemotherapy (training set: 218 patients; testing set: 110 patients).
Table 1. Selected demographic and clinical characteristics of CRC patients
Association with OS
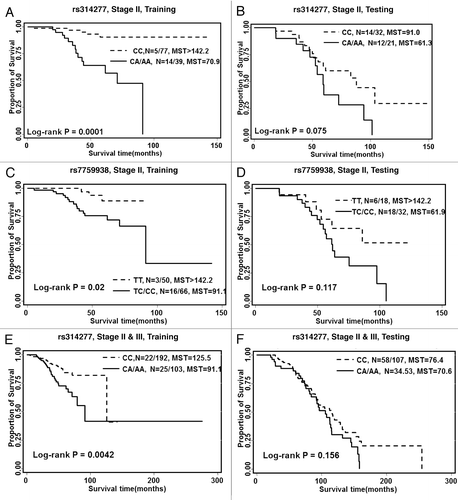
The association of three SNPs (rs314277, rs7759938, and rs61764370) with overall survival is listed in . Among 118 patients with stage II disease in the training set, rs314277 (HR = 6.85; 95% CI = 2.32–20.24) and rs7759938 (HR = 4.27; 95% CI = 1.17–15.62) was associated with a significantly increased risk of death. The effects of both SNPs were also significant in a dose-dependent manner. In the testing set, both SNPs were also associated with an increased risk of death (HR > 1), although the associations were not significant which might be due to the sample size (only 53 patients with stage II disease in the testing set). As shown in , we observed a significantly longer survival for patients carrying a common homozygous genotype than patients carrying variant alleles for rs314277 (median survival time [MST] > 142 vs MST = 70.9, log rank p = 0.0001) and rs7759938 (MST > 142 vs MST = 91.1, log rank p = 0.02) in the training set data. Among the 298 patients with stage II or stage III disease in the training set, the variant containing genotype of rs314277 had a 2.55-fold increased risk of death (95% CI = 1.37–4.75). This association remained borderline significant in the testing set (HR = 1.55; 95% CI = 0.94–2.58). The HR of the combined data set was 1.89 (95% CI = 1.28–2.8). No significant association was observed between the three SNPs and overall survival among patients with stage IV disease.
Table 2. Association of rs314277, rs7759938 and rs61764370 with overall survival
Figure 1. Kaplan Meier curves for overall survival based on the SNP genotype stratified by training and testing data. (A) snp rs314277 in patients with stage II disease in the training data, (B) snp rs314277 in patients with stage II disease in the testing data, (C) snp rs7759938 in patients with stage II disease in the training data, (D) snp rs7759938 in patients with stage II disease in the testing data, (E) snp rs314277 in patients with stage II and III disease in the training data, (F) snp rs314277 in patients with stage II and III disease in the testing data. N, number of event/total number of patients in each genotype group. MST, median survival time.
Association with RFS and PFS
The association of three SNPs (rs314277, rs7759938, and rs61764370) with RFS or PFS is listed in . In the training set, rs314277 showed significant association with risk of recurrence in 118 patients with stage II disease. This variant containing genotype had a 2.65-fold increased risk of recurrence (95% CI = 1.18–5.95) compared with the common homozygous genotype. The variant containing genotype of rs61764370 had a significantly reduced risk of recurrence (HR = 0.28, 95% CI = 0.09–0.92) in the combined stage II and stage III colorectal cancer patients. However, none of these results was replicated in the testing set. There was no significant association between the three SNPs and progression among patients with stage IV disease.
Table 3. Association of rs314277, rs7759938 and rs61764370 with RFS or PFS
Discussion
This study used a two-stage design to assess the association of one SNP in KRAS and two SNPs in LIN28B with the clinical outcomes of colorectal cancer. Both SNPs, rs314277 and rs7759938, in LIN28B exhibited consistent association with overall survival in both training and testing sets for patients with stage II disease and one SNP rs314277 in LIN28B showed significant association with overall survival in the training set and borderline significant association in the testing set for patients with stage II or stage III disease. None of the SNPs showed significant association with clinical outcomes of colorectal cancer for patients with stage IV disease.
In terms of other LIN28B SNPs, we found that rs314280 and rs314276 were in strong linkage with rs7759938, which is genotyped in this study (R2 = 0.62 and 0.897, respectively). Although rs12194974 is not included in our study, its association with ovarian cancer is not that significant based on the reported large sample size (p = 0.015 for 3,987 EOC cases and 4,952 controls). Given the relationship between LIN28B and Let-7 miRNA, we have analyzed two SNPs from let-7 with colorectal cancer clinical outcomes and identified the association of let7f-2:rs17276588 with progression in stage IV patients receiving fluoropyrimidine-based chemotherapy.Citation21 Of note, the currently identified 8q24 region SNPs were distant from the MYC gene region, and these SNPs have not yet been connected with alterations in MYC function and the risk of colorectal cancer.Citation22,Citation23
There are hereditary and sporadic forms of colorectal cancer. The hereditary forms of colorectal cancer include Lynch syndrome (germline mutations in MLH1, MSH2, MSH6, PMS2, EPCAM), familial adenomatous polyposis or FAP (germline mutations in the APC gene), MYH associated polyposis or MAP (germline mutations in the MYH gene), and rarer hamartomatous polyposis-associated syndromes, such as Peutz-Jeghers syndrome, Juvenile Polyposis, and Cowden syndrome. There is no gene polymorphism associated with colorectal cancer outcome that is available currently as a viable biomarker. In our studies, LIN28B SNP rs314277 may fill such a need in that it predicts for reduced survival in stage II colorectal cancer, which is the focus of attention for possible adjuvant therapy. Thus, LIN28B’s functions may be important not only in sporadic colon cancer in fostering tumor cell migration, invasion and metastasis, but portend risk stratification in stage II patients who may benefit from novel adjuvant therapeutic approaches.
Patients and Methods
Study population and data collection
This study included 1103 histologically confirmed colorectal adenocarcinoma patients recruited from January 1990 to June 2008 at the University of Texas MD Anderson Cancer Center. These patients were classified into two sets: the training set included 745 patients that were newly diagnosed (diagnosed within 1 y before referral to MD Anderson Cancer Center) and the replication set included 358 patients with a long history (diagnosed > 1 y) of CRC prior to recruitment. We collected the epidemiological data through a self-administered questionnaire and the clinical information and follow-up data was abstracted from patients’ medical records. A 10 to 20-mL blood sample was collected from each participant for immediate molecular analyses and the isolation of DNA. The study was approved by the Institutional Review Board of MD Anderson Cancer Center and informed consent was signed by each study participant.
Genotyping
The functional SNP in the KRAS 3′ untranslated region (rs61764370), which affects let-7 microRNA complementary site,Citation19 and two SNPs (rs7759938 and rs314277) from LIN28B identified in the GWAS study,Citation20 were genotyped in this study. Genomic DNA was isolated from peripheral blood by the Qiagen kit. Genotyping was performed by using the Taqman method with a 7900 HT sequence detection system (Applied Biosystems). Sample DNA (5 ng), 1 × Taqman buffer A, 200 μM deoxynucleotide triphosphate, MgCl2 (5 mM), AmpliTaq Gold (0.65 units), each primer (900 nM), and each probe (200 nM) constituted the amplification mixes (5 μl). The thermal cycling conditions started with 1 cycle for 10 min at 95°C, followed by 40 cycles of 95°C for 15 sec, and the final 40 cycles of 60°C for 1 min. Each plate also contained a water control, ample internal controls, and previously genotyped samples to guarantee genotyping accuracy.
Statistical analysis
The study endpoints were overall survival (OS) which is the time from pathologic diagnosis until disease from any cause, progression-free survival (PFS), which is the time from pathologic diagnosis until disease progression or death, and recurrence-free survival (RFS), which is defined as the time from the date of pathologic diagnosis until first recurrence or death. To assess the association of demographic and clinical variables with endpoint of interest, chi-square test was used for categorical variables and two-sample t-test was used for continuous variables. Hazard ratios (HRs) and their 95% confidence intervals (CIs) was estimated in the training and testing sets using the multivariate Cox’s proportional hazards model while adjusting for age, gender, ethnicity, clinical stage, histologic grade, and if necessary, treatment modality and chemotherapy regimen. Meta-analysis was performed to combine the results in the training and testing sets. Fixed-effect meta-analysis was used if the Cochrane Q statistics test for the assessment of heterogeneity not significant. Otherwise, the random effect meta-analysis was used. The Kaplan-Meier method and log-rank test were used to compare the differences in OS, PFS, and RFS by genotype status. All P-values were two-sided and a p ≤ 0.05 was considered statistically significant. The above analyses were conducted using the Stata 10.1 software package (Stata Co.).
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Acknowledgments
This work was supported by NIH R01-DK056645 (A.K.R., B.M.M.), the Hansen Foundation (A.K.R.), the National Colon Cancer Research Alliance (A.K.R.), NIH K01-DK093885 (B.M.M.), NIH grant R01 CA098897 (X.W.) and MD Anderson’s Center for Translational and Public Health Genomics.
References
- Moss EG, Lee RC, Ambros V. The cold shock domain protein LIN-28 controls developmental timing in C. elegans and is regulated by the lin-4 RNA. Cell 1997; 88:637 - 46; http://dx.doi.org/10.1016/S0092-8674(00)81906-6; PMID: 9054503
- Mayr F, Schütz A, Döge N, Heinemann U. The Lin28 cold-shock domain remodels pre-let-7 microRNA. Nucleic Acids Res 2012; 40:7492 - 506; http://dx.doi.org/10.1093/nar/gks355; PMID: 22570413
- Nam Y, Chen C, Gregory RI, Chou JJ, Sliz P. Molecular basis for interaction of let-7 microRNAs with Lin28. Cell 2011; 147:1080 - 91; http://dx.doi.org/10.1016/j.cell.2011.10.020; PMID: 22078496
- Piskounova E, Polytarchou C, Thornton JE, LaPierre RJ, Pothoulakis C, Hagan JP, et al. Lin28A and Lin28B inhibit let-7 microRNA biogenesis by distinct mechanisms. Cell 2011; 147:1066 - 79; http://dx.doi.org/10.1016/j.cell.2011.10.039; PMID: 22118463
- Ong KK, Elks CE, Li S, Zhao JH, Luan J, Andersen LB, et al. Genetic variation in LIN28B is associated with the timing of puberty. Nat Genet 2009; 41:729 - 33; http://dx.doi.org/10.1038/ng.382; PMID: 19448623
- Sulem P, Gudbjartsson DF, Rafnar T, Holm H, Olafsdottir EJ, Olafsdottir GH, et al. Genome-wide association study identifies sequence variants on 6q21 associated with age at menarche. Nat Genet 2009; 41:734 - 8; http://dx.doi.org/10.1038/ng.383; PMID: 19448622
- Perry JR, Stolk L, Franceschini N, Lunetta KL, Zhai G, McArdle PF, et al. Meta-analysis of genome-wide association data identifies two loci influencing age at menarche. Nat Genet 2009; 41:648 - 50; http://dx.doi.org/10.1038/ng.386; PMID: 19448620
- He C, Kraft P, Chen C, Buring JE, Paré G, Hankinson SE, et al. Genome-wide association studies identify loci associated with age at menarche and age at natural menopause. Nat Genet 2009; 41:724 - 8; http://dx.doi.org/10.1038/ng.385; PMID: 19448621
- Viswanathan SR, Powers JT, Einhorn W, Hoshida Y, Ng TL, Toffanin S, et al. Lin28 promotes transformation and is associated with advanced human malignancies. Nat Genet 2009; 41:843 - 8; http://dx.doi.org/10.1038/ng.392; PMID: 19483683
- Saiki Y, Ishimaru S, Mimori K, Takatsuno Y, Nagahara M, Ishii H, et al. Comprehensive analysis of the clinical significance of inducing pluripotent stemness-related gene expression in colorectal cancer cells. Ann Surg Oncol 2009; 16:2638 - 44; http://dx.doi.org/10.1245/s10434-009-0567-5; PMID: 19554373
- Peng S, Maihle NJ, Huang Y. Pluripotency factors Lin28 and Oct4 identify a sub-population of stem cell-like cells in ovarian cancer. Oncogene 2010; 29:2153 - 9; http://dx.doi.org/10.1038/onc.2009.500; PMID: 20101213
- Kong D, Banerjee S, Ahmad A, Li Y, Wang Z, Sethi S, et al. Epithelial to mesenchymal transition is mechanistically linked with stem cell signatures in prostate cancer cells. PLoS One 2010; 5:e12445; http://dx.doi.org/10.1371/journal.pone.0012445; PMID: 20805998
- Wang YC, Chen YL, Yuan RH, Pan HW, Yang WC, Hsu HC, et al. Lin-28B expression promotes transformation and invasion in human hepatocellular carcinoma. Carcinogenesis 2010; 31:1516 - 22; http://dx.doi.org/10.1093/carcin/bgq107; PMID: 20525879
- King CE, Wang L, Winograd R, Madison BB, Mongroo PS, Johnstone CN, et al. LIN28B fosters colon cancer migration, invasion and transformation through let-7-dependent and -independent mechanisms. Oncogene 2011; 30:4185 - 93; http://dx.doi.org/10.1038/onc.2011.131; PMID: 21625210
- Helland A, Anglesio MS, George J, Cowin PA, Johnstone CN, House CM, et al, Australian Ovarian Cancer Study Group. Deregulation of MYCN, LIN28B and LET7 in a molecular subtype of aggressive high-grade serous ovarian cancers. PLoS One 2011; 6:e18064; http://dx.doi.org/10.1371/journal.pone.0018064; PMID: 21533284
- Hamano R, Miyata H, Yamasaki M, Sugimura K, Tanaka K, Kurokawa Y, et al. High expression of Lin28 is associated with tumour aggressiveness and poor prognosis of patients in oesophagus cancer. Br J Cancer 2012; 106:1415 - 23; http://dx.doi.org/10.1038/bjc.2012.90; PMID: 22433967
- King CE, Cuatrecasas M, Castells A, Sepulveda AR, Lee JS, Rustgi AK. LIN28B promotes colon cancer progression and metastasis. Cancer Res 2011; 71:4260 - 8; http://dx.doi.org/10.1158/0008-5472.CAN-10-4637; PMID: 21512136
- Permuth-Wey J, Kim D, Tsai YY, Lin HY, Chen YA, Barnholtz-Sloan J, et al, Ovarian Cancer Association Consortium. LIN28B polymorphisms influence susceptibility to epithelial ovarian cancer. Cancer Res 2011; 71:3896 - 903; http://dx.doi.org/10.1158/0008-5472.CAN-10-4167; PMID: 21482675
- Chin LJ, Ratner E, Leng S, Zhai R, Nallur S, Babar I, et al. A SNP in a let-7 microRNA complementary site in the KRAS 3′ untranslated region increases non-small cell lung cancer risk. Cancer Res 2008; 68:8535 - 40; http://dx.doi.org/10.1158/0008-5472.CAN-08-2129; PMID: 18922928
- Widén E, Ripatti S, Cousminer DL, Surakka I, Lappalainen T, Järvelin MR, et al. Distinct variants at LIN28B influence growth in height from birth to adulthood. Am J Hum Genet 2010; 86:773 - 82; http://dx.doi.org/10.1016/j.ajhg.2010.03.010; PMID: 20398887
- Lin M, Gu J, Eng C, Ellis LM, Hildebrandt MA, Lin J, et al. Genetic polymorphisms in MicroRNA-related genes as predictors of clinical outcomes in colorectal adenocarcinoma patients. Clin Cancer Res 2012; 18:3982 - 91; http://dx.doi.org/10.1158/1078-0432.CCR-11-2951; PMID: 22661538
- Cicek MS, Slager SL, Achenbach SJ, French AJ, Blair HE, Fink SR, et al. Functional and clinical significance of variants localized to 8q24 in colon cancer. Cancer Epidemiol Biomarkers Prev 2009; 18:2492 - 500
- Huppi K, Pitt JJ, Wahlberg BM, Caplen NJ. The 8q24 gene desert: an oasis of non-coding transcriptional activity. Front Genet 2012; 3:69; http://dx.doi.org/10.3389/fgene.2012.00069; PMID: 22558003
- Rustgi AK. The geneticcs of hereditary colon cancer. Genes Dev 2007; 21:2525 - 38; http://dx.doi.org/10.1101/gad.1593107; PMID: 17938238