Abstract
Overexpression of high-mobility group box 2 (HMGB2) is recently reported in several malignant cancers and was correlated with poor response to preoperative chemoradiotherapy of colorectal cancer patients. To enhance the chemoradiotherapy efficacy, the biological function of HMGB2 was investigated with respect to radiation response. HMGB2 gene knockdown cells were constructed by infecting shRNA expressing lentivirus and clonogenic assay was performed to count the radiosensitivity. HMGB2 knockdown sensitized HCT-116 and HT-29 colorectal cancer cells to ionizing radiation. This could be due to an increased DNA damage and an inefficient DNA damage repair in HMGB2 knockdown cells. In addition, an exposure to radiation downregulated HMGB2 expression in colorectal cancer cells with an intact TP53 gene. HMGB2 gene expression of TP53-mutant cell was not affected by irradiation. p53-mediated downregulation of HMGB2 was confirmed by direct activation of p53 using Nutlin-3 or by inducing p53 expression using Tet-On system. Luciferase reporter assay showed that HMGB2 promoter activity was inversely correlated with the amount p53 cotransfected. Our study revealed that HMGB2 is necessary to protect colorectal cancer cells from DNA damage and efficient DNA repair and p53-mediated downregulation is a critical mechanism of modulating HMGB2 expression.
Keywords: :
Introduction
High-mobility group box (HMGB) proteins are one of the HMG protein families, containing characteristic HMG box motif. Four members of the HMGB family are currently known in mammals and characterized by two tandem basic HMG box domains followed by a long acidic tail.Citation1 The HMGB1 and HMGB2 are highly conserved nuclear proteins, acting as chromatin-binding factor that bends DNA and promotes access to transcriptional protein assemblies on specific targets.Citation2,Citation3 In addition, HMGB1 functions as an extracellular signaling molecule during inflammation, cell differentiation, cell migration and tumor metastasis.Citation3,Citation4 It is also reported that HMGB2 is released to extracellular space in various inflammatory states.Citation5,Citation6 and binds to receptors like Receptor for Advanced Glycation End products (RAGE).Citation7 HMGB2 is highly expressed during embryogenesis and in adults it is mainly expressed in testicles and lymphoid organs. Mice lacking HMGB2 gene are viable, but knockout males have reduced fertility.Citation8 Further, HMGB2 is highly expressed in chondrocytes of the superficial zone in joints and knockout mice develop osteoarthritis earlier than wild-type mice.Citation9 Recently, a novel function of HMGB2 in mesenchymal stem cell differentiation is revealed.Citation10 HMGB1 overexpression has been reported in a variety of human cancers, including melanoma,Citation11 pancreatic cancer, prostate cancer,Citation12 colorectal cancer,Citation13 and breast cancer.Citation14 HMGB2 have been significantly upregulated in glioblastoma tissues, compared with a low level of expression in normal human brain tissues.Citation15 In hepatocellular carcinoma, HMGB2 overexpression has been significantly correlated with shorter overall survival time.Citation16 Despite extensive characterization of the diverse roles of HMGB1, relatively less is known of the specific biological function of HMGB2.
In the management of locally advanced colorectal cancer, preoperative chemoradiotherapy has been established as a standard component of multimodal treatment.Citation17 Several studies have performed microarray for predicting preoperative chemoradiotherapy responses in patients with advanced rectal cancer.Citation18-Citation20 We also identified differentially expressed genes between non-responder and responder to chemoradiotherapy and clustering analysis was performed (unpublished). Among them, HMGB2 expression was 2.66‑fold higher in non-responder group than in responder group (p = 0.0058). This lead us to hypothesize that HMGB2 might regulate cellular response to radiation in colorectal cancer. To prove this, we studied the biological function of HMGB2 with respect to radiation in greater detail through in vitro analysis.
Results
Knockdown of HMGB2 sensitized colorectal cancer cells to radiation
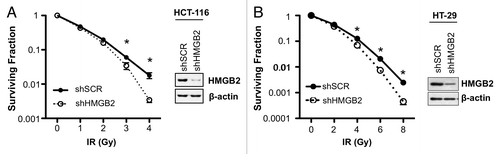
To study the biological function of HMGB2 protein in radiation response, stable HMGB2 knockdown cell lines were established using pLKO.1 lentivirus system continuously expressing shRNA targeting HMGB2 gene. Established shHMGB2 cells were confirmed by checking the decreased level of HMGB2 mRNA (data not shown) and protein expression compared with shSCR control cells (, right blot). Clonogenic assay showed that HMGB2 depletion made both HCT-116 cells () and HT-29 () colorectal cancer cells more sensitive to radiation. These results suggested that HMGB2 was required for proper DNA damage responses to irradiation. Furthermore, coincident with microarray analysis results of human colorectal cancer sample, HMGB2 expression level could be a biomarker predicting radioresponse of colorectal cancer cell.
Figure 1. Knockdown of HMGB2 sensitized colorectal cancer cells to radiation. HMGB2 knockdown HCT-116 (A) and HT-29 (B) cells were established by infecting lentivirus expressing shRNA targeting HMGB2 and were confirmed by western blot analysis. A clonogenic assay was performed after exposure to the indicated dose of radiation in shRNA expressing cells. Values are means ± standard deviation for triplicate wells for each dose point (*p < 0.05 compared with control).
HMGB2 knockdown enhanced DNA damage and delayed DNA damage repair
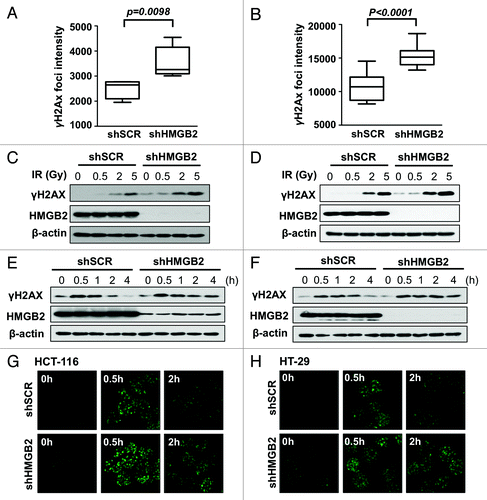
HMGB2 is a non-histone chromatin protein and reported to be involved in DNA damage repair.Citation3,Citation4 To study the effect of HMGB2 knockdown on DNA damage response, immunofluorescence staining of γH2AX was performed. γH2AX is a well-established DNA damage marker. DNA double strand breaks caused by DNA damage like ionizing radiation lead to the phosphorylation of neighboring histone H2A subtype X at C-terminal Ser-139 residue, which is important for recruiting DNA repair proteins. When γH2AX foci intensity per cell was quantified, γH2AX intensity of HMGB2 knockdown cell was significantly higher than that of shSCR control cells, which suggested that more DNA stand breaks are generated in exponentially growing HMGB2 knockdown cells without any external DNA damage induction both in HCT‑116 and HT‑29 cells (). This immunofluorescence staining result was well match with western blot analysis showing higher level of γH2AX in shHMGB2 cells (). Not only the basal level of DNA damage, HMGB2 knockdown cells were more sensitive to irradiation, which was clearly observed in cells exposed to relatively low dose (0.5 Gy) of ionizing radiation. When the DNA damage repair kinetics compared after exposure to 2 Gy of radiation (), HMGB2 knockdown delayed the DNA damage repair, which suggested that HMGB2 are required for efficient DNA breaks repair. Coincident with this western blot result, immunofluorescence staining () showed that γH2AX phosphorylation peaked after 30 min and DNA damages were significantly repaired after 2 h in shSCR control cell. However, γH2AX phosphorylation was sustained in HMGB2 knockdown cells. These results suggested that HMGB2 is required for protecting cells from DNA damage and efficient DNA damage repair process, which could be a major contributor of increasing sensitivity to radiation of HMGB2 knockdown cells.
Figure 2. Knockdown of HMGB2 increased DNA damage and delayed DNA damage repair. Exponentially growing HMGB2 knockdown HCT-116 (A) and HT-29 (B) cells were cultured on slide glass and stained with anti-γH2AX antibody. γH2AX foci intensity per cell was calculated with Image Pro software and statistically analyzed. HMGB2-knockdown HCT-116 (C) and HT-29 (D) cells were exposed to the indicated dose of radiation, harvested after 1 h and examined by western blot analysis. After exposure to 2 Gy of radiation, HCT-116 (E) and HT-29 (F) cells were harvested at the indicated time and examined by western blot analysis. Protein extracts were quantified and equal amounts of total protein were subjected to SDS-PAGE and detected with anti-γH2AX, -HMGB2 and -β-actin antibodies. At the same condition, immunofluorescence staining with anti-γH2AX antibody was performed to measure the amount of DNA damage in HCT-116 (G) and HT-29 (H) cells.
HMGB2 expression was downregulated by ionizing radiation in colorectal cancer cells with functional TP53 gene
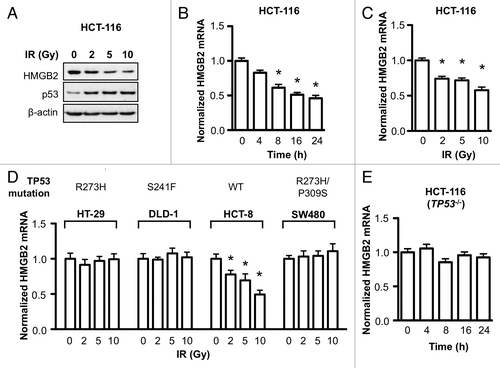
Expression of many DNA damage response genes is known to be induced by external DNA damage stresses to protect cells from death by activating cell cycle checkpoints and repairing DNA damage. If the DNA damage is too severe to be recovered, genes regulating apoptotic cell death was also induced.Citation23 Because HMGB2 was required for protection cell from DNA damage and its knockdown sensitized cell to irradiation, we hypothesized HMGB2 might be induced by irradiation to protect cells from radiation induced death. So the effect of ionizing radiation on HMGB2 gene expression was investigated. Contrary to our expectation, irradiation decreased HMGB2 protein level in a dose-dependent manner (). Increase of p53 protein resulted from stabilization by irradiation shown as an experimental control. In order to reveal at what step radiation regulated HMGB2 level, first, qRT-PCR was performed. As HMGB2 mRNA level was decreased in a time- and dose-dependent manner after irradiation in HCT-116 cell (), we can conclude this downregulation of HMGB2 by irradiation is occurred at transcription level. However, in contrast to HCT-116, HMGB2 transcription in HT-29 cell was not affected by irradiation (). To figure out the different response to radiation between two cell lines, HMGB2 expression after irradiation was investigated in three more colorectal cancer cell lines. HMGB2 mRNA level in DLD-1 and SW480 cell was not changed after irradiation similar to HT-29. However, HMGB2 transcription was downregulated by irradiation in HCT-8 cell (). Considering HT-29, DLD-1 and SW480 has a mutant TP53 tumor suppressor gene (R273H, S241F and R273H/P309S respectively), we hypothesized functional p53 may be required for HMGB2 downregulation by radiation. To prove this, HCT-116 cell with deleted TP53 gene (HCT-116 (p53−/−)) was irradiated and HMGB2 mRNA level was checked. Compared with HCT-116 with functional p53 (), HMGB2 mRNA level in HCT-116 (p53−/−) cell was not changed after irradiation (). From these results, HMGB2 expression was downregulated by ionizing radiation in colorectal cancer cells with functional TP53 gene.
Figure 3. HMGB2 expression was downregulated by ionizing radiation in colorectal cancer cells with functional TP53 gene. (A) Two days after exposure to the indicated dose of radiation, HCT-116 cells were collected and HMGB2 protein level was examined by immunoblot analysis. p53 protein level was detected as an experimental control. (B) After exposure to 10 Gy of radiation, HCT-116 cells were harvested at the indicated of time and HMGB2 mRNA level was examined by qRT-PCR (*p < 0.01 compared with control). (C) HCT-116 cells were exposed to the indicated dose of radiation and HMGB2 mRNA level was examined after 16 h (* p < 0.01 compared with control). (D) HT-29, DLD-1, HCT-8 and SW480 colorectal cancer cells were exposed to the indicated dose of radiation and HMGB2 mRNA level was examined by qRT-PCR (* p < 0.01 compared with control). TP53 gene status is shown above. (E) HCT-116 (p53−/−) cells were exposed to 10 Gy of radiation and were harvested at the indicated of time. HMGB2 transcript level was examined by qRT-PCR and normalized with reference genes. Error bars represent standard error.
p53 played a crucial role in downregulation of HMGB2 transcription
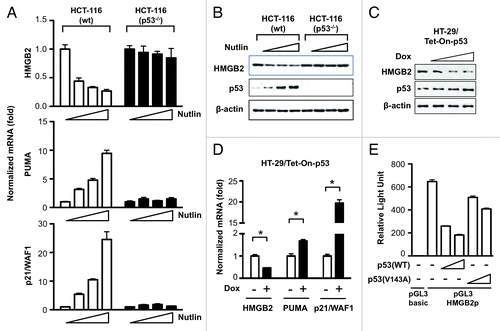
To examine whether p53 directly regulates HMGB2 expression, p53 was activated by treating HCT-116 cells (both wt and p53−/−) with Nutlin-3, a p53 activator by inhibiting the interaction between MDM2 and p53, leading to p53 stabilization. As shown in , Nutlin-3 treatment induced p53 downstream target PUMA and p21/WAF1 in a dose-dependent manner only in HCT-116 cell with wild type p53. In this condition, HMGB2 mRNA level was inversely correlated with Nutlin-3 concentration treated. p53 accumulation and concomitant decrease of HMGB2 protein was confirmed by western blot analysis (). As expected, not only HMGB2 but also PUMA and p21/WAF gene expression in HCT-116 cell with null p53 did not response to Nutlin-3 treatment. Next, conditional inducible system of functional p53 was introduced in HT-29 cell. Induction of p53 protein was not as dramatic due to overexpression of endogenous mutant p53 protein in HT-29 cell, however, HMGB2 protein level was decreased in a dose dependent manner (). qRT-PCR result also clearly demonstrated that p53 induction by doxycycline caused downregulation of HMGB2 and concomitantly induced p53 target genes, PUMA and p21/WAF1 (). Finally, to verify whether p53 downregulated HMGB2 at the transcription level, a luciferase reporter assay was performed. pGL3 plasmid containing HMGB2 promoter region DNA (pGL3-HMGB2p) was cotransfected with an increasing amount of pCMV-p53 plasmids and normalized luciferase activity was compared (). Transfection of pGL3-HMGB2p showed 650-fold higher luciferase activity compared with the empty pGL3-basic plasmid. Coincide with all previous results, HMGB2 promoter activity was inversely correlated with the amount of p53 wt plasmid cotransfected. However, cotransfection with p53 harboring mutation (V143A) could not inhibit HMGB2 promoter efficiently, which confirmed that the presence of the CMV promoter did not affect reporter assay significantly. All these result proved that p53 is a key molecule in downregulating HMGB2 transcription.
Figure 4. p53 played a crucial role in downregulation of HMGB2 expression at transcription level. (A) HCT-116 wild type and p53−/− cells were treated with 0, 2.5, 5 and 10 μM of Nutlin-3 for 16 h and HMGB2 mRNA level was examined by qRT-PCR. PUMA and p21/WAF1 mRNA analysis was included as a p53-dependent transcription control. (B) Immunoblot assay was performed the same condition as in (A) except treatment was last for 2 d. HT-29 cells harboring tet-inducible p53 expression system were treated with doxycyclin for 2 d and western blot (C) and qRT-PCR (D) analysis were performed. * p < 0.01 compared with untreated group. (E) pGL3 vector harboring HMGB2 promoter DNA was cotransfected with an increasing amount of p53 expression plasmid into 293T cell and transcription activity was measured with dual luciferase assay. Error bars represent standard error. Nutlin, Nutlin-3; Dox, doxycyclin; HMGB2p, HMG2 promoter.
Discussion
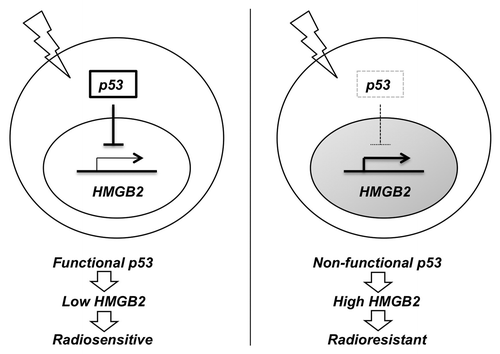
The purpose of this study was to check the possibility of HMGB2, a ubiquitous nuclear regulator of chromatin, as a mediator of DNA damage signaling and a radioresponse regulator. Initially, HMGB2 was identified as a differentially expressed marker gene predicting the response to preoperative chemoradiotherapy in colorectal cancer patients. In this study, we showed that HMGB2 protects cell from DNA damage () and deficiency of HMGB2 sensitizes cell to radiation in colorectal cancer (). So, any disorder in HMGB2 downregulation induced by DNA damage signaling could confer radioresistance to cancer cells. We identified p53 as a key regulator of HMGB2 expression in response to DNA damage ( and ) and colorectal cancer cells expressing mutant p53 did not show HMGB2 downregulation (). So, fractionated radiation or DNA damaging agents could downregulate HMGB2 expression by activating p53, which could make cell more sensitive to the following application of chemoradiotherapy. p53 tumor suppressor protein plays a role in genomic stability, cell cycle progression and apoptosis. Approximately half of all colorectal cancers show p53 gene mutation, with higher frequencies observed in distal colon and rectal tumors.Citation24,Citation25 It is also established that the presence of mutant p53 is associated with decreased local control following radiotherapy.Citation26,Citation27 However, further study is required to validate the contribution of HMGB2 to chemoradioresistance of p53 mutant cell. A biological implication of p53-mediated downregulation of HMGB2 in chemoradiotherapy is summarized in .
Figure 5. A proposed diagram suggesting biological implication of p53-mediated downregulation of HMGB2 in chemoradiotherapy. Cancer cell with wild type p53 could inhibit the transcription of HMGB2 gene in responds to radiation, which might sensitize cell to the following chemoradiotherapy. In contrast, sustained expression of HMGB2 in p53 mutant cancer might contribute to chemoradioresistancy.
With respect to DNA binding proteins, HMGB2 are closely related to the susceptibility of cells to DNA damage-induced cell death. A transient overexpression of HMGB2 conferred resistance to cisplatin-induced cell death, however, elicited significantly less apoptosis in response to etoposide in hepatocellular carcinoma cells.Citation16 It has been reported that multiprotein complex including HMGB2 detects changes in DNA structure caused by incorporation of nonnatural nucleosides and determines cellular sensitivity to chemotherapy.Citation28 Knockdown of each component of this protein complex resulted in 8-fold to 50-fold increased chemoresistance to cytarabine, probably by delaying p53 phosphorylation after exposure to genotoxic stress.Citation29 HMGB1 have also been reported to recognize aberrant or damaged DNA in multiple in vitro experiments. Prasad et al. directly demonstrated accumulation of HMGB1 at sites of oxidative DNA damage in live cells thus defining HMGB1 as a component of an early DNA damage response.Citation30 Lange et al. also demonstrated that mammalian cells lacking HMGB1 are hypersensitive to DNA damage induced by psoralen plus UV irradiation and nucleotide excision repair efficiency is significantly decreased.Citation31 Recently it was reported that overexpression of HMGB1 or HMGB2 using adenoviral system elevated the radiosensitivity of breast cancer cells.Citation32,Citation33 Taken together, HMGB2 is of particular relevance in DNA damage repair and radioresponse regulation, although differential regulation mechanism must be resided in depending on the genotoxic stresses introduced in cancer cells.
In addition, there are several possible mechanisms of radiosensitization mediated by HMGB2 deficiency. First, HMGB2 is necessary for cell survival, proliferation and growth signal transduction to nucleus. HMGB2 inhibition by antisense RNA repressed cell cycle progression in COS-1 cellsCitation34 and HMGB2 knockdown by siRNA inhibited cell proliferation in rat HCC cells.Citation16 Additionally, IHC study of HCC showed that positivity of HMGB2 was correlated with PCNA, a known marker of cell proliferation.Citation35 We also observed that HMGB2 gene knock down significantly delayed cell proliferation (Data not shown). Second, HMGB2 regulates chromatin structure and induces genome-wide changes in gene expression. HMGBs can interact directly with nucleosomes, loosen the wrapped DNA and so enhance accessibility to chromatin-remodeling complexes and to transcription factors.Citation36 Recently, Franklin et al. demonstrate that the HMGB2 (but not HMGB1) controls a gene expression program responsible for hypertrophic cell growth. They suggest general principles for global shifts in chromatin accessibility: altered linker to core histone ratio; differing abundance of chromatin structural proteins; and reprogrammed histone post-translational modifications.Citation37 Finally, like HMGB1, HMGB2 might be involved in regulation of telomerase activity and lead to telomere dysfunction. Knockout of the HMGB1 gene in mouse embryonic fibroblasts results in chromosomal abnormalities, enhanced colocalization of γH2AX foci at telomeres and a moderate shortening of telomere lengths.Citation38
In conclusion, our data suggest that HMGB2 plays a significant role in cellular response and protection to DNA damage stress and p53-mediated downregulation of HMGB2 transcription is a key regulatory mechanism.
Methods and Materials
Cell culture
The human colorectal adenocarcinoma cancer cell lines HCT-116, HT-29, DLD-1, HCT-8 and SW480 were obtained from the Korean Cell Line Bank (KCLB) and HCT116 (p53−/−) derivatives were supplied by Dr. B. Vogelstein.Citation21 All cells were maintained in RPMI-1640 medium (PAA Laboratories) supplemented with 10% heat-inactivated fetal bovine serum (PAA Laboratories) or and 100 U/ml of penicillin and streptomycin (PAA Laboratories) except HCT-8 cell with horse serum (WelGene) instead of FBS.
shRNA-mediated HMGB2 gene silencing
The lentiviral shRNA expression plasmids targeting HMGB2 were generated by cloning oligonucleotides into the pLKO.1 puro vector (Addgene, plasmid #8453). The oligonucleotide used for targeting HMGB2 is 5′-GCAAAGGAGAAGTCGAAGTTTCTCGAGAAACTTCGACTTCTCCTTTGC -3′ (TRCN0000019010) and 5′-CCATCTGCCTTCTTCCTGTTTCTCGAGAAACAGGAAGAAGGCAGATGG -3′ (TRCN0000019013). The oligonucleotides for HMGB2 were annealed and subcloned to pLKO.1-puro vector after digestion with AgeI and EcoRI. The control scramble shRNA expressing plasmid (shSCR) was obtained from Addgene (plasmid #17920) with following hairpin sequence: 5′-CCTAAGGTTAAGTCGCCCTCGCTCGAGCGAGGGCGACTTAACCTTAGG-3′. To produce lentivirus particle, 293T cells were seeded to 3 x 106 onto 100-mm dishes and 16 h later were transiently transfected with 3 µg of pLKO.1 lentiviral vector, 2.66 µg of pCMV-dR8.2 dvprC (Addgene plasmid #8455) and 0.33 µg of pCMV-VSV-G (Addgene plasmid #8454) using JetPEI (Polyplus). Supernatants were collected 48 and 72 h post transfection followed by concentration of the virus using Centricon 100K (Millipore) and used for infections in the presence of 8 µg/ml polybrene. One day after infection, puromycin (Sigma) was applied to 1 µg/ml for 7 d to eliminate uninfected cells.
Clonogenic assay
Clonogenic survival was defined as the ability of cells to maintain clonogenic capacity and to form colonies.Citation22 Briefly, cells were harvested from exponential phase cultures by trypsinization, counted and were seeded for colony formation. On following day, γ radiation was delivered using a dual-source 137Cs unit at a dose rate of 3.2 Gy/min with a GC-3000 Elan irradiator (MDS Nordion). After 14 d, colonies were stained with 0.4% crystal violet (Sigma). The colonies were counted with a counter and > 30 cells were considered as colonies. The plating efficiency (PE) represents the percentage of cells seeded that grow into colonies under a specific culture condition of a given cell line. The survival fraction, expressed as a function of irradiation, was calculated as follows: Survival fraction = colonies counted/(cells seeded × PE/100).
Immunofluorescence analysis
Cells cultured on 18-mm cover glass were fixed with 3.7% paraformaldehyde for 10 min, permeabilized with 1% Triton X-100 for 5 min and then blocked with blocking buffer (10% FBS in PBS) for 30 min. Anti-γH2AX antibody (Millipore) was diluted to 1:500 and applied overnight at 4°C and subsequent incubation with fluorescein-conjugated secondary antibodies (Invitrogen) was conducted for 1 h at room temperature. Immunofluorescence images were captured with a laser-scanning confocal microscope (LSM 710, Carl Zeiss MicroImaging GmbH). To calculate γH2AX foci intensity per cell, images were processed and measured using Image-Pro Plus software (Media Cybernetics Inc.) and then divided by the number of nucleus.
Western blot analysis
Cells were lysed with RIPA buffer (20 mM TRIS-HCl (pH 7.5), 150 mM NaCl, 1 mM EDTA, 1 mM EGTA, 1% NP-40, 1% sodium deoxycholate and protease inhibitors) and briefly sonicated. Protein content was measured with Coomassie (Bradford) Protein Assay Kit (Thermo Fisher Scientific, Inc.). Equal amounts of cell lysate were separated by SDS-polyacrylamide gels, transferred to nitrocellulose membrane (Bio-Rad), immunoblotted and detected by chemiluminescence using ECL detection reagents. Antibodies were obtained as follow. Anti-HMGB2 was from Abcam, anti-γH2AX was from Millipore, anti-p53, anti-β-actin and Horseradish peroxidase-conjugated secondary antibodies were from Santa Cruz Biotechnology, Inc.
Quantitative realtime polymerase chain reaction (qRT-PCR)
Total RNA from cells was isolated by using the Hybrid-R Total RNA Purification Kit (GeneAll). A total of 1 µg of total RNA preparation was reverse transcribed with PrimeScript RT Master Mix (Takara). After dilution to 1:10, 2 µl of cDNA was used as template in 20 µl PCR reaction. qRT-PCR was performed using SYBR Premix Ex TaqII (Takara) or qPCR Master Mix (MBiotech) on a CFX96TM realtime PCR system (Bio-Rad). The following primers were used for qRT-PCR: HMGB2 (5′-CTTGGCACGATATGCAGCAA-3′, 5′-CAGCCAAAGATAAACAACCATATGA-3′); p21/WAF1 (5′-CTGCGCCAGCTGAGGTGTGAG-3′, 5′-GCCGCATGGGTTCTGACGGA-3′); BBC3/PUMA (5′-CCTGGAGGGTCCTGTACAATCT-3′, 5′GCACCTAATTGGGCTCCATCT-3); PPIA/CYPA (5′-CCCACCGTGTTCTTCGACAT-3′, 5′-CCAGTGCTCAGAGCACGAAA-3′); RPL13A (5′-CCTGGAGGAGAAGAGGAAAGAGA-3′, 5′-TTGAGGACCTCTGTGTATTTGTCAA-3′); B2M (5′-TGCTGTCTCCATGTTTGATGTATCT-3′, 5′-TCTCTGCTCCCCACCTCTAAGT-3′). The relative quantification of gene expression was calculated with the ΔΔCt method using CFX manager software (Bio-Rad) with multiple reference genes (PPIA, RPL13A and B2M). PCR program was as follows: after an initial pre-incubation step at 95°C for 10 min, there were 45 cycles, each consisting of 95°C for 5 sec and 60°C for 30 sec. The last amplification cycle was followed by a melt curve analysis to make sure about the specificity of the PCR amplification.
Establishment of tetracycline inducible p53 expressing HT-29 cell
Tet-inducible p53 expressing HT-29 cell was made using Retro-XTM Ter-On advanced inducible expression system (Clontech Laboratories) and retroviruses were generated in Plat-GP packaging cell line (Cell Biolabs).To make a rtTA-expressing cell, HT-29 cell was infected with retrovirus generated with pRetroX-Tet-On Advanced vector and selected 500 µg/ml of G418 for 7 d. To make a tet-inducible p53 construct, XbaI fragment of wildtype TP53 gene (Addgene plasmid #16434) was subcloned into pRetroX-Tight-pur vector. Retrovirus was introduced to previously made rtTA-expressing HT-29 cell and selected with puromycin (1 µg/ml) for 7 d. To induce p53 expression, doxycycline (Sigma) was treated to 500 µg/ml for 2 d.
Construction of HMGB2 luciferase reporter plasmid and luciferase assay
Human HMGB2 promoter region covering upstream 1,000 bp and downstream795 bp from the transcription start site of the reference sequence (NM_002129.3) was PCR amplified with primers (5′-AAACTCCTCGAGGTAGGACGAGACAGTGAATGC -3′ and 5′-AATGCCAAGCTTGTTGACAGATCCGCGTCCAC-3′) and subcloned into a position that was 5′ to the luciferase gene between the XhoI and HindIII sites of the pGL3-Basic luciferase reporter vector (Promega). To assay the HMGB2 promoter activity, 5 x 104 293T cells were transfected with 100 ng of pGL3-basic or pGL3-HMGB2p and 2 ng of pRL-SV40 as a transfection efficiency control. To examine the effect of p53 on HMGB2 promoter activity, 20 or 100 ng of pCMV-Neo-Bam p53 wt (Addgene plasmid #16434) or V143A mutant (Addgene plasmid #16435) plasmid was cotransfected. Cells were then allowed to recover for 24 h at 37°C and harvested and analyzed by a Dual-Luciferase® Reporter Assay System (Promega) and luminescence was measured using GloMax 20/20 Luminometer (Promega).
| Abbreviations: | ||
| HMGB2 | = | high-mobility group box 2 |
| SCR | = | scrambled |
| qRT-PCR | = | quantitative realtime polymerase chain reaction |
| Tet-On | = | tetracycline inducible |
| Dox | = | doxycycline |
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Acknowledgments
We acknowledge Drs. Bob Weinberg, Sheila Stewart and Bert Vogelstein for providing plasmids. This work was supported by the National Nuclear R and D Program of the Ministry of Education, Science and Technology, Republic of Korea.
References
- Bianchi ME, Agresti A. HMG proteins: dynamic players in gene regulation and differentiation. Curr Opin Genet Dev 2005; 15:496 - 506; http://dx.doi.org/10.1016/j.gde.2005.08.007; PMID: 16102963
- Agresti A, Bianchi ME. HMGB proteins and gene expression. Curr Opin Genet Dev 2003; 13:170 - 8; http://dx.doi.org/10.1016/S0959-437X(03)00023-6; PMID: 12672494
- Tang D, Kang R, Zeh HJ 3rd, Lotze MT. High-mobility group box 1 and cancer. Biochim Biophys Acta 2010; 1799:131 - 40; http://dx.doi.org/10.1016/j.bbagrm.2009.11.014; PMID: 20123075
- Lotze MT, Tracey KJ. High-mobility group box 1 protein (HMGB1): nuclear weapon in the immune arsenal. Nat Rev Immunol 2005; 5:331 - 42; http://dx.doi.org/10.1038/nri1594; PMID: 15803152
- Yamada S, Inoue K, Yakabe K, Imaizumi H, Maruyama I. High mobility group protein 1 (HMGB1) quantified by ELISA with a monoclonal antibody that does not cross-react with HMGB2. Clin Chem 2003; 49:1535 - 7; http://dx.doi.org/10.1373/49.9.1535; PMID: 12928240
- Yamoah K, Brebene A, Baliram R, Inagaki K, Dolios G, Arabi A, et al. High-mobility group box proteins modulate tumor necrosis factor-alpha expression in osteoclastogenesis via a novel deoxyribonucleic acid sequence. Mol Endocrinol 2008; 22:1141 - 53; http://dx.doi.org/10.1210/me.2007-0460; PMID: 18218727
- Pusterla T, de Marchis F, Palumbo R, Bianchi ME. High mobility group B2 is secreted by myeloid cells and has mitogenic and chemoattractant activities similar to high mobility group B1. Autoimmunity 2009; 42:308 - 10; http://dx.doi.org/10.1080/08916930902831845; PMID: 19811285
- Ronfani L, Ferraguti M, Croci L, Ovitt CE, Schöler HR, Consalez GG, et al. Reduced fertility and spermatogenesis defects in mice lacking chromosomal protein Hmgb2. Development 2001; 128:1265 - 73; PMID: 11262228
- Taniguchi N, Caramés B, Kawakami Y, Amendt BA, Komiya S, Lotz M. Chromatin protein HMGB2 regulates articular cartilage surface maintenance via beta-catenin pathway. Proc Natl Acad Sci U S A 2009; 106:16817 - 22; http://dx.doi.org/10.1073/pnas.0904414106; PMID: 19805379
- Taniguchi N, Caramés B, Hsu E, Cherqui S, Kawakami Y, Lotz M. Expression patterns and function of chromatin protein HMGB2 during mesenchymal stem cell differentiation. J Biol Chem 2011; 286:41489 - 98; http://dx.doi.org/10.1074/jbc.M111.236984; PMID: 21890638
- Pardo M, García A, Thomas B, Piñeiro A, Akoulitchev A, Dwek RA, et al. The characterization of the invasion phenotype of uveal melanoma tumour cells shows the presence of MUC18 and HMG-1 metastasis markers and leads to the identification of DJ-1 as a potential serum biomarker. Int J Cancer 2006; 119:1014 - 22; http://dx.doi.org/10.1002/ijc.21942; PMID: 16570276
- Mayer C, Stich N, Palkovits R, Bauer G, Pittner F, Schalkhammer T. High-throughput assays on the chip based on metal nano-cluster resonance transducers. J Pharm Biomed Anal 2001; 24:773 - 83; http://dx.doi.org/10.1016/S0731-7085(00)00545-8; PMID: 11248470
- Balasubramani M, Day BW, Schoen RE, Getzenberg RH. Altered expression and localization of creatine kinase B, heterogeneous nuclear ribonucleoprotein F and high mobility group box 1 protein in the nuclear matrix associated with colon cancer. Cancer Res 2006; 66:763 - 9; http://dx.doi.org/10.1158/0008-5472.CAN-05-3771; PMID: 16424007
- He Q, Liang CH, Lippard SJ. Steroid hormones induce HMG1 overexpression and sensitize breast cancer cells to cisplatin and carboplatin. Proc Natl Acad Sci U S A 2000; 97:5768 - 72; http://dx.doi.org/10.1073/pnas.100108697; PMID: 10811891
- Balani P, Boulaire J, Zhao Y, Zeng J, Lin J, Wang S. High mobility group box2 promoter-controlled suicide gene expression enables targeted glioblastoma treatment. Mol Ther 2009; 17:1003 - 11; http://dx.doi.org/10.1038/mt.2009.22; PMID: 19240692
- Kwon JH, Kim J, Park JY, Hong SM, Park CW, Hong SJ, et al. Overexpression of high-mobility group box 2 is associated with tumor aggressiveness and prognosis of hepatocellular carcinoma. Clin Cancer Res 2010; 16:5511 - 21; http://dx.doi.org/10.1158/1078-0432.CCR-10-0825; PMID: 20851854
- Colorectal Cancer Collaborative Group. Adjuvant radiotherapy for rectal cancer: a systematic overview of 8,507 patients from 22 randomised trials. Lancet 2001; 358:1291 - 304; http://dx.doi.org/10.1016/S0140-6736(01)06409-1; PMID: 11684209
- Ghadimi BM, Grade M, Difilippantonio MJ, Varma S, Simon R, Montagna C, et al. Effectiveness of gene expression profiling for response prediction of rectal adenocarcinomas to preoperative chemoradiotherapy. J Clin Oncol 2005; 23:1826 - 38; http://dx.doi.org/10.1200/JCO.2005.00.406; PMID: 15774776
- Watanabe T, Komuro Y, Kiyomatsu T, Kanazawa T, Kazama Y, Tanaka J, et al. Prediction of sensitivity of rectal cancer cells in response to preoperative radiotherapy by DNA microarray analysis of gene expression profiles. Cancer Res 2006; 66:3370 - 4; http://dx.doi.org/10.1158/0008-5472.CAN-05-3834; PMID: 16585155
- Kim IJ, Lim SB, Kang HC, Chang HJ, Ahn SA, Park HW, et al. Microarray gene expression profiling for predicting complete response to preoperative chemoradiotherapy in patients with advanced rectal cancer. Dis Colon Rectum 2007; 50:1342 - 53; http://dx.doi.org/10.1007/s10350-007-277-7; PMID: 17665260
- Bunz F, Dutriaux A, Lengauer C, Waldman T, Zhou S, Brown JP, et al. Requirement for p53 and p21 to sustain G2 arrest after DNA damage. Science 1998; 282:1497 - 501; http://dx.doi.org/10.1126/science.282.5393.1497; PMID: 9822382
- Munshi A, Hobbs M, Meyn RE. Clonogenic cell survival assay. Methods Mol Med 2005; 110:21 - 8; PMID: 15901923
- Rashi-Elkeles S, Elkon R, Shavit S, Lerenthal Y, Linhart C, Kupershtein A, et al. Transcriptional modulation induced by ionizing radiation: p53 remains a central player. Mol Oncol 2011; 5:336 - 48; http://dx.doi.org/10.1016/j.molonc.2011.06.004; PMID: 21795128
- Iacopetta B. TP53 mutation in colorectal cancer. Hum Mutat 2003; 21:271 - 6; http://dx.doi.org/10.1002/humu.10175; PMID: 12619112
- Rodrigues NR, Rowan A, Smith ME, Kerr IB, Bodmer WF, Gannon JV, et al. p53 mutations in colorectal cancer. Proc Natl Acad Sci U S A 1990; 87:7555 - 9; http://dx.doi.org/10.1073/pnas.87.19.7555; PMID: 1699228
- Vousden KH, Lu X. Live or let die: the cell’s response to p53. Nat Rev Cancer 2002; 2:594 - 604; http://dx.doi.org/10.1038/nrc864; PMID: 12154352
- Cuddihy AR, Bristow RG. The p53 protein family and radiation sensitivity: Yes or no?. Cancer Metastasis Rev 2004; 23:237 - 57; http://dx.doi.org/10.1023/B:CANC.0000031764.81141.e4; PMID: 15197326
- Krynetski EY, Krynetskaia NF, Bianchi ME, Evans WE. A nuclear protein complex containing high mobility group proteins B1 and B2, heat shock cognate protein 70, ERp60 and glyceraldehyde-3-phosphate dehydrogenase is involved in the cytotoxic response to DNA modified by incorporation of anticancer nucleoside analogues. Cancer Res 2003; 63:100 - 6; PMID: 12517784
- Krynetskaia NF, Phadke MS, Jadhav SH, Krynetskiy EY. Chromatin-associated proteins HMGB1/2 and PDIA3 trigger cellular response to chemotherapy-induced DNA damage. Mol Cancer Ther 2009; 8:864 - 72; http://dx.doi.org/10.1158/1535-7163.MCT-08-0695; PMID: 19372559
- Prasad R, Liu Y, Deterding LJ, Poltoratsky VP, Kedar PS, Horton JK, et al. HMGB1 is a cofactor in mammalian base excision repair. Mol Cell 2007; 27:829 - 41; http://dx.doi.org/10.1016/j.molcel.2007.06.029; PMID: 17803946
- Lange SS, Mitchell DL, Vasquez KM. High mobility group protein B1 enhances DNA repair and chromatin modification after DNA damage. Proc Natl Acad Sci U S A 2008; 105:10320 - 5; http://dx.doi.org/10.1073/pnas.0803181105; PMID: 18650382
- Jiao Y, Wang HC, Fan SJ. Growth suppression and radiosensitivity increase by HMGB1 in breast cancer. Acta Pharmacol Sin 2007; 28:1957 - 67; http://dx.doi.org/10.1111/j.1745-7254.2007.00669.x; PMID: 18031610
- Wang LL, Meng QH, Jiao Y, Xu JY, Ge CM, Zhou JY, et al. High-mobility group boxes mediate cell proliferation and radiosensitivity via retinoblastoma-interaction-dependent and -independent mechanisms. Cancer Biother Radiopharm 2012; 27:329 - 35; http://dx.doi.org/10.1089/cbr.2012.1199; PMID: 22655796
- Yamazaki F, Nagatsuka Y, Shirakawa H, Yoshida M. Repression of cell cycle progression by antisense HMG2 RNA. Biochem Biophys Res Commun 1995; 210:1045 - 51; http://dx.doi.org/10.1006/bbrc.1995.1762; PMID: 7763232
- Stros M, Ozaki T, Bacikova A, Kageyama H, Nakagawara A. HMGB1 and HMGB2 cell-specifically down-regulate the p53- and p73-dependent sequence-specific transactivation from the human Bax gene promoter. J Biol Chem 2002; 277:7157 - 64; http://dx.doi.org/10.1074/jbc.M110233200; PMID: 11748232
- Travers AA. Priming the nucleosome: a role for HMGB proteins?. EMBO Rep 2003; 4:131 - 6; http://dx.doi.org/10.1038/sj.embor.embor741; PMID: 12612600
- Franklin S, Chen H, Mitchell-Jordan S, Ren S, Wang Y, Vondriska TM. Quantitative analysis of the chromatin proteome in disease reveals remodeling principles and identifies high mobility group protein B2 as a regulator of hypertrophic growth. Mol Cell Proteomics 2012; 11:M111 - , 014258; http://dx.doi.org/10.1074/mcp.M111.014258; PMID: 22270000
- Polanská E, Dobšáková Z, Dvořáčková M, Fajkus J, Štros M. HMGB1 gene knockout in mouse embryonic fibroblasts results in reduced telomerase activity and telomere dysfunction. Chromosoma 2012; 121:419 - 31; http://dx.doi.org/10.1007/s00412-012-0373-x; PMID: 22544226