Abstract
p19INK4d (CDKN2D) is a negative regulator of the cell cycle. Little is known of its role in cancer development and prognosis. We aimed to evaluate the clinical significance of p19INK4d expression in ovarian carcinomas with respect to the TP53 accumulation status, as well as the frequency of CDKN2D mutations. p19INK4d and TP53 expression was evaluated immunohistochemically in 445 ovarian carcinomas: 246 patients were treated with platinum–cyclophosphamide (PC/PAC), while 199 were treated with taxane–platinum agents (TP). CDKN2D gene expression (mRNA) was examined in 106 carcinomas, while CDKN2D mutations in 68 tumors. Uni- and multivariate statistical analyses (logistic regression and the Cox proportional hazards model) were performed for patient groups divided according to the chemotherapeutic regimen administered, and in subgroups with and without TP53 accumulation. High p19INK4d expression increased the risk of death, but only in patients with the TP53-negative carcinomas (HR 1.61, P = 0.049 for PC/PAC-treated patients, HR 2.00, P = 0.015 for TP-treated patients). This result was confirmed by the mRNA analysis (HR 4.24, P = 0.001 for TP-treated group). High p19INK4d protein expression associated with adverse clinicopathological factors. We found no alterations in the CDKN2D gene; the c.90C>G (p.R30R; rs1968445) polymorphism was detected in 10% of tumors. Our results suggest that p19INK4d expression is a poor prognostic factor in ovarian cancer patients. Analyses of tumor groups according to the TP53 accumulation status facilitate the identification of cancer biomarkers.
Introduction
Ovarian cancer is the most lethal gynecological malignancy. In the last decade, taxanes combined with cisplatin or its analogs (TP regimen) have been considered as a standard first-line treatment for ovarian cancer.Citation1,Citation2 This therapy replaced platinum–cyclophosphamide (PC) or other protocols based on DNA-damaging agents (PAC, platinum–doxorubicin–cyclophosphamide, or monotherapy). Although the introduction of taxanes significantly improved treatment results, the overall survival rates are still poor in patients with advanced disease.Citation2-Citation4 Analysis of molecular prognostic factors may reveal novel potential targets of molecular therapy.
Inhibitors of cyclin-dependent kinases (INK4) play a key role in the cell cycle arrest under cellular stress caused, for example, by chemotherapeutic drugs. The INK4 family includes four members, p16INK4a, p15INK4b, p18INK4c, and p19INK4d, that specifically bind to and inhibit the kinase activity of CDK4/6 by preventing their interaction with cyclin D. The CDK4/6-cyclin D complex phosphorylates the retinoblastoma protein (pRB), thereby enabling the cell to enter S phase of the cell cycle.Citation5 One member of INK4 family, p16INK4a, is homozygously deleted at a high frequency in a wide variety of human tumor-derived cell lines and several specific types of primary tumors.Citation6 Inactivation of other INK4 family genes (p15INK4b or p18INK4c) was observed in some cell lines, whereas there is no evidence of their inactivation in solid tumors; even less is known on the role of p19INK4d protein in tumorigenesis.Citation7
p19INK4d was identified in yeast in 1995.Citation8 This 166 amino acid protein is encoded by the CDKN2D (cyclin-dependent kinase inhibitor 2D) gene located on chromosome 19p13. p19INK4d is highly similar to other INK4 family members, but in contrast to p16INK4a, it has five ankyrin-like repeats that are thought to mediate protein–protein interactions.Citation9 Data obtained from mouse NIH 3T3 cells showed that p19INK4d protein, like other INK4s, inhibits CDK4–cyclin D1 activity in vivo, and induces G1 phase arrest.Citation8 Recent discoveries suggest that p19INK4d also plays an important role in the DNA damage response pathway dependent on ATM/ATR kinases. After genotoxic stress caused by agents such as cisplatin, p19INK4d enters the nucleus.Citation10 Interestingly, p19INK4d activities in DNA repair and cell cycle arrest seem to be independent. The protein with mutated phosphorylation sites is unable to enter the nucleus but can still stop cell cycle progression. These data suggest that p19INK4d status may interfere with cellular responses to cytotoxic agents.
p19INK4d is expressed ubiquitously in proliferating cultured cells and in normal mouse tissues.Citation8,Citation11,Citation12 Its expression shows variations during organ development and cell cycle progression.Citation13 p19INK4d expression in cancers has been examined in only a few studies and, to date, it has not been linked to cancer development.Citation14-Citation16
TP53 accumulation is one of the most frequently observed aberrations in ovarian carcinomas.Citation17 TP53 dysfunction may enhance response of ovarian cancer to taxane–platinum treatment.Citation4,Citation18,Citation19 Regarding cisplatin, impaired TP53 protein function may contribute to resistance to this drug, as observed in cell lines and some clinical studies.Citation20-Citation23 The results obtained in recent years suggest that the TP53 status may influence the clinical importance of other molecular factors.Citation24-Citation26
We aimed to evaluate the prognostic and predictive value of p19INK4d expression at the protein and mRNA level with respect to TP53 status in a large group of ovarian cancer patients treated with two different chemotherapy modules. We also searched for CDKN2D gene alterations in these tumors.
Results
Analysis of p19INK4d protein expression
p19INK4d expression was observed in both the nuclei and the cytoplasm of ovarian cancer cells, but predominantly in the former (). High p19INK4d expression (overall score ≥4) was found in 65% (288 of 446) of tumors. Rates for tumors with score category 0, 1, 2, 3, 4, 5, and 6 were 0%, 2%, 7%, 27%, 41%, 23%, and 1%, respectively ().
Figure 1. p19INK4d protein expression in two different ovarian carcinomas (400× , hematoxylin counterstain): (A) low p19INK4d expression, (B) high p19INK4d expression.
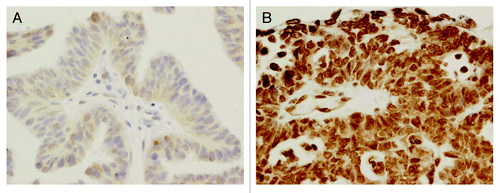
Table 1. p19INK4d and TP53 expression in ovarian carcinomas
The increased p19INK4d immunostaining was detected in 218 serous (218 of 342, 69%), 16 endometrioid (16 of 22; 73%), 8 clear cell (8 of 15, 53%), 25 undifferentiated (25 of 33, 76%), and in 18 tumors of other histological types (18 of 31, 58%). There was no association between p19INK4d expression and tumor histology. High p19INK4d expression was more common in poorly differentiated (86 of 120, 72%) than in moderately and well differentiated carcinomas (172 of 269, 64%; 29 of 56, 52%, respectively; P = 0.035). It was also generally more frequent in higher clinical stages than in lower ones (P = 0.037 in linear trend). High expression of p19INK4d was found in 39 of 50 (78%) carcinomas in FIGO stage IV, 185 of 284 (65%) in stage IIIC, 45 of 84 (54%) in stages III A, B; unexpectedly, p19INK4d expression was also high in FIGO stages IIB and C—in 18 of 27 (67%) tumors. High p19INK4d expression was significantly more frequent in cases with residual tumor size (RT) >2 cm (148 of 211, 70%) than in those with RT 0.5–2 cm (89 of 145, 61%) or complete debulking (50 of 89, 56%) (P = 0.044).
We investigated the potential relationship between the intensity of p19INK4d immunostaining and patients outcomes with respect to TP53 accumulation status.
p19INK4d expression in the platinum/cyclophosphamide-treated group
In the group of patients treated with PC/PAC regimen (n = 246) there was an association between high p19INK4d expression and complete remission of the disease (CR). Patients who reached CR had lower frequency of high p19INK4d expression (78 of 143; 55% patients) than those who had other responses (81 of 112; 72% patients). In the univariate analysis, high p19INK4d expression negatively influenced the probability of complete remission (OR 0.53, P = 0.022; ). This result was not confirmed by the multivariate analysis.
Table 2. Associations of p19INK4d expression with clinical endpoints in ovarian cancer* (univariate Cox proportional hazards and logistic regression models)
In the subgroup of patients without TP53 accumulation in their cancers (n = 101), median overall survival time was significantly shorter in case of high p19INK4d protein expression than in case of low level of p19INK4d (20 vs. 29 mo, P = 0.035). In both the univariate and multivariate analysis, high p19INK4d expression increased the risk of death (HR 1.59, P = 0.035, ; ; HR 1.61, P = 0.049, , respectively). Overall survival also showed an association with FIGO stage and debulking status (). Disease-free survival, complete remission and platinum sensitivity did not associate with p19INK4d expression in the TP53− subgroup.
Figure 2. Prognostic significance of p19INK4d (CDKN2D) expression. Patients treated with cisplatin/cyclophosphamide (A) and taxane–platinum (B) regimens with high p19INK4d expression had a significantly worse overall survival than patients with low p19INK4d expression (Kaplan–Meier curve). CDKN2D gene expression at the mRNA level correlated with enhanced risk of death in taxane–platinum-treated group (C).
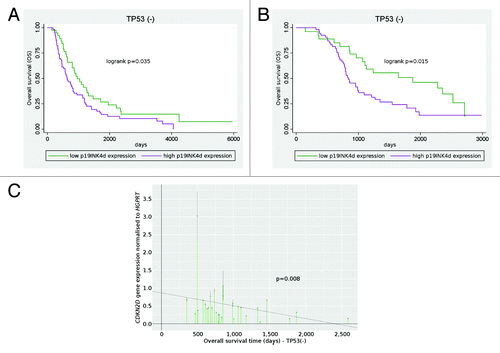
Table 3. Associations of p19INK4d expression and clinico-pathological factors with overall survival in ovarian cancer patients* (multivariate Cox proportional hazards models)
In the TP53+ subgroup (n = 145), there were no associations between clinical endpoints and p19INK4d protein expression.
p19INK4d expression in the taxane/platinum-treated group
In the entire group of patients treated with the TP regimen (n = 199) and in the TP53+ subgroup (n = 110), there were no associations between clinical endpoints and the expression of the p19INK4d protein.
Similarly to the above-described findings for the PC/PAC-treated group, in the subgroup of patients without TP53 accumulation in their tumors (n = 81), median overall survival was significantly shorter for patients with high level of p19INK4d protein than for those with low level of p19INK4d (27 vs. 42.5 mo, P = 0.015). In both the univariate and multivariate analysis, high p19INK4d expression increased the risk of death (HR 2.03, P = 0.015, ; ; HR 2.00, P = 0.015, , respectively). Disease-free survival, complete remission and platinum sensitivity did not associate with p19INK4d expression.
Analysis of CDKN2D gene expression
We examined whether there was an association between high CDKN2D mRNA level and patients’ outcome.
In accordance with the IHC results, CDKN2D mRNA expression showed an association with overall survival in the TP53− subgroup of patients treated with TP regimen (n = 29). High CDKN2D mRNA levels associated with an enhanced risk of death in both the univariate and multivariate analysis (HR 2.94, P = 0.008, ; HR 4.24, P = 0.001, , respectively).
Table 4. Associations of CDKN2D mRNA expression and clinico-pathological factors with overall survival in taxane-platinum-treated group* (uni- and multivariate Cox proportional hazards model)
In the entire patients group (n = 74) and in the TP53+ subgroup (n = 45), as well as in the group of patients treated with the PC/PAC regimen (only 11 TP53-negative carcinomas), there were no associations between CDKN2D gene expression and clinical endpoints.
Analysis of gene mutations and polymorphisms
We found no gene alterations except for one synonymous substitution. The c.90C>G (p.R30R; rs1968445) polymorphism in exon 1 of the CDKN2D gene was present in seven of 68 cases (10.3%).
Discussion
This study has revealed a new prognostic factor in solid tumors, i.e., the p19INK4d from the family of genes encoding cyclin-dependent kinase inhibitors. Its expression at the mRNA and/or protein level was found to negatively influence overall survival in a large group of ovarian cancer patients. This association was clearly demonstrated in patients treated either with platinum–cyclophosphamide (PC/PAC) or taxane–platinum (TP) regimens, but only in the subgroup without TP53 accumulation in ovarian carcinomas.
In the literature there are few data on p19INK4d expression in human tumors and its role in tumorigenesis. p19INK4d expression in paraffin-embedded tissue was first analyzed by Thullberg et al.Citation27 in 2000. In agreement with our results, they observed the p19INK4d protein in the nucleus and cytoplasm. It is noteworthy that the mouse monoclonal antibody (clone DCS-100), used in that and the current study, does not give any cross-reactions with other cellular proteins present in whole-cell lysates of human cell lines. Bartkova et al.Citation14 also observed p19INK4d expression in both nuclei and cytoplasm in normal human testis, but not in testicular cancer. However, there are some studies on cell lines demonstrating that p19INK4d has only a cytoplasmic location.Citation8,Citation11,Citation28 Ceruti et al.Citation28 observed in human neuroblastoma cell line SH-SY5Y that p19INK4d protein translocated into the nucleus after UV irradiation. It appears that the translocation is not a constitutive process but occurs in response to cell cycle regulatory signals and external stimuli such as UV or cisplatin treatment.Citation10,Citation28
The results obtained by our group show that high p19INK4d protein expression associates with adverse clinicopathological factors, i.e., high tumor grade, advanced FIGO stage and bulky residual tumor. Despite the aggressive tumor features, high p19INK4d protein expression seems to be an independent negative prognostic factor, but this was not consistent in all patients evaluated.
There are two other analyses of clinical importance of p19INK4d immunohistochemical expression in cancer patients: Bartkova et al.Citation14,Citation15 studied 71 testicular cancers but these tumors did not express the p19INK4d protein. Zhang et al.Citation16 did not find any prognostic value of p19INK4d protein expression (and also CDKN2D mRNA expression) in patients with nasopharyngeal carcinoma. Thus, the current study and that of Zhang et al.Citation16 are the only ones evaluating CDKN2D mRNA expression in cancer cells and its impact on clinical endpoints. The negative results of these two analyses are in agreement with each other and generally with the lack of clinical importance of p19INK4d protein expression in patients studied.
Our analysis revealed the negative impact of p19INK4d expression (at the protein and mRNA level) only in the subgroup with TP53-negative carcinomas. In the majority of tumors, TP53 immunohistochemical accumulation reflects a TP53 gene missense mutation and TP53 dysfunction, while the lack of TP53 accumulation reflects preserved TP53 function or loss of function due to a nonsense mutation. As previously shown for several proteins, the TP53 immunohistochemical status may determine their clinical significance, particularly if they are regulated by, or interfere with TP53 during the control of tumor cell proliferation or apoptosis.Citation24-Citation26,Citation29
In the literature there are not yet any data connecting the TP53 protein to the p19INK4d protein; nevertheless, they participate in the regulation of the same processes, i.e., proliferation, apoptosis and DNA repair. p19INK4d is activated in response to stress caused by some genotoxic agents such as cisplatin.Citation30 Some experimental results indicate that the p19INK4d overexpression enhances DNA repair and cell viability.Citation30,Citation31 In cell lines deprived of p19INK4d, repair of DNA lesions was impaired and apoptosis was enhanced.Citation30-Citation32 In a previous study, p19INK4d-deficient mice did not develop cancers, and apoptosis was found to be increased.Citation33 Therefore, low levels of p19INK4d protein expression may decrease the efficiency of DNA repair processes, which may result in a higher level of lesions and the activation of apoptosis. Some studies indicate that p19INK4d may promote cell viability: the expression of p19INK4d enhances cell survival after UV treatment, while silencing of its expression increases sensitivity to genotoxic agents in comparison with cells overexpressing p19INK4d.Citation31 Our results suggest that the effect of p19INK4d action may be particularly efficient in cells with functional TP53, which is when it becomes clinically visible. Further studies are necessary to elucidate whether there is any interaction between p19INK4d and TP53.
Mutations in CDKN2D gene were also described in various tumor types. The analysis of 67 patients with osteosarcomas showed alterations of the CDKN2D gene, including one frameshift mutation, and five gene rearrangements.Citation34 A frameshift mutation in exon 1 (10%) was also reported in two ovarian cell lines but in no other sample in a large series of cell lines and tumors.Citation35,Citation36 In our study we did not observe any deletions or point mutations in the CDKN2D gene in a group of 68 patients with ovarian cancer. Only one genetic polymorphism at codon 30 (c.90C>G) in exon 1 of the CDKN2D gene was identified in 10% of samples. Other studies did not report the presence of that alteration in a variety of neoplasms, including hematopoietic neoplasms, cervical, breast and prostate cancer.Citation34,Citation37,Citation38
In conclusion, our results show for the first time that high p19INK4d expression associates with aggressive clinicopathological cancer features and enhances the risk of death in patients with TP53-negative ovarian carcinomas. This confirms our previous observations that analyses of uniform groups, such as with regard to the TP53 accumulation status, facilitates the identification of cancer biomarkers.
Materials and Methods
Patients and tumors
The study was performed on 445 archival samples of ovarian carcinomas taken during staging laparotomy, before administration of chemotherapy. Medical records were critically reviewed by at least two clinicians. The patients were treated with standard PC (cisplatin−cyclophosphamide or carboplatin−cyclophosphamide) or PAC chemotherapy (PC and doxorubicin) (246 patients), or with taxane–platinum chemotherapy (TP: paclitaxel or docetaxel with cisplatin or carboplatin) (199 patients).Citation4 The material was carefully selected out of a total of 899 cases submitted to meet additional criteria: adequate staging procedure, International Federation of Gynaecologists and Obstetricians (FIGO) stage IIB to IV disease.Citation39 tumor tissue from the first laparotomy available, moderate (G2) or poor tumor differentiation (G3 and G4), availability of clinical data, including residual tumor size and follow-up.
All tumors were uniformly reviewed histopathologically, classified according to the criteria of the World Health OrganizationCitation40 and graded in a four-grade scale, according to the criteria given by Broders.Citation41 Clinicopathological characteristics are presented in .
Table 5. Patients’ characteristics
For the PC/PAC-treated group, the follow-up time ranged from 4.4 to 198.3 mo (median: 27.5); the respective values for the TP-treated group were: 4.8 to 103.4 mo (median: 35.5). Short follow-up times resulted from early patient deaths. All surviving patients had at least a 6 mo of follow-up. Response to chemotherapy was evaluated retrospectively according to the World Health Organization response evaluation criteria.Citation42 The evaluation was based on data from medical records describing the patient’s clinical condition and CA125 levels in 3–4 week intervals. Complete remission (CR) was defined as the disappearance of all clinical and biochemical symptoms of ovarian cancer evaluated after completion of first-line chemotherapy and confirmed at four weeks. Within the CR group, we identified a platinum-sensitive group (PS, disease-free survival longer than six months). Other tumors were described as platinum-resistant.Citation43
The study was approved by the bioethics committee of the Institute of Oncology (39/2007).
Immunohistochemical analysis
Immunohistochemical stainings were performed on paraffin-embedded material after heat-induced epitope retrieval (HIER). We used a mouse monoclonal anti-p19INK4d antibody (1/175, Sigma-Aldrich). TP53 protein was detected with the use of PAb1801 monoclonal antibody (1/3000, Sigma-Genosys), as described previously.Citation24 The antigens were retrieved by heating the sections in 0.01 M citrate buffer (pH 6.0): 2 h at 99 °C in a water bath for p19INK4d and 2 × 5 min for TP53 at 700 W in a microwave oven. Non-specific tissue and endogenous peroxidase reactivities were blocked with 10% BSA and 3% H2O2, respectively. The sections were incubated with primary antibodies overnight, at 4 °C. Biotinylated secondary goat anti-mouse IgG (1/1500), peroxidase-conjugated streptavidin (1/500) (all from Immunotech), and DAB were used as a detection system. As a positive control for TP53 accumulation, we used a tumor with a defined TP53 gene missense mutation.Citation44 Normal mouse IgG of the same subclasses and at the concentrations of the relevant primary antibodies served as negative controls.
The evaluation of immunohistochemical stainings
p19INK4d expression was scored independently for nuclear and cytoplasmic staining. Light microscopic evaluation at 400× magnification was used to count tumor cells within the areas of the strongest staining. p19INK4d expression was categorized according to the staining intensity (0 or weak to strong: 1 to 3). An overall protein expression score was calculated by adding the nuclear and cytoplasmic scores (overall score range, 0–6). This overall score for each case was further simplified by dichotomizing it to low (overall score of ≤3) or high (score of ≥4). Two independent assessors (AFG and AR) concurred in 80% of the cases, and reached consensus in the remaining cases. TP53 protein accumulation was assessed as present (more than 10% positive cells) or absent, as previously described.Citation4
CDKN2D expression at the mRNA level
Real-time PCR experiments were conducted on 74 of 199 ovarian cancer samples from patients treated with a taxane–platinum regimen and on 32 of 426 samples from patients treated with a platinum/cyclophosphamide regimen. Only tumors containing less than 5% of non-tumor tissue were analyzed at mRNA level. Total RNA was extracted from frozen tissues with the Nucleo Spin RNA kit (Mecherey Nagel) according to the manufacturer’s protocol. RNA quantity was measured by UV spectrophotometry, and its quality was assessed by the 260/280 ratio and in a 1% agarose gel. One microgram of total RNA was transcribed to cDNA using the Super Script III First Strand kit (Invitrogen,). Quantitative RT-PCR (Q-PCR) was run on the 7500 Fast Real-Time PCR System (Applied Biosystems), with the use of the TaqMan Gene Expression Assays (Hs00176481_m1 for CDKN2D and 4326321E for the reference gene HGPRT). All real-time PCR reactions were performed in triplicate in the final volume 10 μl for 40 cycles according to the following protocol: each cycle at 95 °C for the initial 10 min, then at 95 °C for 15 s, and 60 °C for 1 min. The obtained results were averaged, and gene expression levels were normalized to the HGPRT expression. A standard curve, used in all experiments, was prepared from serial dilutions of cDNA from one of the tumors analyzed.
Analysis of the CDKN2D gene mutations and polymorphisms
The analysis of genetic alterations of the CDKN2D gene was performed for 68 ovarian tumors. Genomic DNA was isolated from frozen tissues using the QIAamp DNA Extraction Kit (Qiagen) following the kit’s instructions. Molecular analyses were performed by the PCR-SSCP and sequencing methods. The first exon of CDKN2D was amplified with primers (1F) 5′-TTTGCAGGCC GCCAGTGTC-3′, and (1R) 5′-GTCTCGATCC TCATCCCGCT TAGCC-3′. The second exon was analyzed as two fragments with primers 2a: (2a-F) 5′-CTGATCCTCT GTCCCTCACA C-3′ and (2a-R) 5′-TGAACTGCCA GATGGATTGG AAGT-3′, and 2b: (2b-F) 5′-ATTCCTGGAC ACCCTGAAG GTCCT-3′, and (2b-R) 5′-GGGCAGGAGA AACAAGAAGA GAAAG-3′. The CDKN2D DNA sequence accession number AF518878.1 (GenBank) was used to design the primers sequences. The PCR mixture was prepared according to the protocol provided with AmpliTaq Gold PCR kit (Life Technologies) with the addition of 5 mM betaine (Sigma-Aldrich). Thermal cycling conditions were as follows: an initial denaturation at 94 °C for 10 min; followed by 36 cycles of denaturing at 94 °C for 30 s, annealing at 60 °C (exon 1 and fragment 2b) or 55 °C (fragment 2a) for 30 s, and extension at 72 °C for 30 s, with a final 7 min extension at 72 °C. Electrophoresis for SSCP analysis was performed as previously described.Citation45 DNA from the samples with bands shifts detected by SSCP were sequenced with ABI dye terminator sequencing kit according to the manufacturer's recommendations in an ABI PRISM 3100 sequencer (Life Technologies).
Statistical analysis
The associations between p19INK4d expression and clinicopathological variables were assessed using the chi-square test. Overall survival (OS) and disease-free survival (DFS) were estimated using the Kaplan–Meier method and a log-rank test. Overall and disease-free survival time analyses were performed using multivariate Cox proportional hazard models. Tumor response to chemotherapy (probability of CR, PS, or HPS) was evaluated using a multivariate logistic regression model.
Statistical analyses included the following independent variables: age of patients (median: 53 y), the FIGO stage, histopathological type, grade, residual tumor size and TP53 accumulation status. The p19INK4d expression was analyzed as a categorical variable (cut-off point at median: 3). Important factors were selected using a backward selection technique, where factors not significant at 0.1 were removed stepwise from the model.
The analyses were performed in the two groups of patients treated with different chemotherapeutic regimens, and additionally in the TP53– and TP53+ subgroups. All tests were two-sided. A P value < 0.05 was considered significant. All calculations were performed using STATA 5 software.
| Abbreviations: | ||
| CR | = | complete remission |
| DFS | = | disease-free survival |
| HR | = | hazard ratio |
| OR | = | odds ratio |
| OS | = | overall survival |
| PC/PAC | = | platinum–cyclophosphamide chemotherapy |
| PS | = | platinum sensitivity |
| TP | = | taxane-platinum chemotherapy |
Acknowledgments
This study was supported by grants 2 PO5A 068 27 and N N401 236134 of the Polish Ministry of Science and Higher education.
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
References
- McGuire WP, Hoskins WJ, Brady MF, Kucera PR, Partridge EE, Look KY, Clarke-Pearson DL, Davidson M. Cyclophosphamide and cisplatin versus paclitaxel and cisplatin: a phase III randomized trial in patients with suboptimal stage III/IV ovarian cancer (from the Gynecologic Oncology Group). Semin Oncol 1996; 23:Suppl 12 40 - 7; PMID: 8941409
- Trimble EL, Wright J, Christian MC. Treatment of platinum-resistant ovarian cancer. Expert Opin Pharmacother 2001; 2:1299 - 306; http://dx.doi.org/10.1517/14656566.2.8.1299; PMID: 11584998
- Stuart GCE. First-line treatment regimens and the role of consolidation therapy in advanced ovarian cancer. Gynecol Oncol 2003; 90:S8 - 15; http://dx.doi.org/10.1016/S0090-8258(03)00472-4; PMID: 13129490
- Kupryjańczyk J, Kraszewska E, Ziółkowska-Seta I, Mądry R, Timorek A, Markowska J, Stelmachow J, Bidzinski M, Polish Ovarian Cancer Study Group (POCSG). TP53 status and taxane-platinum versus platinum-based therapy in ovarian cancer patients: a non-randomized retrospective study. BMC Cancer 2008; 8:27; http://dx.doi.org/10.1186/1471-2407-8-27; PMID: 18230133
- Roussel MF. The INK4 family of cell cycle inhibitors in cancer. Oncogene 1999; 18:5311 - 7; http://dx.doi.org/10.1038/sj.onc.1202998; PMID: 10498883
- Ruas M, Peters G. The p16INK4a/CDKN2A tumor suppressor and its relatives. Biochim Biophys Acta 1998; 1378:F115 - 77; PMID: 9823374
- Ortega S, Malumbres M, Barbacid M. Cyclin D-dependent kinases, INK4 inhibitors and cancer. Biochim Biophys Acta 2002; 1602:73 - 87; PMID: 11960696
- Hirai H, Roussel MF, Kato J-Y, Ashmun RA, Sherr CJ. Novel INK4 proteins, p19 and p18, are specific inhibitors of the cyclin D-dependent kinases CDK4 and CDK6. Mol Cell Biol 1995; 15:2672 - 81; PMID: 7739547
- Kalus W, Baumgartner R, Renner Ch, Noegel A, Chan FKM, Winoto A, Holak TA. NMR structural characterization of the CDK inhibitor p19INK4d. FEBS Lett 1997; 401:127 - 32; http://dx.doi.org/10.1016/S0014-5793(96)01465-2; PMID: 9013872
- Marazita MC, Ogara MF, Sonzogni SV, Martí M, Dusetti NJ, Pignataro OP, Cánepa ET. CDK2 and PKA mediated-sequential phosphorylation is critical for p19INK4d function in the DNA damage response. PLoS One 2012; 7:e35638; http://dx.doi.org/10.1371/journal.pone.0035638; PMID: 22558186
- Guan K-L, Jenkins CW, Li Y, O’Keefe CL, Noh S, Wu X, Zariwala M, Matera AG, Xiong Y. Isolation and characterization of p19INK4d, a p16-related inhibitor specific to CDK6 and CDK4. Mol Biol Cell 1996; 7:57 - 70; PMID: 8741839
- Han J, Ito Y, Yeo JY, Sucov HM, Maas R, Chai Y. Cranical neural crest-derived mesenchymal proliferation is regulated by Msx1-mediated p19INK4d expression during odontogenesis. Dev Bio 2003; l61:183 - 96; http://dx.doi.org/10.1016/S0012-1606(03)00300-2
- Thullberg M, Bartek J, Lukas J. Ubiquitin/proteasome-mediated degradation of p19INK4d determines its periodic expression during the cell cycle. Oncogene 2000; 19:2870 - 6; http://dx.doi.org/10.1038/sj.onc.1203579; PMID: 10851091
- Bartkova J, Thullberg M, Rajpert-De Meyts E, Skakkebaek NE, Bartek J. Lack of p19INK4d in human testicular germ-cell tumours contrasts with high expression during normal spermatogenesis. Oncogene 2000; 19:4146 - 50; http://dx.doi.org/10.1038/sj.onc.1203769; PMID: 10962575
- Bartkova J, Rajpert-De Meyts E, Skakkebaek NE, Lukas J, Bartek J. Deregulation of the G1/S-phase control in human testicular germ cell tumours. APMIS 2003; 111:252 - 65, discussion 265-6; http://dx.doi.org/10.1034/j.1600-0463.2003.1110129.x; PMID: 12760379
- Zhang W, Zeng Z, Zhou Y, Xiong W, Fan S, Xiao L, Huang D, Li Z, Li D, Wu M, et al. Identification of aberrant cell cycle regulation in Epstein-Barr virus-associated nasopharyngeal carcinoma by cDNA microarray and gene set enrichment analysis. Acta Biochim Biophys Sin (Shanghai) 2009; 41:414 - 28; http://dx.doi.org/10.1093/abbs/gmp025; PMID: 19430707
- Kmet LM, Cook LS, Magliocco AM. A review of p53 expression and mutation in human benign, low malignant potential, and invasive epithelial ovarian tumors. Cancer 2003; 97:389 - 404; http://dx.doi.org/10.1002/cncr.11064; PMID: 12518363
- Smith-Sørensen B, Kaern J, Holm R, Dørum A, Tropé C, Børresen-Dale A-L. Therapy effect of either paclitaxel or cyclophosphamide combination treatment in patients with epithelial ovarian cancer and relation to TP53 gene status. Br J Cancer 1998; 78:375 - 81; http://dx.doi.org/10.1038/bjc.1998.502; PMID: 9703286
- Ueno Y, Enomoto T, Otsuki Y, Sugita N, Nakashima R, Yoshino K, Kuragaki C, Ueda Y, Aki T, Ikegami H, et al. Prognostic significance of p53 mutation in suboptimally resected advanced ovarian carcinoma treated with the combination chemotherapy of paclitaxel and carboplatin. Cancer Lett 2006; 241:289 - 300; http://dx.doi.org/10.1016/j.canlet.2005.10.035; PMID: 16459017
- Herod JJO, Eliopoulos AG, Warwick J, Niedobitek G, Young LS, Kerr DJ. The prognostic significance of Bcl-2 and p53 expression in ovarian carcinoma. Cancer Res 1996; 56:2178 - 84; PMID: 8616869
- Bellamy CO. p53 and apoptosis. Br Med Bull 1997; 53:522 - 38; http://dx.doi.org/10.1093/oxfordjournals.bmb.a011628; PMID: 9374035
- Jones NA, Turner J, McIlwrath AJ, Brown R, Dive C. Cisplatin- and paclitaxel-induced apoptosis of ovarian carcinoma cells and the relationship between bax and bak up-regulation and the functional status of p53. Mol Pharmacol 1998; 53:819 - 26; PMID: 9584207
- Shimada M, Kigawa J, Kanamori Y, Itamochi H, Takahashi M, Kamazawa S, Sato S, Terakawa N. Mechanism of the combination effect of wild-type TP53 gene transfection and cisplatin treatment for ovarian cancer xenografts. Eur J Cancer 2000; 36:1869 - 75; http://dx.doi.org/10.1016/S0959-8049(00)00161-1; PMID: 10974636
- Kupryjańczyk J, Szymańska T, Madry R, Timorek A, Stelmachów J, Karpińska G, Rembiszewska A, Ziółkowska I, Kraszewska E, Debniak J, et al. Evaluation of clinical significance of TP53, BCL-2, BAX and MEK1 expression in 229 ovarian carcinomas treated with platinum-based regimen. Br J Cancer 2003; 88:848 - 54; http://dx.doi.org/10.1038/sj.bjc.6600789; PMID: 12644821
- Kupryjańczyk J, Mądry R, Plisiecka-Hałasa J, Bar J, Kraszewska E, Ziółkowska I, Timorek A, Stelmachów J, Emerich J, Jedryka M, et al. TP53 status determines clinical significance of ERBB2 expression in ovarian cancer. Br J Cancer 2004; 91:1916 - 23; http://dx.doi.org/10.1038/sj.bjc.6602238; PMID: 15545967
- Felisiak-Golabek A, Rembiszewska A, Rzepecka IK, Szafron L, Madry R, Murawska M, Napiorkowski T, Sobiczewski P, Osuch B, Kupryjanczyk J, Polish Ovarian Cancer Study Group (POCSG). Nuclear survivin expression is a positive prognostic factor in taxane-platinum-treated ovarian cancer patients. J Ovarian Res 2011; 4:20; http://dx.doi.org/10.1186/1757-2215-4-20; PMID: 22075440
- Thullberg M, Welcker M, Bartkova J, Kjerulff AA, Lukas J, Högberg J, Bartek J. Monoclonal antibody probes for p21WAF1/CIP1 and the INK4 family of cyclin-dependent kinase inhibitors. Hybridoma 2000; 19:63 - 72; http://dx.doi.org/10.1089/027245700315806; PMID: 10768842
- Ceruti JM, Scassa ME, Fló JM, Varone CL, Cánepa ET. Induction of p19INK4d in response to ultraviolet light improves DNA repair and confers resistance to apoptosis in neuroblastoma cells. Oncogene 2005; 24:4065 - 80; http://dx.doi.org/10.1038/sj.onc.1208570; PMID: 15750620
- Kupryjańczyk J. Why TP53 status does not associate with clinical endpoints in ovarian cancer: facts and hypotheses. Gynecol Oncol 2006; 100:5 - 7; http://dx.doi.org/10.1016/j.ygyno.2005.08.003; PMID: 16169062
- Ceruti JM, Scassa ME, Marazita MC, Carcagno AC, Sirkin PF, Cánepa ET. Transcriptional upregulation of p19INK4d upon diverse genotoxic stress is critical for optimal DNA damage response. Int J Biochem Cell Biol 2009; 41:1344 - 53; http://dx.doi.org/10.1016/j.biocel.2008.12.005; PMID: 19130897
- Scassa ME, Marazita MC, Ceruti JM, Carcagno AL, Sirkin PF, González-Cid M, Pignataro OP, Cánepa ET. Cell cycle inhibitor, p19INK4d, promotes cell survival and decreases chromosomal aberrations after genotoxic insult due to enhanced DNA repair. DNA Repair (Amst) 2007; 6:626 - 38; http://dx.doi.org/10.1016/j.dnarep.2006.12.003; PMID: 17218167
- Zindy F, Quelle DE, Roussel MF, Sherr CJ. Expression of the p16INK4a tumor suppressor versus other INK4 family members during mouse development and aging. Oncogene 1997; 15:203 - 11; http://dx.doi.org/10.1038/sj.onc.1201178; PMID: 9244355
- Zindy F, van Deursen J, Grosveld G, Sherr CJ, Roussel MF. INK4d-deficient mice are fertile despite testicular atrophy. Mol Cell Biol 2000; 20:372 - 8; http://dx.doi.org/10.1128/MCB.20.1.372-378.2000; PMID: 10594039
- Miller CW, Yeon C, Aslo A, Mendoza S, Aytac U, Koeffler HP. The p19INK4D cyclin dependent kinase inhibitor gene is altered in osteosarcoma. Oncogene 1997; 15:231 - 5; http://dx.doi.org/10.1038/sj.onc.1201185; PMID: 9244358
- Gemma A, Takenoshita S, Hagiwara K, Okamoto A, Spillare EA, McMemamin MG, Hussain SP, Forrester K, Zariwala M, Xiong Y, et al. Molecular analysis of the cyclin-dependent kinase inhibitor genes p15INK4b/MTS2, p16INK4/MTS1, p18 and p19 in human cancer cell lines. Int J Cancer 1996; 68:605 - 11; http://dx.doi.org/10.1002/(SICI)1097-0215(19961127)68:5<605::AID-IJC9>3.0.CO;2-2; PMID: 8938142
- Zariwala M, Xiong Y. Lack of mutation in the cyclin-dependent kinase inhibitor, p19INK4d, in tumor-derived cell lines and primary tumors. Oncogene 1996; 13:2033 - 8; PMID: 8934552
- Shiohara M, Spirin K, Said JW, Gombart AF, Nakamaki T, Takeuchi S, Hatta Y, Morosetti R, Tasaka T, Seriu T, et al. Alterations of the cyclin-dependent kinase inhibitor p19 (INK4D) is rare in hematopoietic malignancies. Leukemia 1996; 10:1897 - 900; PMID: 8946928
- Park DJ, Wilczynski SP, Pham EY, Miller CW, Koeffler HP. Molecular analysis of the INK4 family of genes in prostate carcinomas. J Urol 1997; 157:1995 - 9; http://dx.doi.org/10.1016/S0022-5347(01)64917-6; PMID: 9112579
- Creasman WJ. Announcement, FIGO stages 1988, revisions. Gynecol Oncol 1989; 35:125 - 7
- Tavassoli FA, Devilee P. Edited: pathology and genetics of tumours of the breast and female genital organs World Health Organisation classification of tumours. IARC Press Lyon 2003.
- Barber HR, Sommers SC, Synder R, Kwon TH. Histologic and nuclear grading and stromal reactions as indices for prognosis in ovarian cancer. Am J Obstet Gynecol 1975; 121:795 - 807; PMID: 1092171
- Miller AB, Hoogstraten B, Staquet M, Winkler A. Reporting results of cancer treatment. Cancer 1981; 47:207 - 14; http://dx.doi.org/10.1002/1097-0142(19810101)47:1<207::AID-CNCR2820470134>3.0.CO;2-6; PMID: 7459811
- Christian MC, Trimble EL. Salvage chemotherapy for epithelial ovarian carcinoma. Gynecol Oncol 1994; 55:S143 - 50; http://dx.doi.org/10.1006/gyno.1994.1354; PMID: 7835799
- Dansonka-Mieszkowska A, Ludwig AH, Kraszewska E, Kupryjańczyk J. Geographical variations in TP53 mutational spectrum in ovarian carcinomas. Ann Hum Genet 2006; 70:594 - 604; http://dx.doi.org/10.1111/j.1469-1809.2006.00257.x; PMID: 16907706
- Kolasa IK, Rembiszewska A, Janiec-Jankowska A, Dansonka-Mieszkowska A, Lewandowska AM, Konopka B, Kupryjańczyk J. PTEN mutation, expression and LOH at its locus in ovarian carcinomas. Relation to TP53, K-RAS and BRCA1 mutations. Gynecol Oncol 2006; 103:692 - 7; http://dx.doi.org/10.1016/j.ygyno.2006.05.007; PMID: 16793127