Abstract
The ability of eukaryotes to alter chromatin structure and function is modulated, in part, by histone-modifying enzymes and the post-translational modifications they create. One of these enzymes, PR-Set7/Set8/KMT5a, is the sole histone methyltransferase responsible for the monomethylation of histone H4 lysine 20 (H4K20me1) in higher eukaryotes. Both PR-Set7 and H4K20me1 were previously found to be tightly cell cycle regulated suggesting that they play an important, although unknown, role in cell cycle progression. Several recent reports reveal that PR-Set7 abundance is dynamically regulated during different cell cycle phases by distinct enzymes including cdk1/cyclinB, Cdc14, SCFSkp2, CRL4cdt2 and APCcdh1. Importantly, these reports demonstrate that inappropriate levels of PR-Set7 result in profound cell cycle defects including the inability to initiate S phase, the re-replication of DNA and the improper timing of mitotic progression. Here, we summarize the significance of these new findings, raise some important questions that require further investigation and explore several possibilities of how PR-Set7 and methylated H4K20 may likely function as novel regulators of the cell cycle.
Introduction
Eukaryotic genomes are assembled within the nucleus in the form of chromatin—a complex of DNA and associated histone and non-histone proteins. While chromatin functions to package and organize DNA, it also creates a physical barrier that inhibits fundamental DNA-templated processes including transcription, replication and repair. Therefore, the ability to modulate chromatin in response to a constant barrage of ever changing cellular cues is essential. To this end, eukaryotes have developed several sophisticated mechanisms to regulate chromatin structure and function. One such mechanism involves the post-translational modification of the DNA-associated histone proteins whereby specific amino acids within histones are targets for certain enzyme-containing protein complexes resulting in their acetylation, phosphorylation, ubiquitination or methylation. Modified histones can directly alter chromatin structure and/or create high affinity binding sites for other proteins that participate in the specific DNA-templated process.Citation1,Citation2
Previous studies demonstrated that a specific histone modification, the monomethylation of H4 lysine 20 (H4K20me1) and the enzyme responsible for the modification, PR-Set7/Set8/KMD5a, are tightly regulated during the mammalian cell cycle suggesting that they directly function in the control of cell division.Citation3,Citation4 New reports by our group and others have elucidated the mechanisms responsible for the dynamic regulation of PR-Set7 during different cell cycle phases and demonstrated that its orchestrated proteolysis mediated by specific ubiquitin ligases is essential for proper mammalian cell cycle progression.Citation5–Citation9 Here we review the importance of these recent findings, speculate how PR-Set7 and H4K20me1 may function as key cell cycle regulators and present new questions that will require further investigation to answer.
PR-Set7 and H4K20me1
Although histone H4 methylation was one of the first histone post-translational modifications to be discovered nearly half a century ago, the identification of the enzymes responsible for H4K20 methylation were only recently identified.Citation10 While lower eukaryotes typically utilize a single enzyme to catalyze all three degrees of H4K20 methylation (mono-, di- and tri-methylation),Citation11 higher eukaryotes use distinct enzymes for each degree of methylation.Citation12,Citation13 Here, the Suv4-20 enzymes (KMD5b and KMD5c) are responsible for global di- and trimethylation of H4K20 (H4K20me2/3), whereas PR-Set7 solely catalyzes the monomethylation of H4K20 in a nucleosomal-specific context.Citation3,Citation14–Citation17
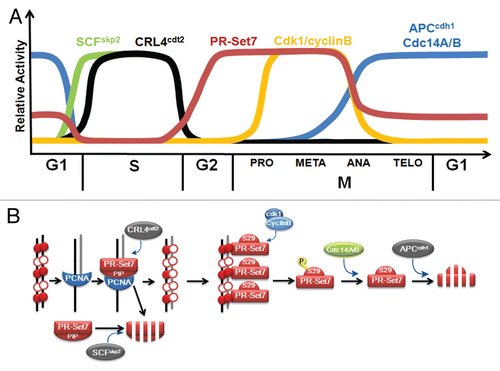
In contrast to H4K20me2/3, we previously discovered that PR-Set7 and H4K20me1 are tightly cell cycle regulated.Citation4,Citation18 Both were nearly absent during G1 and S phases but were robustly elevated during G2 and mitosis in human cells (). As cells exited mitosis, PR-Set7 levels rapidly declined whereas H4K20me1 slowly declined. These findings strongly suggested that PR-Set7 and H4K20me1 play an essential, although unknown, role in the mammalian cell cycle. Consistent with this, studies in various animal systems revealed that the depletion of PR-Set7 resulted in growth and cell cycle defects. Drosophila lacking the PR-Set7 orthologue displayed chromosome condensation defects and growth arrest.Citation19,Citation20 As expected, PR-Set7-/- mice were lethal with embryos failing to progress past the eight cell stage.Citation21,Citation22 The lack of PR-Set7 and H4K20me1 results predominantly in a G2 arrest with cells failing to condense chromosomes.Citation18 These collective findings indicated that PR-Set7 and H4K20me1 are required for mitotic entry.
PR-Set7 during Mitosis
PR-Set7 was previously demonstrated to be phosphorylated specifically during mitosis in Xenopus laevis embryos coincident with the observed increased abundance of PR-Set7.Citation23 These findings suggested that this phosphorylation event may be important for PR-Set7 function during mitosis. Recent findings by our group showed that a specific residue of PR-Set7, serine 29 (S29), was the predominant site of PR-Set7 phosphorylation during mitosis.Citation9 We further discovered that the cdk1/cyclinB complex was responsible for S29 phosphorylation specifically from prophase to anaphase. Importantly, we found that this phosphorylation event has two functional consequences. First, phosphorylation of PR-Set7 results in its removal from mitotic chromosomes and relocation to the extrachromosomal space, subsequent to global accumulation of H4K20me1 in G2. Second, phosphorylation of PR-Set7 stabilizes PR-Set7 levels by directly inhibiting its ubiquitination and degradation by the APCcdh1 E3 ubiquitin ligase. During early anaphase, we found that PR-Set7 is dephosphorylated by the Cdc14 phosphatases resulting in the APCcdh1-mediated ubiquitination and degradation of PR-Set7 consistent with the observed decrease in PR-Set7 levels at G1. Importantly, we demonstrated that a constitutively phosphorylated PR-Set7 mutant resulted in a significant metaphase delay. These findings strongly suggest that degradation of PR-Set7 is required for anaphase entry and proper cell cycle progression.
These findings raise several interesting and important questions for further investigation. For example, how does the phosphorylation of PR-Set7 S29 so dramatically alter its localization? Does phosphorylated S29 function directly to physically decrease the affinity of PR-Set7 for mitotic chromosomes? Does S29 phosphorylation indirectly inhibit PR-Set7 from interacting with an unknown protein complex responsible for recruiting PR-Set7 to chromatin? Alternatively, does S29 phosphorylation create a high affinity binding site for an unidentified protein complex that actively transports PR-Set7 away from chromosomes? These possibilities are not necessarily mutually exclusive and may function cooperatively to relocate PR-Set7. Furthermore, why does PR-Set7 need to be removed from mitotic chromosomes? Does the continued presence and activity of chromatin-bound PR-Set7 during mitosis inhibit mitotic progression? New findings suggest that the removal of PR-Set7 may be required to expose H4K20me1 to permit the binding of factors of the condensin II complex, although this has yet to be validated.Citation24 Finally, why does the persistence of a degradation-resistant PR-Set7 mutant result in a substantial metaphase delay? Does the degradation of PR-Set7 serve as a checkpoint for progression to anaphase and could this event be required for proper chromosome segregation?
PR-Set7 during S Phase
PR-Set7 was recently reported to be recruited to DNA replication foci via interaction with the proliferating cell nuclear antigen (PCNA) and importantly, this recruitment was shown to be required for proper DNA replication.Citation21,Citation25,Citation26 Interestingly, the PR-Set7 protein is nearly undetectable during S phase despite relatively high levels of PR-Set7 transcription indicating that PR-Set7 must be rapidly degraded following its recruitment to replication foci.Citation9 Several new reports demonstrate that the CRL4cdt2 ubiquitin ligase complex is specifically responsible for the degradation of chromatin-bound PR-Set7 during S phase ().Citation5–Citation8 CRL4cdt2 was previously shown to play an essential role in S phase and coordinating DNA replication origin licensing in G1.Citation27–Citation29 It is a unique E3 ubiquitin ligase that specifically recognizes substrates bound to PCNA on chromatin via a short highly conserved motif called the PIP (PCNA-interacting protein) box or degron.Citation30 For example, CRL4cdt2 targets the replication licensing protein Cdt1 and the cyclin-dependent kinase inhibitor p21 for degradation during S phase in a process that requires the PIP box and PCNA as a cofactor.Citation31–Citation33 Sequence analysis of PR-Set7 revealed that it also contains two putative PIP boxes. Although both PIP boxes can bind PCNA, only the second PIP box (aa 148–155) was found to be required for PCNA-dependent ubiquitination and degradation of PR-Set7 by CRL4cdt2 during S phase.
The biological significance of S phase-coupled PR-Set7 degradation appears to be twofold. First, the destruction of PR-Set7 was necessary to prevent aberrant H4K20me1 and premature chromatin compaction during DNA replication. We previously demonstrated that catalytically active PR-Set7 was required for chromatin condensation during mitosis, most likely by methylating H4K20.Citation18 Similarly, expression of a degradation-resistant PR-Set7 mutant lacking the PIP box (PR-Set7ΔPIP) resulted in the inappropriate accumulation of H4K20me1 and chromatin condensation coincident with a delay in S phase progression and checkpoint-mediated G2 arrest.Citation6 Second, the timely degradation of PR-Set7 is required to prevent replication-relicensing. The inability to degrade PR-Set7, by expressing the PR-Set7ΔPIP mutant or forced overexpression of wild type PR-Set7, resulted in re-replicated DNA during the G2 arrest.Citation8 Consistent with our previous findings, a catalytically active PR-Set7 was required to induce all observed aberrant phenotypes.
PR-Set7 and Methylated H4K20 as Putative Replication Licensing Factors
The striking similarity of PCNA-mediated degradation between PR-Set7 and Cdt1 during S phase suggests that PR-Set7 and, more importantly, methylated H4K20 may function as novel replication licensing factors. Consistent with this, the targeting of catalytically active PR-Set7 to an integrated reporter was sufficient to nucleate pre-replication complex (RC) components.Citation8 Furthermore, H4K20me1 was enriched at certain replication origins at the onset of replication licensing during mitosis but was reduced at G1 and S phases. Since H4K20me1 levels peak at mitosis and plummet during G1/S, both globally and at loci not associated with replication origins, the significance of these observations remain unclear.Citation18 However, the absence of PR-Set7 and H4K20me1 did result in a reduction of chromatin-bound pre-RC components.Citation8 Collectively, these findings suggest that PR-Set7 and H4K20me1 may participate in the activation of origin licensing and/or may function to prevent premature origin firing during mitosis and G1.
It is interesting to note that an unbiased SILAC-based histone peptide pull-down experiment revealed that several origin of replication components specifically bound H4K20me2 but not H4K20me1.Citation7 These findings suggest that the nucleation of pre-RC components at origins could be dependent on a pathway that also involves H4K20me2. Several previous studies provided evidence that H4K20me1 may be the preferred substrate for H4K20me2/3 by the Suv4-20 enzymes.Citation34–Citation36 Therefore, PR-Set7-mediated H4K20me1 may function to recruit the Suv4-20 enzymes to induce H4K20me2 at origins resulting in the binding of pre-RC components. It currently remains unknown if H4K20me2 is enriched at replication origins. If H4K20me2 is involved in this pathway, then one may predict that Suv4-20 h1/h2 knock-out mice would exhibit a similar severe preimplantation arrest as observed in PR-Set7 knock-out mice, however, this was not the case.Citation21,Citation22,Citation35 Since there are several possible reasons for this discrepancy, further studies are needed to determine if Suv4-20 h1, Suv4-20 h2 and H4K20me2/3 play a role in regulating replication origins.
PR-Set7 during G1
As cells enter G1 following division, PR-Set7 protein levels are diminished by proteolysis in an APCcdh1-dependent manner as described above. It was recently reported that endogenous PR-Set7 levels during G1 are sustained, although greatly reduced compared to G2/M, until cells begin to transition to S phase ().Citation37 Prior to S phase entry, PR-Set7 is ubiquitinated by the SCFSkp2 ubiquitin ligase resulting in the degradation of PR-Set7. SCFSkp2 remains active during DNA replication suggesting that it may also participate in PR-Set7 degradation during S phase. Indeed, elevated levels of soluble PR-Set7, but not chromatin-bound PR-Set7, were observed in Skp2 depleted cells indicating that both CRL4cdt2 and SCFSkp2 function in S phase to degrade chromatin-bound and chromatin-free PR-Set7, respectively.Citation7 Another interesting finding from this study was that a sizeable portion of PR-Set7 proximal to the catalytic SET domain (aa 51–194) could bind the N-terminal tail of histone H4.Citation37 When ectopically expressed, this fragment of PR-Set7 was resistant to degradation, despite containing the PIP box and resulted in the dramatic inhibition of S phase entry. These findings suggest that SCFSkp2-mediated ubiquitination and degradation of PR-Set7 is required for initiation of DNA replication, in part, by inhibiting PR-Set7 binding to chromatin. Further studies are required to validate this proposed model.
PR-Set7 may also participate in controlling the initiation of DNA replication by regulating the expression of a subset of E2F-target genes. We and others previously demonstrated that PR-Set7 negatively regulates transcription of certain genes whereby PR-Set7-mediated H4K20me1 can recruit and bind the L3MBTL1 repressor protein resulting in chromatin condensation and reduced gene expression.Citation38–Citation43 Several E2F-target genes, including cyclin E1, were previously found to be enriched with H4K20me1 and L3MBTL1 and, importantly, depletion of L3MBTL1 resulted in increased expression of these genes.Citation43 These findings suggest that PR-Set7 may prevent S phase entry by directly repressing genes required for the initiation of DNA replication. Consistent with this, a recent report demonstrated that inhibiting PR-Set7 degradation by expressing the PR-Set7ΔPIP mutant resulted in the repression of most E2F-regulated genes, which also required its catalytic activity.Citation5 These findings strongly suggest that one important function of PCNA-dependent degradation of PR-Set7 during DNA replication is to relieve the transcriptional repression of genes required for cell cycle progression. Since the PR-Set7ΔPIP mutant altered expression of genes and global H4K20 methylation levels, these findings also indicate that PR-Set7 is recruited to chromatin in a PCNA-independent manner. Further investigation is required to determine how and where PR-Set7 is recruited to chromatin in both the PCNA-dependent and independent pathways.
Summary
Based on several new independent reports, it is now clear that the precise modulation of PR-Set7 levels is essential for proper cell cycle progression in higher eukaryotes (). The degradation of PR-Set7, mediated by the SCFSkp2 ubiquitin ligase, appears to be required for the initiation of S phase by removing PR-Set7 from chromatin and derepressing the expression of E2F-target genes.Citation5,Citation37 The rapid and sustained degradation of PR-Set7 during S phase, mediated by the CRL4cdt2 ubiquitin ligase, is required to prevent aberrant chromatin condensation and DNA re-replication.Citation6–Citation8 The elevated levels of PR-Set7 observed during late S and G2, presumably a result of the decreased activity of the ubiquitin ligases, are required for mitotic entry.Citation18 And the degradation of PR-Set7 during mitosis, mediated by the APCcdh1 ubiquitin ligase, is required for the timely transition from metaphase to anaphase.Citation9 The inability to control PR-Set7 levels results in severe cell cycle defects. It is important to note that, in all cases, the methyltransferase activity of PR-Set7 was required to prevent the observed cell cycle defects. These findings strongly suggest that it is the regulation of H4K20me1 during distinct cell cycle phases that is required for proper progression and not PR-Set7 per se. How H4K20me1 is regulated and how it functions to promote cell cycle progression are just beginning to emerge and remain active areas of investigation. As with other histone-modifying enzymes, substrates other than histone H4 can also be methylated by PR-Set7 and may function in these pathways.Citation44 Therefore, it will be necessary to continue to identify these substrates and evaluate the contribution of their PR-Set7-mediated methylation in the cell cycle.
Figures and Tables
Figure 1 (A) Relative abundance of PR-Set7 and activity of the various enzymes that regulate PR-Set7 abundance (y-axis) during different cell cycle phases (x-axis). (B) Model of PR-Set7 regulation through the cell cycle. During G1, PR-Set7 is targeted to specific loci resulting in H4K20me1 (closed red circles). Prior to S phase, PR-Set7 is ubiquitinated by SCFSkp2 and degraded coincident with S phase entry. PR-Set7 is nearly undetectable during DNA replication as (1) PR-Set7 binds chromatin-bound PCNA via the PIP box resulting in ubiquitination by CRL4cdt2 and subsequent degradation and (2) chromatin-free PR-Set7 is ubiquitinated by SCFSkp2 and degraded. During G2, PR-Set7 levels dramatically rise coincident with decreased SCFSkp2 and CRL4cdt2 activity resulting in H4K20me1 of unmodified nucleosomes (open red circles) on the newly replicated daughter strand (gray line). Following H4K20me1-associated chromatin condensation, chromosome-bound PR-Set7 is phosphorylated on serine 29 (S29) by cdk1/cyclinB during prophase to anaphase resulting in removal of PR-Set7 from mitotic chromosomes. This degradation-resistant form of PR-Set7 is dephosphorylated in anaphase by the Cdc14 phosphatases resulting in APCcdh1-mediated ubiquitination and degradation of PR-Set7 as cell progress to G1.
Acknowledgements
This work was supported in part by the Pew Charitable Trusts (J.C.R.), the Wang Predoctoral Award (S.W.) and NIH grant GM075094 (J.C.R.).
References
- Shogren-Knaak M, Ishii H, Sun JM, Pazin MJ, Davie JR, Peterson CL. Histone H4-K16 acetylation controls chromatin structure and protein interactions. Science 2006; 311:844 - 847
- Taverna SD, Li H, Ruthenburg AJ, Allis CD, Patel DJ. How chromatin-binding modules interpret histone modifications: lessons from professional pocket pickers. Nat Struct Mol Biol 2007; 14:1025 - 1040
- Nishioka K, Rice JC, Sarma K, Erdjument-Bromage H, Werner J, Wang Y, et al. PR-Set7 is a nucleosome-specific methyltransferase that modifies lysine 20 of histone H4 and is associated with silent chromatin. Mol Cell 2002; 9:1201 - 1213
- Rice JC, Nishioka K, Sarma K, Steward R, Reinberg D, Allis CD. Mitotic-specific methylation of histone H4 Lys 20 follows increased PR-Set7 expression and its localization to mitotic chromosomes. Genes Dev 2002; 16:2225 - 2230
- Abbas T, Shibata E, Park J, Jha S, Karnani N, Dutta A. CRL4(Cdt2) regulates cell proliferation and histone gene expression by targeting PR-Set7/Set8 for degradation. Mol Cell 2010; 40:9 - 21
- Centore RC, Havens CG, Manning AL, Li JM, Flynn RL, Tse A, et al. CRL4(Cdt2)-mediated destruction of the histone methyltransferase Set8 prevents premature chromatin compaction in S phase. Mol Cell 2010; 40:22 - 33
- Oda H, Hubner MR, Beck DB, Vermeulen M, Hurwitz J, Spector DL, et al. Regulation of the Histone H4 Monomethylase PR-Set7 by CRL4(Cdt2)-Mediated PCNA-Dependent Degradation during DNA Damage. Mol Cell 2010; 40:364 - 376
- Tardat M, Brustel J, Kirsh O, Lefevbre C, Callanan M, Sardet C, et al. The histone H4 Lys 20 methyltransferase PR-Set7 regulates replication origins in mammalian cells. Nat Cell Biol 2010; 12:1086 - 1093
- Wu S, Wang W, Kong X, Congdon LM, Yokomori K, Kirschner MW, et al. Dynamic regulation of the PR-Set7 histone methyltransferase is required for normal cell cycle progression. Genes Dev 2010; 24:2531 - 2542
- Murray K. The occurrence of e-N-methyl lysine in histones. Biochemistry 1964; 3:10 - 15
- Rice JC, Allis CD. Histone methylation versus histone acetylation: new insights into epigenetic regulation. Current Opinion in Cell Biology 2001; 13:263 - 273
- Sanders SL, Portoso M, Mata J, Bahler J, Allshire RC, Kouzarides T. Methylation of histone H4 lysine 20 controls recruitment of Crb2 to sites of DNA damage. Cell 2004; 119:603 - 614
- Sautel CF, Cannella D, Bastien O, Kieffer S, Aldebert D, Garin J, et al. SET8-mediated methylations of histone H4 lysine 20 mark silent heterochromatic domains in apicomplexan genomes. Mol Cell Biol 2007; 27:5711 - 5724
- Couture JF, Collazo E, Brunzelle JS, Trievel RC. Structural and functional analysis of SET8, a histone H4 Lys-20 methyltransferase. Genes Dev 2005; 19:1455 - 1465
- Fang J, Feng Q, Ketel CS, Wang H, Cao R, Xia L, et al. Purification and functional characterization of SET8, a nucleosomal histone H4-lysine 20-specific methyltransferase. Curr Biol 2002; 12:1086 - 1099
- Schotta G, Lachner M, Sarma K, Ebert A, Sengupta R, Reuter G, et al. A silencing pathway to induce H3-K9 and H4-K20 trimethylation at constitutive heterochromatin. Genes Dev 2004; 18:1251 - 1262
- Xiao B, Jing C, Kelly G, Walker PA, Muskett FW, Frenkiel TA, et al. Specificity and mechanism of the histone methyltransferase Pr-Set7. Genes Dev 2005; 19:1444 - 1454
- Houston SI, McManus KJ, Adams MM, Sims JK, Carpenter PB, Hendzel MJ, et al. Catalytic function of the PR-Set7 histone H4 lysine 20 monomethyltransferase is essential for mitotic entry and genomic stability. J Biol Chem 2008; 283:19478 - 19488
- Karachentsev D, Sarma K, Reinberg D, Steward R. PR-Set7-dependent methylation of histone H4 Lys 20 functions in repression of gene expression and is essential for mitosis. Genes Dev 2005; 19:431 - 435
- Sakaguchi A, Steward R. Aberrant monomethylation of histone H4 lysine 20 activates the DNA damage checkpoint in Drosophila melanogaster. J Cell Biol 2007; 176:155 - 162
- Huen MS, Sy SM, van Deursen JM, Chen J. Direct interaction between SET8 and PCNA couples H4-K20 methylation with DNA replication. J Biol Chem 2008; 283:11073 - 11077
- Oda H, Okamoto I, Murphy N, Chu J, Price SM, Shen MM, et al. Monomethylation of histone H4-lysine 20 is involved in chromosome structure and stability and is essential for mouse development. Mol Cell Biol 2009; 29:2278 - 2295
- Stukenberg PT, Lustig KD, McGarry TJ, King RW, Kuang J, Kirschner MW. Systematic identification of mitotic phosphoproteins. Curr Biol 1997; 7:338 - 348
- Liu W, Tanasa B, Tyurina OV, Zhou TY, Gassmann R, Liu WT, et al. PHF8 mediates histone H4 lysine 20 demethylation events involved in cell cycle progression. Nature 2010; 466:508 - 512
- Jorgensen S, Elvers I, Trelle MB, Menzel T, Eskildsen M, Jensen ON, et al. The histone methyltransferase SET8 is required for S-phase progression. J Cell Biol 2007; 179:1337 - 1345
- Tardat M, Murr R, Herceg Z, Sardet C, Julien E. PR-Set7-dependent lysine methylation ensures genome replication and stability through S phase. J Cell Biol 2007; 179:1413 - 1426
- Jin J, Arias EE, Chen J, Harper JW, Walter JC. A family of diverse Cul4-Ddb1-interacting proteins includes Cdt2, which is required for S phase destruction of the replication factor Cdt1. Mol Cell 2006; 23:709 - 721
- Nishitani H, Sugimoto N, Roukos V, Nakanishi Y, Saijo M, Obuse C, et al. Two E3 ubiquitin ligases, SCF-Skp2 and DDB1-Cul4, target human Cdt1 for proteolysis. EMBO J 2006; 25:1126 - 1136
- Zhong W, Feng H, Santiago FE, Kipreos ET. CUL-4 ubiquitin ligase maintains genome stability by restraining DNA-replication licensing. Nature 2003; 423:885 - 889
- Havens CG, Walter JC. Docking of a specialized PIP Box onto chromatin-bound PCNA creates a degron for the ubiquitin ligase CRL4Cdt2. Mol Cell 2009; 35:93 - 104
- Abbas T, Sivaprasad U, Terai K, Amador V, Pagano M, Dutta A. PCNA-dependent regulation of p21 ubiquitylation and degradation via the CRL4Cdt2 ubiquitin ligase complex. Genes Dev 2008; 22:2496 - 2506
- Arias EE, Walter JC. PCNA functions as a molecular platform to trigger Cdt1 destruction and prevent re-replication. Nat Cell Biol 2006; 8:84 - 90
- Kim Y, Starostina NG, Kipreos ET. The CRL4Cdt2 ubiquitin ligase targets the degradation of p21Cip1 to control replication licensing. Genes Dev 2008; 22:2507 - 2519
- Pesavento JJ, Yang H, Kelleher NL, Mizzen CA. Certain and Progressive Methylation of Histone H4 at Lysine 20 During the Cell Cycle. Mol Cell Biol 2007;
- Schotta G, Sengupta R, Kubicek S, Malin S, Kauer M, Callen E, et al. A chromatin-wide transition to H4K20 monomethylation impairs genome integrity and programmed DNA rearrangements in the mouse. Genes Dev 2008; 22:2048 - 2061
- Yang H, Pesavento JJ, Starnes TW, Cryderman DE, Wallrath LL, Kelleher NL, et al. Preferential dimethylation of histone H4-lysine 20 by Suv4-20. J Biol Chem 2008; 283:12085 - 12092
- Yin Y, Yu VC, Zhu G, Chang DC. SET8 plays a role in controlling G1/S transition by blocking lysine acetylation in histone through binding to H4 N-terminal tail. Cell Cycle 2008; 7:1423 - 1432
- Congdon LM, Houston SI, Veerappan CS, Spektor TM, Rice JC. PR-Set7-mediated monomethylation of histone H4 lysine 20 at specific genomic regions induces transcriptional repression. J Cell Biochem 2010; 110:609 - 619
- Kalakonda N, Fischle W, Boccuni P, Gurvich N, Hoya-Arias R, Zhao X, et al. Histone H4 lysine 20 monomethylation promotes transcriptional repression by L3MBTL1. Oncogene 2008; 27:4293 - 4304
- Min J, Allali-Hassani A, Nady N, Qi C, Ouyang H, Liu Y, et al. L3MBTL1 recognition of mono- and dimethylated histones. Nat Struct Mol Biol 2007; 14:1229 - 1230
- Sims JK, Houston SI, Magazinnik T, Rice JC. A trans-tail histone code defined by monomethylated H4 Lys-20 and H3 Lys-9 demarcates distinct regions of silent chromatin. J Biol Chem 2006; 281:12760 - 12766
- Sims JK, Rice JC. PR-Set7 establishes a repressive trans-tail histone code that regulates differentiation. Mol Cell Biol 2008; 283:19478 - 19488
- Trojer P, Li G, Sims RJ 3rd, Vaquero A, Kalakonda N, Boccuni P, et al. L3MBTL1, a histone-methylation-dependent chromatin lock. Cell 2007; 129:915 - 928
- Shi X, Kachirskaia I, Yamaguchi H, West LE, Wen H, Wang EW, et al. Modulation of p53 function by SET8-mediated methylation at lysine 382. Mol Cell 2007; 27:636 - 646