The hypoxia-inducible factor (HIF) family of bHLH-PAS (basic helix-loop-helix per/arnt/sim domain) transcription factors are crucial for responding to changes in cellular oxygen levels.Citation1 Mitochondria are the powerhouse of oxygen consumption in eukaryotic cells, generating chemical energy in the form of adenosine triphosphate (ATP). Recent studies have highlighted the importance of mitochondria for relaying hypoxic signals to regulate HIF function in mammalian cells.Citation2 In addition, other studies have described a role for multiple HIF-1 transcriptional targets (e.g., pyruvate dehydrogenase kinase 1, BNIP3, lactate dehydrogenase A, complex IV and miRNA210) in controlling cellular oxygen consumption and metabolism by regulating mitochondrial function.Citation3 In this way, HIF transcriptional activity indirectly controls mitochondrial function, and mitochondria indirectly regulate HIF transcriptional function in response to hypoxia. Despite more than a decade of advances in HIF research, the precise molecular mechanisms for how mitochondria interface with the cellular HIF/oxygen-sensing machinery are yet to be unravelled.
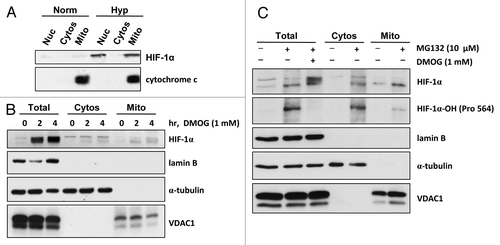
We envisage that yet-undiscovered molecular mechanisms exist that allow mitochondria to rapidly respond to fluctuations in cellular oxygen levels, enabling cells to continuously control metabolic output and conserve energy. Mitochondriamediated responses to changes in cellular oxygen may not wholly depend on transcriptional events in the nucleus, which could take several hours to relay. Indeed, we have been exploring the possibility that HIFs themselves are associated with mitochondria, since a previous study described the association of HIF-1α protein with mitochondria in cultured (rat) cardiac myocytes in response to hypoxia preconditioning.Citation4 Confirming these observations using subcellular fractionation analyses and purified mitochondria, we have found that endogenous HIF-1α protein localizes with both nuclear and mitochondrial fractions in response to hypoxia (1% O2) and is detected in the mitochondrial fractions within 2 h of exposure (). Moreover, exposure of cells to the 2-oxoglutarate analog dimethyloxalylglycine (DMOG), which leads to the rapid stabilization of HIF-1α protein by blockade of the prolyl hydroxylase domain enzymes, also promotes the localization of HIF-1α with mitochondrial fractions in normoxia (). We have estimated that probably less than 5% of the total HIF-1α protein stabilized overall in the cell is associated with mitochondrial fractions, presenting a challenge for evaluating endogenous mitochondria-associated HIF-1α protein in situ in cells using standard immunostaining techniques. Of course, taking particular care to purify mitochondria by subcellular fraction and sucrose gradient and assessing appropriate subcellular marker proteins to validate protein localization is essential. Interestingly, we have found that HIF-1α protein stabilized in normoxia by inhibition of proteasomal degradation using MG132 is also detected in mitochondrial fractions and is hydroxylated at Pro564 (). Collectively, our data suggest several exciting possibilities; that HIF-1α protein is (1) escorted to mitochondria and/or (2) imported into mitochondria and/or (3) localized and stabilized within mitochondria.
Although HIF-1α is best known as the regulatory component of the HIF-1 transcription factor, clearly the HIFs may have other functions outside the nucleus. The possibility that the HIFs and components of the cellular oxygen-sensing machinery interface directly with mitochondria presents a complementary mechanism for controlling mitochondrial function and metabolism when cellular oxygen levels fluctuate.
Figures and Tables
Figure 1 HIF-1α protein localizes with mitochondria. (A) HCT116 cells were incubated in normoxia (Norm) or hypoxia (1% O2, Hyp) for 8 h. Subcellular fractionation was performed, and the fractions (Nuc, nuclear; Cytos, cytosolic; Mito, mitochondrial) were analyzed by protein gel blot for HIF-1α. Cytochrome c and α-tubulin were used as subcellular fractionation controls. (B and C) HL-1 mouse cardiomyocytes were exposed to (B) DMOG (1 mM) for the times indicated or (C) MG132 (10 µM) and DMOG (1 mM), as indicated, for 4 h. Subcellular fractionation was performed and fractions (Total; Cytos, cytosolic; Mito, mitochondrial) were analyzed by protein gel blot for HIF-1α protein and hydroxylated (Pro564) HIF-1α protein. Analysis of lamin B (nuclear marker), α-tubulin (cytoplasmic marker) and voltage-dependent anion-selective channel protein 1, VDAC1 (mitochondrial marker) was performed to assess the efficiency of subcellular fractionation.
Acknowledgments
We thank Dr. Luke Thomas for helpful discussions and Dr. Andrew Hall for suggesting use of VDAC1 as a mitochondrial marker (both at University College London; London UK). The work was funded by Cancer Research UK grants C7358/A4420 and C7358/A11223.
References
- Majmundar AJ, et al. Mol Cell 2010; 40:294 - 309; PMID: 20965423; http://dx.doi.org/10.1016/j.molcel.2010.09.022
- Chandel NS. Adv Exp Med Biol 2010; 661:339 - 354; PMID: 20204741; http://dx.doi.org/10.1007/978-1-60761-500-2_22
- Semenza GL, et al. Biochim Biophys Acta 2011; 1813:1263 - 1268; PMID: 20732359; http://dx.doi.org/10.1016/j.bbamcr.2010.08.006
- Rane S, et al. Circ Res 2009; 104:879 - 886; PMID: 19265035; http://dx.doi.org/10.1161/CIRCRESAHA.108.193102