Abstract
Aurora family kinases play pivotal roles in several steps during mitosis. Specifically, Aurora A kinase is an important regulator of bipolar mitotic spindle formation and chromosome segregation. Like other members of the Aurora family, Aurora A kinase is also regulated by post-translational modifications. Here, we show that a previously undescribed E3 ligase component belonging to the SCF (Skp-Cullin1-F-box protein) E3 ligase family, SCFFBXL7, impairs cell proliferation by mediating Aurora A polyubiquitination and degradation. Both Aurora A and FBXL7 co-localize within the centrosome during spindle formation. FBXL7 ectopic expression led to G2/M phase arrest in transformed epithelia, resulting in the appearance of tetraploidy and mitotic arrest with circular monopolar spindles and multipolar spindle formation. Interestingly, FBXL7 specifically interacts with Aurora A during mitosis but not in interphase, suggesting a regulatory role for FBXL7 in controlling Aurora A abundance during mitosis.
Introduction
One of the most critical events during cellular mitosis is mitotic spindle formation and proper chromosome segregation.Citation1,Citation2 During this process, spindle checkpoint proteins ensure the correct distribution of sister chromatids in anaphase before the completion of cytokinesis by abscission.Citation3,Citation4 Among the network of regulatory proteins involved in spindle formation, several serine/threonine protein kinases play an essential role in phosphorylating structural and motor proteins required for bipolar spindle formation.Citation5 Due to their important roles in mitosis, Aurora family members Aurora A and Aurora B were extensively studied in the last decade.Citation6 Aurora B kinase is a multi-role protein that localizes at different cell organelles. Aurora B has been shown to localize at the mitotic chromosome, specifically associating with kinetochores in metaphase, but dislocates at anaphase and later accumulates on the midzone and midbody.Citation7,Citation8 Aurora A, however, only localizes to centrosomes and the mitotic spindle during mitosis.Citation9 The function of Aurora A is also drastically different from Aurora B; previous studies have shown that the major role of Aurora A is mitotic spindle formation, chromosome alignment and separation.Citation10 Mammalian cells are extremely sensitive to Aurora A modulation, as chemical inhibition or siRNA knockdown of Aurora A lead to similar phenotypes, including chromosomal misalignment and cytokinesis failure.Citation11–Citation13
Aurora A is modulated and regulated by a variety of post-translational modifications, such as phosphorylation, dephosphorylation and, perhaps most importantly, ubiquitination.Citation14 Unlike Aurora B kinase, which is degraded by the proteasome via many ubiquitin E3 ligases, including the anaphase-promoting complex (APC, Cdc27) pathway, BCR (KLHL9-KLHL13-KLHL21) E3 ubiquitin ligase, CUL3/KLHL E3 ligase complex and BRCA1-associated ring domain protein E3 ligase (BRAD1), little is known regarding ubiquitin-mediated disposal of Aurora A.Citation7,Citation15–Citation19 There are many studies on SCF E3 ligase (Skp-Cullin1-F-box protein) complex functions during mitosis, especially with regard to centrosome expression, raising the possibility that this E3 ligase family targets Aurora A. For example, the SCFβ-trcp complex has roles in centrosome stability and mitotic progression;Citation20,Citation21 SCFFBXL17 complex is involved in cell cycle regulation during male gametogenesis in Arabidopsis thaliana;Citation22 SCFcyclinF complex controls centrosome homeostasis and mitotic fidelity through CP110 degradation,Citation23 and SCFFBXW5 E3-ubiquitin ligase targets HsSAS-6 to control centrosome duplication.Citation24 However, thus far, only one specialized ubiquitin ligase, APC/Cdh1, has been shown to ubiquitinate Aurora A. There are two signal motifs within the Aurora A primary sequence: the destruction box (D-box) and the three residue KEN signatures, both of which are well-characterized APC targeting signals.Citation25 The SCF machinery has a catalytic core complex consisting of Skp1, Cullin1 and the E2 ubiquitin-conjugating (Ubc) enzyme.Citation26,Citation27 The SCF complex also contains an adaptor subunit, termed F-box protein, that targets hundreds of substrates through phosphospecific domain interactions.Citation28 F-box proteins have two domains: an NH2-terminal F-box motif and a C-terminal leucine-rich repeat (LRR) motif or WD repeat motif. The SCF complex uses the F-box motif to bind Skp1, whereas the leucine-rich/WD repeat motif is used for substrate recognition.Citation29 Of the nearly 70 F-box proteins described, only a few of them have defined roles in cellular processes.Citation30 In fact, our recent study showed that another novel F-box protein, FBXL2, regulates CDK11p58 kinase, PLK4 and Aurora A kinase localization on the centrosome.Citation31 However, a single F-box protein appears to target many substrates, and a single protein can be targeted by multiple E3 ligases that could potentially modulate mitotic progression.
Herein, we uncovered an orphan protein, FBXL7, which displays E3 ligase activity. We demonstrate that FBXL7 over-expression leads to Aurora A ubiquitination and its subsequent degradation in a SV40 tumorogenic cell line, resulting in G2/M arrest. Interestingly, FBXL7 colocalizes with Aurora A throughout the cell cycle but only binds and ubiquitinates Aurora A during mitosis. FBXL7-mediated ubiquitination and depletion of centrosomal Aurora A disrupts normal mitotic spindle formation, thereby resulting in mitotic arrest and apoptosis.
Results
Ectopic expression of FBXL7 induces the polyubiquitination and degradation of Aurora A in the proteasome.
We first screened 18 novel F-box proteins that might be involved in Aurora A degradation. As shown in and B, 18 randomly selected F-box proteins (FBXL, FBXW family members) were ectopically expressed in a transformed murine lung epithelial (MLE) cell line. Twenty-four h later, cells were collected, and cell lysates were analyzed for expression of F-box protein and endogenous Aurora A. Interestingly, compared with all other F-box proteins, only expression of FBXL7 decreased Aurora A steady-state levels. We next examined Aurora A half-life after FBXL7 knockdown. Knockdown of FBXL7 markedly increased Aurora A t1/2 (). We also observed extended Aurora A t1/2 upon proteasomal inhibitor MG132 treatment (). To further evaluate this, cells were transfected with increasing amounts of FBXL7 plasmid, as shown in . Aurora A was degraded in a dose-dependent manner in correlation with FBXL7 overexpression. FBXL7 was also expressed in MLE cells using a doxycycline-inducible plasmid; Aurora A was degraded in a time-dependent manner in correlation with doxycycline treatment (). To confirm the specificity of FBXL7 targeting, we performed an in vitro ubiquitination assay using purified Aurora A as a substrate. Importantly, inclusion of purified SCFFBXL7 with the full complement of E1 and E2 enzymes plus ubiquitin was sufficient to generate polyubiquitinated Aurora A species in vitro (, upper left). The level of Aurora A ubiquitination also displayed increases in a dose-dependent manner with purified SCFFBXL7 (, lower left). As specificity controls, FBXL7 did not induce polyubiquitination of Aurora B (, upper right). Also, a randomly selected F-box protein FBXL18 did not ubiquitinate Aurora A under similar experimental conditions (, lower right).
Overexpression of FBXL7 induces G2/M-phase accumulation and polyploidy.
First, MLE cells were transfected with FBXL7, then cells were labled with BrdU and harvested for processing by two-color FACS ( and B). The results indicate a significant increase in a cell poplulation within the G2/M phase that accumulated over time (35% vs. 15% in control cells). Interestingly, overexpression of FBXL7 tended to reduce the diploidal cell population and increase the number of polyploidal cells (, red arrow). We also observed that ectopically expressed FBXL7 induced binucleate cell formation (). Specifically, upon FBXL7 overexpression, ∼30% of cells contained two or more nuclei 24 h after quantitative analysis.
Overexpression of FBXL7 induces apoptosis and decreases proliferation.
We investigated FBXL7 effects on cell proliferation first using human adenocarcinoma cells (A549) and then extended studies using transformed MLE cells. A plasmid encoding FBXL7 was first overexpressed in A549 cells in a dose-dependent manner; cells were collected and stained with Annexin V, a marker of apoptosis, prior to FACS analysis ( and C). The results indicate that expression of FBXL7 induces a 3-fold increase in apoptotic cells (40% vs. 13% in control). To further test this, pTRE-FBXL7 and pTET-advanced plasmids were co-transfected in A549 cells; 24 h after transfection, cells were treated with doxycycline for an additional 24 h. Cells were then collected and stained with Annexin V, a marker of apoptosis prior to FACS analysis ( and D). The results indicate that expression of FBXL7 induces a 3-fold increase in apoptotic cells (44% vs. 12% in control). We also performed a proliferation study, where MLE cells were transfected with either empty plasmid or a plasmid encoding FBXL7. Viable cells were counted using trypan blue staining at indicated time points and quantified, showing that overexpression of FBXL7 markedly inhibited the growth of cells in culture ().
FBXL7 and Aurora A colocalize within the centrosome during mitosis.
To confirm the specific localization of Aurora A and FBXL7 in cells, we first transfected MLE cells with only 2 µg of FBXL7-YFP plasmid (to limit mitotic arrest), then co-stained synchronized cells with antibodies to Aurora A and the centrosomal marker, γ-tubulin. Cells were also counterstained with DAPI to visualize DNA. Aurora A antibody decorated cells in a punctate pattern but with specific localization within the centrosome throughout mitosis. Interestingly, YFP-labeled FBXL7 decorated cells with specific localization within the centrosome from prometaphase to telophase. Hence, Aurora A and FBXL7 colocalize on the centrosome during key phases of the mitotic program ().
FBXL7 binds and depletes Aurora A on the centrosome.
Next, we examined whether FBXL7 engages Aurora A specifically during mitosis. CFP-Aurora A (green) and YFP-FBXL7 (red) were co-transfected into cells. Cells were analyzed by fixing followed by irreversible photobleaching using FRET ( and B). FRET is indicative of protein interaction between two partners when the donor emission (CFP) signal increases after a nearby acceptor fluorophore (YFP) is inactivated by irreversible photobleaching. The emission fluorescence of both the donor CFP-Aurora A and acceptor YFP-FBXL7 before and after acceptor photobleaching, are shown; the region of interest around the centrosome is marked (red arrow) ( and B, upper images and lower plots). The results from three independent experiments indicate that upon bleaching, there is decreased acceptor fluorescence (YFP-FBXL7) coupled with increased donor emission fluorescence (CFP-Aurora A), consistent with FBLX7 interaction with Aurora A while the cell is in mitosis (i.e., metaphase) (). However, when cells are in interphase, although both CFP-Aurora A and YFP-FBXL7 still colocalize within the centrosome, we do not observe the increase in donor emission fluorescence (CFP-Aurora A) upon photobleaching the acceptor YFP-FBXL7 (). Together, these results suggest that molecular interactions between FBXL7 and Aurora A during mitosis may regulate FBXL7-mediated Aurora A ubiquitination and degradation. These single cell interaction studies were supplemented with co-immunoprecipitation experiments. MLE cells were lysed and subjected to FBXL7 immunoprecipitation (i.p.), and Aurora A was detected in FBXL7 immunoprecipitates by immunoblotting (). The results suggest that FBXL7 interacts with Aurora A in cells. Last, we investigated effects of FBXL7 overexpression on Aurora A labeling on the centrosome. MLE cells were transfected with either empty plasmid or a plasmid encoding FBXL7; cells were then immunostained with Aurora A and γ-tubulin. Overexpression of FBXL7 specifically depleted Aurora A within the centrosome (). Quantification of Aurora A fluorescent intensity on the centrosome indicated that FBXL7 expression decreased Aurora A levels by ∼65% ().
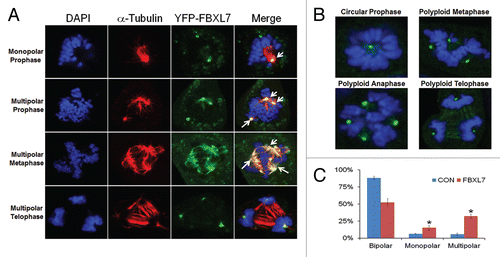
Overexpression of FBXL7 causes centrosomal and mitotic aberrations.
Next, we examined the integrity of the centrosome and mitotic spindles after expression of FBXL7. We first investigated the subcellular localization of FBXL7 upon vigorous transfection (6 µg of FBXL7-YFP plasmid). MLE cells were co-stained with antibody to α-tubulin to visualize the mitotic spindle. Interestingly, we observed that YFP-labeled FBXL7 not only localizes within the centrosome throughout mitosis, but also specifically appears on the tubulin spindle structure during metaphase (). Abnormal cells with several representative mitotic abnormalities were also detected after FBXL7 expression (). We observed the circular prophase configurations (, upper left part). One classic phenotype of the circular chromosome configuration is that circular figures are arranged on monopolar spindles around large centrosomes. We also observed polyploidy in cells during metaphase and anaphase, where multiple centrosomes were identified after FBXL7 expression (, upper part). Events were quantified in which numbers of mitotic cells with both monopolar or multipolar spindles with lagging chromosomes significantly increased after FBXL7 expression ().
Discussion
Cellular mitosis requires highly concerted actions within a network of regulatory proteins involved in cell cycle progression and proteolysis to ensure proper chromosomal segregation. As dysregulated mitotic events are linked to neoplasia, it is reasonable that one of these regulatory proteins might serve as a potential target for chemotherapeutic intervention. Due to the important role of Aurora A during mitosis, its robust expression in many types of cancer with high proliferative rates and its link to poor outcomes in patients with cancer, results of these studies suggest a potential role for Aurora A small-molecule inhibitors.Citation32–Citation34 One advantage of using Aurora A inhibitors with chemotherapeutic agents is their selectively toward dividing cells, therefore reducing the severity of adverse side effects compared with traditional anticancer agents. The results from this study provide the first evidence that overexpression of F-box protein FBXL7 appears to impair mitosis and thus might serve as a key growth inhibitory signal. We demonstrate that one mechanism whereby FBXL7 might induce mitotic arrest is by Aurora A depletion, leading to absence of its interactions with critical downstream effectors that assemble within the centrosomal-mitotic apparatus. Thus, FBXL7 small-molecule mimics might eventually provide a new strategy in neoplastic therapy.
During mitosis, there are many proteins involved in the spindle assembly checkpoint, which ensures the correct distribution of sister chromatids in anaphase before completion of cytokinesis by abscission.Citation35,Citation36 Our data show for the first time that both Aurora A and FBXL7 are localized within the centrosome from prometaphase to telophase during mitosis (), and that expression of FBXL7 reduces Aurora A expression within this organelle ( and E). Interestingly, our FRET experiment suggests that FBXL7 only engages Aurora A during mitosis but not in interphase ( and B). Thus the SCFFBXL7 complex targeting Aurora A during mitosis might be an exquisite mechanism to balance Aurora A levels, thereby regulating cell division. To further support this hypothesis, we analyzed mRNA levels of FBXL7 in cancer subjects through Genecards (www.genecards.org). Of note, in many types of cancer (hepatic, lung, ovary and testicular), FBXL7 expression is strongly suppressed in affected subjects compared with matched controls (data not shown). These observations further implicate FBXL7 as an authentic regulator of the mitotic program and provide one possible explanation for elevated immunoreactive Aurora A levels in many types of cancer.
Thus far, prior studies have not evaluated the F-box protein, FBXL7 and its potential substrates. Here, FBXL7 has a specifc target, Aurora A, within the centrosome. Although Aurora B shares a certain degree of sequence similarity with Aurora A, we did not observe Aurora B ubiquitination by SCFFBXL7 (). Interestingly, our previous studies suggest that FBXL2 colocalizes with cyclin D3 within the centrosome;Citation31 however, we did not observe Aurora A degradation upon FBXL2 overexpression either. These studies suggest that SCF-based E3 ligases have very specific substrates. Aurora A exhibits an extended half-life upon FBXL7 knockdown, which further supports that this F-box protein controls local concentrations of Aurora A in cells. However, we also observe that FBXL7 localizes within the midbody region at the end of telophase (, last part). Moreover, overexpressed FBXL7 is associated with the microtubular bundle within the mitotic spindle (). Thus, we cannot rule out that FBXL7 might have other important substrates during mitotic events.
In conclusion, our data suggest that the SCF-based E3 ligase subunit FBXL7 localizes at the centrosome during mitosis. This E3 ligase component ubiquitinates and degrades Aurora A in vitro and likely within the centrosome during cellular division; further depletion of Aurora A by FBXL7 overexpression leads to G2/M-phase arrest and mitotic abnormalities. Although exactly how FBXL7 translocates to the centrosome, its recognition signals and regulation will require investigation, this study provides a springboard for additional work on the molecular regulation of Aurora A in mitosis. We speculate that Aurora A targeting by the SCFFBXL7 complex during the transition to mitosis might be a fundamental mechanism to balance Aurora A levels, thereby regulating cell proliferation.
Methods and Materials
Materials.
The sources of the transformed murine lung epithelial (MLE) cell line and A549 cell line were described previously in references Citation37 and Citation38. Purified SCFFBXL7 was purchased from Abnova. MG132, purified ubiquitin, E1, E2 were purchased from Calbiochem. Purifed Aurora A protein, γ-tubulin, α-tubulin and Aurora A antibodies were from Abcam. Lipofectamine 2000, mouse monoclonal V5 antibody, DAPI nuclear staining kits, pcDNA3.1D cloning kit, E. coli One Shot Top10 competent cells, pENTR Directional TOPO cloning kits and Gateway mammalian expression system were from Invitrogen. FACS kit was purchased from BD Biosciences. pTRE, pTET-advanced, YFP plasmid and doxycycline were from Clontech. The F-box proteins cDNA was purchased from OpenBiosystems. Nucleofector transfection kits were from Amaxa. Cell viability based on Annexin V staining was assayed using an Annexin V kit from Roche. All DNA sequencing was performed by the University of Pittsburgh DNA Core Facility.
Cell culture.
MLE cells were cultured in Dulbecco's modified Eagle medium-F12 (Gibco) and supplemented with 10% fetal bovine serum (DMEM-F12 10%). A549 cells were cultured in MEM (Gibco) supplemented with 10% fetal bovine serum (MEM-10). Cells in some studies were synchronized using serum starvation (DMEM-F12) for 48 h or treatment with nocodazole or aphidicolin. Cells lysates were prepared by brief sonication in 150 mM NaCl, 50 mM Tris, 1.0 mM EDTA, 2 mM dithiothreitol, 0.025% sodium azide and 1 mM phenylmethylsulfonyl fluoride (Buffer A) at 4°C.
In vitro ubiquitin conjugation assay.
The ubiquitination of purified Aurora A was performed in a volume of 25 µl containing 50 mM Tris pH 7.6, 5 mM MgCl2, 0.6 mM DTT, 2 mM ATP, 1.5 ng/µl E1, 10 ng/µl Ubc5, 10 ng/µl Ubc7, 1 µg/µl ubiquitin (Calbiochem), 1 µM ubiquitin aldehyde, 4–16 µl of purified Cullin1, Skp1, Rbx1 and FBXL7. Reaction products were then processed for Aurora A immunoblotting.
Expression of recombinant protein.
All plasmids were delivered into cells using nucleofection or lipofectamine 2000.Citation39,Citation40 Cellular expression of green fluorescent-tagged plasmids using this device was achieved at > 90% efficiency.
Immunostaining.
Cells (2 × 105) were plated at 70% confluence on 35 mm MatTek glass-bottom culture dishes. Immunofluorescent cell imaging was performed on a Nikon A1 confocal microscope using 405 nm, 458 nm, 488 nm, 514 nm or 647 nm wavelengths. All experiments were done with a 60x oil differential interference contrast objective lens. Cells were washed with PBS and fixed with 4% paraformaldehyde for 20 min then exposed to 2% BSA, 1:500 primary antibodies and 1:1,000 Alexa 488 or Alexa 647 labeled goat anti-mouse or rabbit secondary antibody sequentially for immunostaining.
Fluorescence resonance energy transfer (FRET) analysis.
Cells were plated and co-transfected with CFP-Aurora A and YFP-FBXL7 plasmids. Interactions were detected at the single-cell level using a combination laser-scanning microscope system (Nikon A1 confocal). Interphase or mitotic cells were located where YFP fluorophore were specifically photobleached around the centrosome area.Citation38
Cell cycle and apoptosis analysis.
Transfected cells were incubated with BrdU (20 µM) for 40 min, fixed and stained following manufacturer's protocols (BD Biosciences). FACS samples were analyzed with the AccuriC6 system. DNA content was analyzed using FCS3 express software (De Novo Software). Cells were counted, and the percentage of cells with 2N, 4N and 8N DNA content was expressed as a percentage of total cells. Cells were also stained with Annexin V for 15 min following the manufacturer's protocol (Roche). For in vitro proliferation assays, MLE cells were transfected with empty plasmid or a plasmid encoding FBXL7. Cells were cultured in 35 mm dishes for up to 48 h; at each indicated time point, cells were collected and stained with trypan blue. Viable cells were then counted and quantified.
Statistical analysis.
Statistical comparisons were performed with the Prism program, version 4.03 (GraphPad Software, Inc.) using an ANOVA 1 or an unpaired two-tailed t-test with p < 0.05 indicative of significance.
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Figures and Tables
Figure 1 Ectopic expression of FBXL7 induces the polyubiquitination and degradation of Aurora A. (A and B) MLE cells were transfected with 18 V5 tagged F-box proteins including 9 of each FBXL and FBXW family members. Cells were collected and cell lysates were analyzed for V5, Aurora A and β-actin immunoblotting (n = 2). (C) Aurora A protein half-life determination after FBXL2 knockdown using FBXL7 siRNA or scrambled RNA [CON] (n = 2). (D) Aurora A protein half-life determination after MG132 treatment (n = 2). (E) MLE cells were transfected with increasing amounts of FBXL7 plasmid. Cells were collected and cell lysates were analyzed for V5, Aurora A and β-actin by immunoblotting (n = 3). (F) MLE cells were transfected with an inducible FBXL7 plasmid under control of exogenous doxycycline. Cells were treated with doxycycline for various times, cells were then collected and cell lysates were analyzed for FBXL7, Aurora A and β-actin by immunoblotting. (G) In vitro ubiquitination assays. Purified SCFFBXL7 or purified SCFFBXL18 were incubated with purified Aurora A or purified Aurora B and the full complement of ubiquitination reaction components.
![Figure 1 Ectopic expression of FBXL7 induces the polyubiquitination and degradation of Aurora A. (A and B) MLE cells were transfected with 18 V5 tagged F-box proteins including 9 of each FBXL and FBXW family members. Cells were collected and cell lysates were analyzed for V5, Aurora A and β-actin immunoblotting (n = 2). (C) Aurora A protein half-life determination after FBXL2 knockdown using FBXL7 siRNA or scrambled RNA [CON] (n = 2). (D) Aurora A protein half-life determination after MG132 treatment (n = 2). (E) MLE cells were transfected with increasing amounts of FBXL7 plasmid. Cells were collected and cell lysates were analyzed for V5, Aurora A and β-actin by immunoblotting (n = 3). (F) MLE cells were transfected with an inducible FBXL7 plasmid under control of exogenous doxycycline. Cells were treated with doxycycline for various times, cells were then collected and cell lysates were analyzed for FBXL7, Aurora A and β-actin by immunoblotting. (G) In vitro ubiquitination assays. Purified SCFFBXL7 or purified SCFFBXL18 were incubated with purified Aurora A or purified Aurora B and the full complement of ubiquitination reaction components.](/cms/asset/9647362f-1f54-4d69-8e6e-93369d16a91f/kccy_a_10919171_f0001.gif)
Figure 2 Overexpression of FBXL7 induces G2/M phase arrest and polyploidy. (A–C) Cell cycle analysis. MLE cells were transfected with 5 µg of control plasmid or FBXL7 plasmid for 24 h, cells were analyzed by BrdU uptake and 7-AA D staining, 2n, 4n and 8n DNA contents were then quantitated and graphed after expression of FBXL7. Red arrow indicates 8N DNA. Results from (A–C) are representative of 1 experiment, where consistency was seen 3 times (n = 3). (D) Multi-nucleate cells (arrows, middle part) after FBXL7 overexpression were quantified and graphed (right). White scale bar indicates 25 µm. (n = 150 cells, *p < 0.05 vs. con).
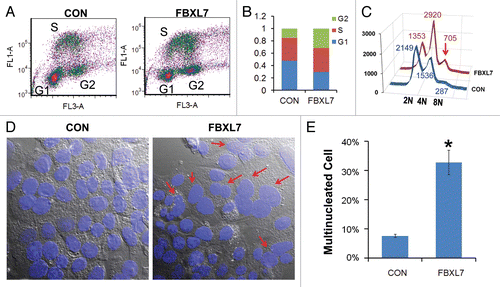
Figure 3 Overexpression of FBXL7 induces apoptosis and decreases cell proliferation. (A–C) FACS analysis of A549 cells transfected with either empty vector or FBXL7 plasmid in a dose dependent manner, viable cells were quantified and graphed in C (n = 3). (B) A549 cells were transfected with an inducible FBXL7 plasmid under control of doxycycline (DOX) for 24 h. Cells were treated with or without doxycycline for additional 24 h. Cells were then collected and viable cells were quantified and graphed in D (n = 3). (E) Proliferation assay. MLE cells were first transfected with 5 µg of either control or FBXL7 plasmid, and at each time point, cells were collected and stained with trypan blue; live cells were quantified and graphed (n = 3).
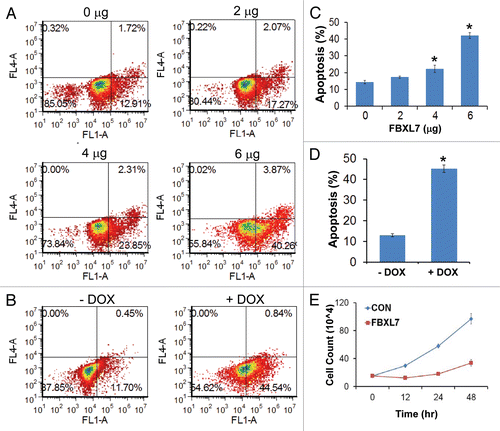
Figure 4 FBXL7 and Aurora A colocalize within the centrosome during mitosis. MLE cells (2 × 105) were first transfected with 2 µg of YFP-FBXL7 plasmid (green) before being plated for 48 h; cells were then washed with PBS and fixed with 4% paraformaldehyde for 20 min. Cells were immunostained for γ-tubulin (blue) and Aurora A (red). Nuclei were counterstained using DAPI (white). Green: FBXL7, Red: Aurora A, Blue: γ-tubulin and White: DAPI. White scale bar indicates 10 µm.
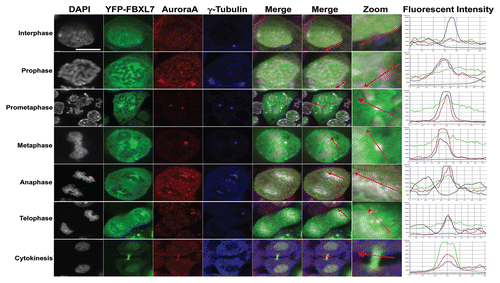
Figure 5 FBXL7 binds and depletes Aurora A on the centrosome. (A and B) MLE cells were co-transfected with CFP-Aurora A and YFP-FBXL7 for 48 h, cells were washed with PBS and fixed with 4% paraformaldehyde for 20 min and observed using a confocal microscope. FBXL7-Aurora A interaction at the single cell level was imaged using laser-scanning microscopy before and after photobleaching ((A) Mitosis. (B) Interphase). Shown in the upper sets of parts is single cell imaging before and after acceptor photobleaching fluorescence with intensities of YFP and CFP in parts. Bottom, the same FRET in each part was confirmed quantitatively and shown graphically. (C) MLE cells were collected and lysed followed by co-immunoprecipitation of endogenous FBXL7 and then Aurora A immunoblotting. (D) MLE cells were transfected with FBXL7 for 48 h. Cells were then washed with PBS and fixed with 4% paraformaldehyde for 20 min. Cells were co-immunostained for γ-tubulin and Aurora A. Cells were counterstained with DAPI to visualize the nucleus. White scale bar indicates 10 µm. (E) Fluorescent intensity of centrosomal Aurora A levels within cells was quantified using imageJ software and graphed (n = 25).
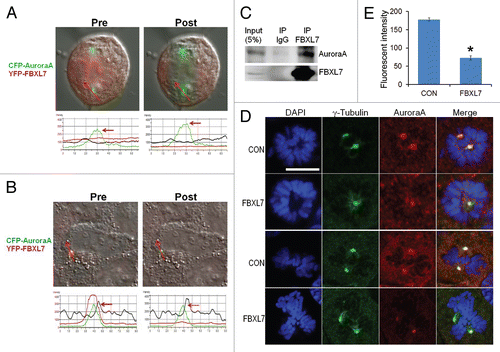
Figure 6 Overexpression of FBXL7 causes centrosomal and mitotic aberrations. (A) MLE cells (2 × 105) were plated and transfected with YFP-FBXL7 (6 µg plasmid) for 48 h. Cells were then immunostained for α-tubulin and counterstained with DAPI to visualize the nucleus. White arrows represent cells in mitotic arrest with abberant centrosomes. (B) MLE cells (2 × 105) were plated and transfected with FBXL7 (6 µg plasmid) for 48 h. Cells were then immunostained for γ-tubulin and counterstained with DAPI to visualize the nucleus. Specific chromosomal anomalies are presented. (C) 100 cells from three individual experiments were counted from experiments in (B) for abnormal centrosomal phenotypes and are presented graphically. White scale bar indicates 10 µm. *p < 0.05 vs. CON.
Acknowledgements
This material is based upon work supported, in part, by the US Department of Veterans Affairs, Veterans Health Administration, Office of Research and Development, Biomedical Laboratory Research and Development. This work was supported by a Merit Review Award from the US Department of Veterans Affairs and National Institutes of Health R01 grants HL096376, HL097376 and HL098174 (to R.K.M.). The contents presented do not represent the views of the Department of Veterans Affairs or the United States Government.
References
- Margolis RL. Tetraploidy and tumor development. Cancer Cell 2005; 8:353 - 354; PMID: 16286243; http://dx.doi.org/10.1016/j.ccr.2005.10.017
- Fujiwara T, Bandi M, Nitta M, Ivanova EV, Bronson RT, Pellman D. Cytokinesis failure generating tetraploids promotes tumorigenesis in p53-null cells. Nature 2005; 437:1043 - 1047; PMID: 16222300; http://dx.doi.org/10.1038/nature04217
- Gortchakov AA, Eggert H, Gan M, Mattow J, Zhimulev IF, Saumweber H. Chriz, a chromodomain protein specific for the interbands of Drosophila melanogaster polytene chromosomes. Chromosoma 2005; 114:54 - 66; PMID: 15821938; http://dx.doi.org/10.1007/s00412-005-0339-3
- Cimini D, Wan X, Hirel CB, Salmon ED. Aurora kinase promotes turnover of kinetochore microtubules to reduce chromosome segregation errors. Curr Biol 2006; 16:1711 - 1718; PMID: 16950108; http://dx.doi.org/10.1016/j.cub.2006.07.022
- Hoar K, Chakravarty A, Rabino C, Wysong D, Bowman D, Roy N, et al. MLN8054, a small-molecule inhibitor of Aurora A, causes spindle pole and chromosome congression defects leading to aneuploidy. Mol Cell Biol 2007; 27:4513 - 4525; PMID: 17438137; http://dx.doi.org/10.1128/MCB.02364-06
- Giet R, Petretti C, Prigent C. Aurora kinases, aneuploidy and cancer, a coincidence or a real link?. Trends Cell Biol 2005; 15:241 - 250; PMID: 15866028; http://dx.doi.org/10.1016/j.tcb.2005.03.004
- Sumara I, Quadroni M, Frei C, Olma MH, Sumara G, Ricci R, et al. A Cul3-based E3 ligase removes Aurora B from mitotic chromosomes, regulating mitotic progression and completion of cytokinesis in human cells. Dev Cell 2007; 12:887 - 900; PMID: 17543862; http://dx.doi.org/10.1016/j.devcel.2007.03.019
- Sumara I, Peter MA. A Cul3-based E3 ligase regulates mitosis and is required to maintain the spindle assembly checkpoint in human cells. Cell Cycle 2007; 6:3004 - 3010; PMID: 18075312; http://dx.doi.org/10.4161/cc.6.24.5068
- Carmena M, Earnshaw WC. The cellular geography of aurora kinases. Nat Rev Mol Cell Biol 2003; 4:842 - 854; PMID: 14625535; http://dx.doi.org/10.1038/nrm1245
- Giet R, Prigent C. Aurora/Ipl1p-related kinases, a new oncogenic family of mitotic serine-threonine kinases. J Cell Sci 1999; 112:3591 - 3601; PMID: 10523496
- Jiang Y, Zhang Y, Lees E, Seghezzi W. Aurora A over-expression overrides the mitotic spindle checkpoint triggered by nocodazole, a microtubule destabilizer. Oncogene 2003; 22:8293 - 8301; PMID: 14614453; http://dx.doi.org/10.1038/sj.onc.1206873
- Tyler RK, Shpiro N, Marquez R, Eyers PA. VX-680 inhibits Aurora A and Aurora B kinase activity in human cells. Cell Cycle 2007; 6:2846 - 2854; PMID: 18032922; http://dx.doi.org/10.4161/cc.6.22.4940
- Rao B, van Leeuwen IM, Higgins M, Campbel J, Thompson AM, Lane DP, et al. Evaluation of an Actinomycin D/VX-680 aurora kinase inhibitor combination in p53-based cyclotherapy. Oncotarget 2010; 1:639 - 650; PMID: 21317459
- Crane R, Gadea B, Littlepage L, Wu H, Ruderman JV. Aurora A, meiosis and mitosis. Biol Cell 2004; 96:215 - 229; PMID: 15182704; http://dx.doi.org/10.1016/j.biolcel.2003.09.008
- Nguyen HG, Chinnappan D, Urano T, Ravid K. Mechanism of Aurora-B degradation and its dependency on intact KEN and A-boxes: identification of an aneuploidy-promoting property. Mol Cell Biol 2005; 25:4977 - 4992; PMID: 15923616; http://dx.doi.org/10.1128/MCB.25.12.4977-92.2005
- Maerki S, Olma MH, Staubli T, Steigemann P, Gerlich DW, Quadroni M, et al. The Cul3-KLHL21 E3 ubiquitin ligase targets Aurora B to midzone microtubules in anaphase and is required for cytokinesis. J Cell Biol 2009; 187:791 - 800; PMID: 19995937; http://dx.doi.org/10.1083/jcb.200906117
- Ryser S, Dizin E, Jefford CE, Delaval B, Gagos S, Christodoulidou A, et al. Distinct roles of BARD1 isoforms in mitosis: full-length BARD1 mediates Aurora B degradation, cancer-associated BARD1beta scaffolds Aurora B and BRCA2. Cancer Res 2009; 69:1125 - 1134; PMID: 19176389; http://dx.doi.org/10.1158/0008-5472.CAN-08-2134
- Maerki S, Beck J, Sumara I, Peter M. Finding the midzone: the role of ubiquitination for CPC localization during anaphase. Cell Cycle 2010; 9:2921 - 2922; PMID: 20714224; http://dx.doi.org/10.4161/cc.9.15.12740
- Visconti R, Palazzo L, Grieco D. Requirement for proteolysis in spindle assembly checkpoint silencing. Cell Cycle 2010; 9:564 - 569; PMID: 20081372; http://dx.doi.org/10.4161/cc.9.3.10581
- Guardavaccaro D, Frescas D, Dorrello NV, Peschiaroli A, Multani AS, Cardozo T, et al. Control of chromosome stability by the beta-TrCP-REST-Mad2 axis. Nature 2008; 452:365 - 369; PMID: 18354482; http://dx.doi.org/10.1038/nature06641
- Seki A, Coppinger JA, Du H, Jang CY, Yates JR 3rd, Fang G. Plk1-and beta-TrCP-dependent degradation of Bora controls mitotic progression. J Cell Biol 2008; 181:65 - 78; PMID: 18378770; http://dx.doi.org/10.1083/jcb.200712027
- Gusti A, Baumberger N, Nowack M, Pusch S, Eisler H, Potuschak T, et al. The Arabidopsis thaliana F-box protein FBL17 is essential for progression through the second mitosis during pollen development. PLoS One 2009; 4:4780; PMID: 19277118; http://dx.doi.org/10.1371/journal.pone.0004780
- D'Angiolella V, Donato V, Vijayakumar S, Saraf A, Florens L, Washburn MP, et al. SCF(Cyclin F) controls centrosome homeostasis and mitotic fidelity through CP110 degradation. Nature 2010; 466:138 - 142; PMID: 20596027; http://dx.doi.org/10.1038/nature09140
- Puklowski A, Homsi Y, Keller D, May M, Chauhan S, Kossatz U, et al. The SCF-FBXW5 E3-ubiquitin ligase is regulated by PLK4 and targets HsSAS-6 to control centrosome duplication. Nat Cell Biol 2011; 13:1004 - 1009; PMID: 21725316; http://dx.doi.org/10.1038/ncb2282
- Littlepage LE, Ruderman JV. Identification of a new APC/C recognition domain, the A box, which is required for the Cdh1-dependent destruction of the kinase Aurora-A during mitotic exit. Genes Dev 2002; 16:2274 - 2285; PMID: 12208850; http://dx.doi.org/10.1101/gad.1007302
- Zheng N, Schulman BA, Song L, Miller JJ, Jeffrey PD, Wang P, et al. Structure of the Cul1-Rbx1-Skp1-F boxSkp2 SCF ubiquitin ligase complex. Nature 2002; 416:703 - 709; PMID: 11961546; http://dx.doi.org/10.1038/416703a
- Cardozo T, Pagano M. The SCF ubiquitin ligase: insights into a molecular machine. Nat Rev Mol Cell Biol 2004; 5:739 - 751; PMID: 15340381; http://dx.doi.org/10.1038/nrm1471
- Cenciarelli C, Chiaur DS, Guardavaccaro D, Parks W, Vidal M, Pagano M. Identification of a family of human F-box proteins. Curr Biol 1999; 9:1177 - 1179; PMID: 10531035; http://dx.doi.org/10.1016/S0960-9822(00)80020-2
- Ilyin GP, Rialland M, Glaise D, Guguen-Guillouzo C. Identification of a novel Skp2-like mammalian protein containing F-box and leucine-rich repeats. FEBS Lett 1999; 459:75 - 79; PMID: 10508920; http://dx.doi.org/10.1016/S0014-5793(99)01211-9
- Skaar JR, Pagan JK, Pagano M. SnapShot: F-box proteins I. Cell 2009; 137:1160 - 1161; PMID: 19524517; http://dx.doi.org/10.1016/j.cell.2009.05.039
- Chen BB, Glasser JR, Coon TA, Mallampalli RK. FBXL2 is a ubiquitin E3 ligase subunit that triggers mitotic arrest. Cell Cycle 2011; 10:3487 - 3494; PMID: 22024926; http://dx.doi.org/10.4161/cc.10.20.17742
- Tao Y, Zhang P, Girdler F, Frascogna V, Castedo M, Bourhis J, et al. Enhancement of radiation response in p53-deficient cancer cells by the Aurora-B kinase inhibitor AZD1152. Oncogene 2008; 27:3244 - 3255; PMID: 18084327; http://dx.doi.org/10.1038/sj.onc.1210990
- Long ZJ, Xu J, Yan M, Zhang JG, Guan Z, Xu DZ, et al. ZM 447439 inhibition of aurora kinase induces Hep2 cancer cell apoptosis in three-dimensional culture. Cell Cycle 2008; 7:1473 - 1479; PMID: 18418083; http://dx.doi.org/10.4161/cc.7.10.5949
- Lu LY, Wood JL, Ye L, Minter-Dykhouse K, Saunders TL, Yu X, et al. Aurora A is essential for early embryonic development and tumor suppression. J Biol Chem 2008; 283:31785 - 31790; PMID: 18801727; http://dx.doi.org/10.1074/jbc.M805880200
- Ho CC, Hau PM, Marxer M, Poon RY. The requirement of p53 for maintaining chromosomal stability during tetraploidization. Oncotarget 2010; 1:583 - 595; PMID: 21317454
- Vazquez-Martin A, Oliveras-Ferraros C, Menendez JA. The active form of the metabolic sensor: AMP-activated protein kinase (AMPK) directly binds the mitotic apparatus and travels from centrosomes to the spindle midzone during mitosis and cytokinesis. Cell Cycle 2009; 8:2385 - 2398; PMID: 19556893; http://dx.doi.org/10.4161/cc.8.15.9082
- Ray NB, Durairaj L, Chen BB, McVerry BJ, Ryan AJ, Donahoe M, et al. Dynamic regulation of cardiolipin by the lipid pump Atp8b1 determines the severity of lung injury in experimental pneumonia. Nat Med 2010; 16:1120 - 1127; PMID: 20852622; http://dx.doi.org/10.1038/nm.2213
- Chen BB, Mallampalli RK. Calmodulin binds and stabilizes the regulatory enzyme, CTP: phosphocholine cytidylyltransferase. J Biol Chem 2007; 282:33494 - 33506; PMID: 17804406; http://dx.doi.org/10.1074/jbc.M706472200
- Agassandian M, Chen BB, Schuster CC, Houtman JC, Mallampalli RK. 14-3-3zeta escorts CCTalpha for calcium-activated nuclear import in lung epithelia. FASEB J 2010; 24:1271 - 1283; PMID: 20007511; http://dx.doi.org/10.1096/fj.09-136044
- Chen BB, Mallampalli RK. Masking of a nuclear signal motif by monoubiquitination leads to mislocalization and degradation of the regulatory enzyme cytidylyltransferase. Mol Cell Biol 2009; 29:3062 - 3075; PMID: 19332566; http://dx.doi.org/10.1128/MCB.01824-08