Keywords: :
Disease-causing mutations are generally assumed to lie within protein-coding regions of genes and to lead to the production of mutated or truncated proteins. This expectation has often limited the search for mutations in protein-coding exons of genes. Strikingly, in the last years, a few disease-related mutations were found at the polyadenylation site located at the 3' side of genes.Citation1-Citation3 The polyadenylation process is an essential nuclear step of gene expression that allows the maturation of the pre-mRNA into a polyadenylated mature mRNA. Formation of the polyadenylated 3' end of most eukaryotic mRNAs occurs through the recognition of the polyadenylation site (generally AAUAAA) and several neighboring cis-acting elements by a complex polyadenylation machinery.Citation4
Lynch syndrome is a rare autosomal dominant inherited syndrome that confers increased risks of colorectal, endometrial and, to a lesser extent, stomach, small intestine, hepatobillary, kidney and ovarian cancers. Germline inactivating mutations in one of the four mismatch repair (MMR) genes MSH2, MLH1, MSH6 and PMS2 are associated with this syndrome. Inactivation of these MMR genes induces somatic instability, in particular in the microsatellite region (MSI). Mutations in MSH6 account for 10–20% of Lynch syndrome colorectal cancers, and the risk of developing cancer for mutation carriers is far from negligible.Citation5 In many patients suspected to belong to high-risk Lynch syndrome families presenting MSI, no promoter hypermethylation nor mutations in the open reading frame, the exon/intron junctions or the promoter region of MMR genes can be detected, suggesting the presence of mutations in other regions controlling MSH6 expression. This prompted us to investigate the prevalence of variants at the 3' side of the MSH6 open reading frame region.
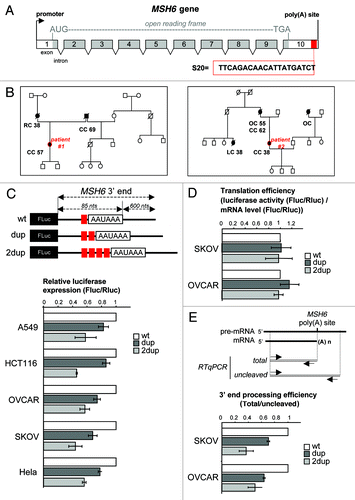
We investigated the presence of variants in the MSH6 last exon in the vicinity of the polyadenylation site for 189 patients fulfilling Amsterdam criteria or selected because of a familial history of colorectal cancer associated to an MSI phenotype in the tumor (described in the Sup. Materials). Our analysis did not reveal any single nucleotide polymorphism (SNP) in the MSH6 3' UTR. For two patients, a 20 bp sequence (TTC AGA CAA CAT TAT GAT CT) (named S20) was found duplicated on two adjacent positions in the vicinity of the polyadenylation site (; Fig. S1). This duplication, highly conserved among higher vertebrates (Fig. S2), could not be identified in a control population composed of 150 patients without any familial history of colon cancer [p < 0.05; power of 86.1% for an exact test (East 5.4 software)]. The two probands carrying these two variants had a strong familial history of colorectal cancers associated with Lynch syndrome (). To investigate whether the duplication of S20 might modify MSH6 expression, we generated an EBV-immortalized lymphoblastoid cell line from patient 1’s lymphocytes (P1 cell line). The level of MSH6 mRNA measured by RT-qPCR tended to be lower in the P1 cell line as compared with three different EBV cell lines established from lymphocytes of patients who did not carry the duplication of S20 (Fig. S3). We next examined whether the duplication of S20 interfered with the process leading to MSH6 mRNA polyadenylation by constructing a reporter vector in which the MSH6 3' end region encompassing the polyadenylation signal and including the 3' flanking region was inserted downstream of the Firefly luciferase (FLuc) open reading frame and two plasmids in which S20 was duplicated (dup) or quadruplicated (2dup) (). Relative luciferase activity showed that the presence of two or four copies of S20 reduced luciferase expression in all the tested cell lines including those (colon, ovarian) belonging to the Lynch syndrome spectrum (). This S20 duplication-dependent decrease in luciferase expression was not due to translation repression (). Furthermore, real-time RT-qPCR analysis of MSH6 mRNA polyadenylation efficiency, measured by the ratio of total RNA to uncleaved RNA, revealed that the presence of two or four copies of S20 reduced MSH6 mRNA polyadenylation ().
Figure 1. (A) Schematic representation of the MSH6 gene. (B) Pedigrees of patient 1’s and patient 2’s families bearing the S20 sequence. Square symbols indicate males, round symbols females. Filled symbols indicate ovarian (OC), rectal (RC), colon (CC) and lung (LC) cancer affected individuals. Ages of diagnosis are indicated below the individuals. (C) Illustration of luciferase reporter vector containing MSH6 polyadenylation site and 3′ end flanking regions inserted downstream from the Firefly luciferase gene (Fluc). Individual clones carrying the region of interest (red square) either wild type (WT), duplicated (dup) or quadruplicated (2dup) are depicted. The ratio of Firefly/Renilla luciferase activities (Fluc/Rluc) was determined using different cells lines (A549, HCT116, OVCAR, SKOV or Hela) cotransfected with WT, dup or 2dup plasmids and an internal control plasmid encoding Renilla luciferase. Bars represented the mean ± SD of the Fluc/Rluc ratio of three different experiments. (D) Translational efficiency of the WT, dup and 2dup MSH6 reporter vectors transfected in the ovarian cancer cell lines, SKOV and OVCAR, was measured by normalization of the relative luciferase activity (Fluc/Rluc) to luciferase RNA levels quantified by real time RT-qPCR analysis. (E) Reverse transcription of RNA from SKOV or OVCAR cells transfected with WT, dup or 2dup plasmids followed by real-time qPCR with primer specific to located on either side of the cleavage site to amplify the uncleaved RNA or upstream of the cleavage site to amplify both the uncleaved and cleaved RNA,Citation7 as depicted. Bars represented the mean ± SD of the 3′ processing efficiency of three different experiments.
In summary, these results indicate that duplication of S20 reduces MSH6 expression by lowering the efficiency of MSH6 mRNA polyadenylation. This duplication-dependent regulation of MSH6 expression strongly suggests that this duplication may be a causal mutation. More generally, these results reinforce the need to search for mutations, outside the protein-coding exons, in the vicinity of the polyadenyaltion regulatory sequences, a region often neglected during the genetic screening of disease-associated candidate genes. It is very likely that, similar to diseases linked to aberrant splicing,Citation6 another nuclear pre-mRNA processing step of gene expression, more examples of diseases caused by mutation in polyadenylation signals will emerge.
Additional material
Download Zip (213.3 KB)Acknowledgments
This work was supported by INSERM and FRM (Equipe FRM, soutenue par la Fondation Recherche Médicale to S.V.) and the Groupe de Recherche de l’Institut Claudius Regaud (F.F., C.T., S.M., S.V., R.G.). A.D. was a recipient of a studentship from ARC (Association pour la recherche sur le cancer) and FRM. The authors sincerely thank Thomas Filleron for his help in statistical analyses, Chantal Darnau, Pauline Dejean and Isabelle Gitlaw for their efficient technical assistance.
References
- Stacey SN, Sulem P, Jonasdottir A, Masson G, Gudmundsson J, Gudbjartsson DF, et al, Swedish Low-risk Colorectal Cancer Study Group. A germline variant in the TP53 polyadenylation signal confers cancer susceptibility. Nat Genet 2011; 43:1098 - 103; http://dx.doi.org/10.1038/ng.926; PMID: 21946351
- Delahaye NF, Rusakiewicz S, Martins I, Ménard C, Roux S, Lyonnet L, et al. Alternatively spliced NKp30 isoforms affect the prognosis of gastrointestinal stromal tumors. Nat Med 2011; 17:700 - 7; http://dx.doi.org/10.1038/nm.2366; PMID: 21552268
- Danckwardt S, Hentze MW, Kulozik AE. 3′ end mRNA processing: molecular mechanisms and implications for health and disease. EMBO J 2008; 27:482 - 98; http://dx.doi.org/10.1038/sj.emboj.7601932; PMID: 18256699
- Millevoi S, Vagner S. Molecular mechanisms of eukaryotic pre-mRNA 3′ end processing regulation. Nucleic Acids Res 2010; 38:2757 - 74; http://dx.doi.org/10.1093/nar/gkp1176; PMID: 20044349
- Baglietto L, Lindor NM, Dowty JG, White DM, Wagner A, Gomez Garcia EB, et al, Dutch Lynch Syndrome Study Group. Risks of Lynch syndrome cancers for MSH6 mutation carriers. J Natl Cancer Inst 2010; 102:193 - 201; http://dx.doi.org/10.1093/jnci/djp473; PMID: 20028993
- Cooper TA, Wan L, Dreyfuss G. RNA and disease. Cell 2009; 136:777 - 93; http://dx.doi.org/10.1016/j.cell.2009.02.011; PMID: 19239895
- Decorsière A, Cayrel A, Vagner S, Millevoi S. Essential role for the interaction between hnRNP H/F and a G quadruplex in maintaining p53 pre-mRNA 3′-end processing and function during DNA damage. Genes Dev 2011; 25:220 - 5; http://dx.doi.org/10.1101/gad.607011; PMID: 21289067